Abstract
Background
We and others recently identified the gene underlying PARK8 linked Parkinson's disease (PD). This gene, LRRK2, contains mutations that cause an autosomal dominant PD, including a mutation, G2019S, which is the most common PD causing mutation identified to date. Common genetic variability in genes that contain PD causing mutations has previously been implicated as a risk factor for typical sporadic disease.
Methods
We undertook a case‐control association analysis of LRRK2 in two independent European PD cohorts using 31 tagging single nucleotide polymorphisms (tSNPs) and five potentially functional SNPs. To assess the structure of this locus in different populations, we have performed linkage disequilibrium (LD) analysis using these variants in a human diversity panel.
Results
We show that common genetic variability in LRRK2 is not associated with risk for PD in the European populations studied here. We also show inter‐population variability in the strength of LD across this locus.
Conclusions
To our knowledge this is the first comprehensive analysis of common variability within LRRK2 as a risk factor for PD.
Keywords: Parkinson's disease, LRRK2, dardarin
Parkinson's disease (PD) is a neurological disease involving the degeneration of dopaminergic nigrostriatal neurons. The degeneration and dysfunction of this neuronal system results in a progressive movement disorder characterised by tremor, rigidity, postural instability, and bradykinesia. The underlying molecular pathogenesis and aetiology of typical PD is largely unknown, but the disease is thought to be caused by a complex interaction of genetics and environment.
We and others have recently identified the gene bearing mutations that underlie PARK8 linked PD.1,2 Leucine rich repeat kinase 2 (gene symbol LRRK2) is a 51 exon gene encoding a 2527 amino acid protein, dardarin. To date numerous missense mutations and one potential splice site mutation within dardarin have been linked to PD. These include a mutation, G2019S, within the highly conserved activation loop of the kinase domain. We and others have shown that this mutation contributes to between 1% and 8% of typical PD and as such is the most common mutation causing PD described to date.3,4,5,6,7,8,9,10
Although a large number of genes have been investigated as risk factor loci for PD, relatively little progress has been made in the identification of common genetic risk factors for this disease. One gene that shows a largely consistent association with the disease is SNCA, encoding α‐synuclein. SNCA contains mutations that cause an autosomal dominant form of parkinsonism and α‐synuclein is a major component of the neuropathological hallmark lesion of PD, the Lewy body.
Given that common variation within genes that contain mutations causing familial forms of disease may be associated with the sporadic forms of this disease, we have undertaken a thorough analysis of the role common variation within LRRK2 plays in risk for PD. We present data here, using a set of 31 tagging single nucleotide polymorphisms (tSNPs) and five potentially functional SNPs in two independent case control PD cohorts. Furthermore, using these SNPs, we tested the level of linkage disequilibrium (LD) across this gene in a series of 1039 samples collected from diverse human populations.
Methods
Subjects
The Finnish cohort of patients and controls has been previously described.11 Briefly, venous blood was taken from 147 patients (87 men, 60 women) with sporadic PD and the patients' spouses, who did not have PD, were used as controls (n = 136, 50 men, 86 women). The mean age at examination of the patients was 67.2 years (range 38–88) and that of controls was 65.8 years (range 37–87). Informed consent from all subjects was obtained. Approval for this study was obtained from the Ethics Committee for the Department of Neurology, Helsinki University Central Hospital (Decision 65/1999) and was updated by the Ethics Committee for Ophthalmology, Otorhinolaryngology, Neurology and Neurosurgery in the Hospital District of Helsinki and Uusimaa (Dnro 439/E9/2000).
A total of 217 PD patients (87 women, 130 men) and 221 healthy controls (97 women, 124 men) age, gender, and ethnicity matched, were included in the Greek cohort. PD patients had a mean (±SD) age of 69.8±8.7 (range 44–95) years at the time of initial examination, while their mean age at onset of disease was 63.3±9.6 (range 30–88) years. Healthy controls had a mean age of 68.3±12.8 (range 32–93) years. The diagnosis of PD was based on established criteria.12 All patients had sporadic PD, based on pedigree analysis. After approval from the hospital scientific committee and informed consent, blood samples were drawn for DNA extraction from patients and controls. Experienced neurologists who were blind to genotyping results performed all clinical assessments (such as PD diagnosis, age at onset, etc).
Molecular genetic analysis
SNPs were selected from 215 SNPs across the LRRK2 locus from a data dump of Caucasian data available through the International HapMap Project web page (www.hapmap.org). A total of 31 tSNPs were identified using Tagit v2.03.13 SNPs with a minor allele frequency of 0.05 or less were excluded from the analysis. The selected SNPs captured 95% of the common genetic variation across the gene. Three apparent blocks of LD were identified containing 17, seven, and seven tSNPs, respectively (table 1).
Table 1 SNPs genotyped in the current study.
| SNP | rs number | Chromosome position | bp to next SNP | Minor allele (frequency) | Tagging/coding |
|---|---|---|---|---|---|
| Block 1 | |||||
| 1 | rs2201144 | 38897130 | 9952 | C (0.093) | Promoter |
| 2 | rs1491941 | 38907082 | 10793 | C (0.373) | tSNP |
| 3 | rs10878244 | 38917875 | 183 | A (0.134) | tSNP |
| 4 | rs10878245 | 38918058 | 309 | T (0.492) | tSNP; L153L |
| 5 | rs10878247 | 38918367 | 9592 | T (0.321) | tSNP |
| 6 | rs10878258 | 38927959 | 3938 | G (0.229) | tSNP |
| 7 | rs1491938 | 38931897 | 4258 | T (0.489) | tSNP |
| 8 | rs10784451 | 38936155 | 2506 | A (0.183) | tSNP |
| 9 | rs2046928 | 38938661 | 126 | G (0.059) | tSNP |
| 10 | rs2723264 | 38938787 | 4705 | T (0.250) | tSNP |
| 11 | rs4293189 | 38943492 | 312 | A (0.427) | tSNP |
| 12 | rs10784661 | 38943804 | 163 | G (0.192) | tSNP |
| 13 | rs7308720 | 38943967 | 3703 | G (0.088) | N551K |
| 14 | rs4768224 | 38947670 | 3240 | A (0.369) | tSNP |
| 15 | rs11564207 | 38950910 | 584 | A (0.107) | tSNP |
| 16 | rs7308193 | 38951494 | 1648 | G (0.347) | tSNP |
| 17 | rs10878299 | 38953142 | 3519 | G (0.058) | tSNP |
| 18 | rs7971935 | 38956661 | 407 | A (0.051) | tSNP |
| 19 | rs4272849 | 38957068 | 1188 | C (0.483) | tSNP |
| Block 2 | |||||
| 20 | rs10878307 | 38958256 | 8952 | G (0.061) | I723V |
| 21 | rs4318033 | 38967208 | 5097 | G (0.067) | tSNP |
| 22 | rs7957754 | 38972305 | 2657 | G (0.449) | tSNP |
| 23 | rs7966550 | 38974962 | 8739 | C (0.150) | tSNP; L953L |
| 24 | rs10784498 | 38983701 | 5477 | A (0.287) | tSNP |
| 25 | rs7133914 | 38989178 | 10990 | A (0.092) | R1398H |
| 26 | rs11564148 | 39000168 | 12027 | A (0.324) | S1647T |
| 27 | rs10878386 | 39012195 | 3079 | G (0.069) | tSNP |
| 28 | rs2404834 | 39015274 | 3723 | T (0.096) | tSNP |
| 29 | rs1427273 | 39018997 | 6763 | C (0.296) | tSNP |
| Block 3 | |||||
| 30 | rs11564147 | 39025760 | 2761 | A (0.069) | tSNP |
| 31 | rs10878405 | 39028521 | 1832 | A (0.324) | tSNP; E2108E |
| 32 | rs4768235 | 39030353 | 689 | A (0.127) | tSNP |
| 33 | rs7303525 | 39031042 | 33 | C (0.196) | tSNP |
| 34 | rs7132187 | 39031075 | 373 | A (0.298) | tSNP |
| 35 | rs7307310 | 39031448 | 3214 | T (0.092) | tSNP |
| 36 | rs890575 | 39034662 | – | G (0.117) | tSNP |
The five non‐synonymous and promoter SNPs that were not part of the tagging set are indicated. Minor allele frequency and chromosome position are based on the NCBI reference sequence in dbSNP build 124.
Five additional SNPs were selected. These included four coding changes we had identified in previous sequencing efforts (rs7308720, rs10878307, rs7133914, and rs11564148) in addition to a SNP within the promoter (rs2201144), predicted to create a transcription binding factor site for VMYB only when the C allele is present.
All 36 SNPs were genotyped in the Finnish cohort, in the Greek cohort, and also in a human diversity panel representing 1051 different DNA samples from 51 populations (SNPs listed in table 1). Genotyping was performed using Assays‐by‐Design SNP Genotyping (Applied Biosystems, Foster City, CA). Data were stored and manipulated for genetic analysis using the internally designed database GERON Genotyping (http://neurogenetics.nia.nih.gov/genotyping/).
Statistical analysis
Based on a minor allele frequency of 0.20, these series possess sufficient (80%) power to detect a difference (p = 0.05) in allele frequency with an effect in the range of OR ⩽0.52 or ⩾1.72 for the Finnish cohort and OR ⩽0.59 or ⩾1.57 for the Greek cohort.
GERON Genotyping was used to perform χ2 tests of association. Haplotype construction and tests of haplotype association were performed using the program Genecounting in association with the module Genecounting Permute. Genecounting implements the EM algorithm for haplotype analysis of unrelated individuals, and Genecounting Permute performs permutation tests for global association and significance of specific haplotypes using Freeman‐Tukey and proportion tests.14 One thousand replications were performed for each analysis using a random number seed. Haplotype associations were performed individually for each of the three previously identified blocks of LD (table 1) including all markers within each block, except for block 1, which had to be split into two sections, 1a (rs2201144 to rs2723264) and 1b (rs4293189 to rs4272849), in order to reduce computation time.
Pair‐wise D′ and r2 measurements were made between SNPs in the Greek and Finnish control cohorts and within the human genome diversity series using the program EMLD (https://epi.mdanderson.org/˜qhuang/Software/pub.htm). The regional distribution of LD across the LRRK2 locus was plotted using the program GOLD.15
Results
A total of 31 tSNPs were identified from the Caucasian HapMap data using Tagit v2.03.13 The selected SNPs captured a minimum of 95% of the common genetic variation across the gene. Three apparent blocks of LD were identified (table 1).
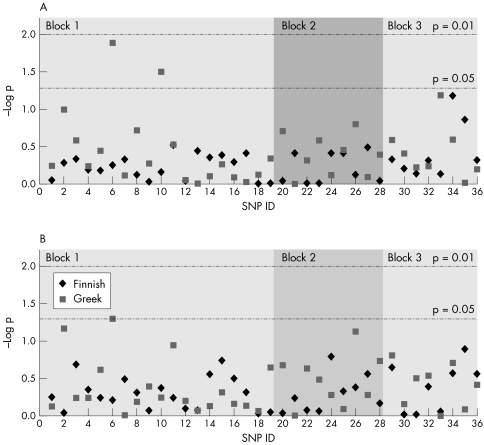
All SNPs were at or near Hardy‐Weinberg equilibrium for all control and disease cohorts (unpublished data), except for rs2046928 which deviated from equilibrium in the Greek case and control cohorts (p = 0.03 and p = 0.002, respectively), rs7132187 which was different in the Greek control cohort (p = 0.04), and rs4293189 which deviated from equilibrium in the Finnish control cohort (p = 0.03). There were, therefore, a total of four deviations from 144 tests (2.7%), a number consistent with chance occurrence at the p value selected (0.05). We identified an association at SNP rs10878258 in the Greek cohort for both allele and genotype frequency (p = 0.05 and p = 0.013, respectively) and at SNP rs2723264 for genotype alone (p = 0.03). Neither of these SNPs were associated with PD in the Finnish cohort. No other SNP showed association with disease (fig 1).
Figure 1 Single marker association analysis for variation within LRRK2 with disease. (A) Genotype. (B) Allele. p values are uncorrected χ2 results.
Global tests of association for each haplotype block in both series failed to show any association (Greek: block 1a χ2 = 84.30, 1013DF, p = 0.685; block 1b χ2 = 51.90, 502DF, p = 0.285; block 2 χ2 = 41.18, 1013DF, p = 0.588; block 3 χ2 = 38.48, 120DF, p = 0.599. Finnish: block 1a χ2 = 30.96, 1013DF, p = 0.826; block 1b χ2 = 38.86, 502DF, p = 0.246; block 2 χ2 = 30.22, 1013DF, p = 0.37; block 3 χ2 = 11.92, 120DF, p = 0.86).
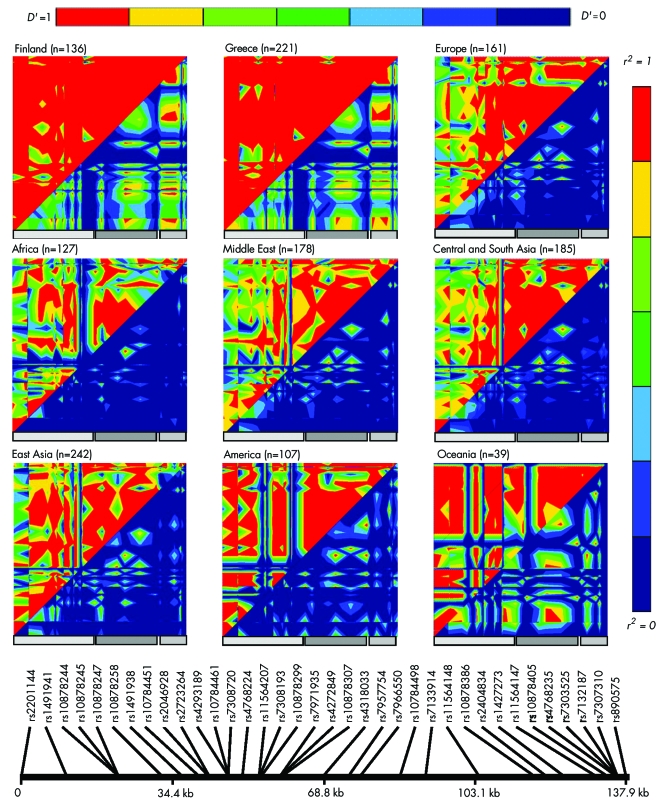
The raw genotypes produced by us for the HGDP‐CEPH Human Genome Diversity Cell Line Panel are available at http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm. A number of genotypes were not informative for several populations. LD across LRRK2 within seven geographically defined populations was visualised using GOLD (fig 2). Consistent with the differences in observed LD patterns across the human diversity series, analysis of HapMap data for Yoruban, Han, and Japanese populations showed markedly different SNPs are required to tag LRRK2 in these populations (data not shown).
Figure 2 Graphical Overview of Linkage Disequilibrium (GOLD) plots showing linkage disequilibrium in diverse ethnic populations. Pairwise LD analysis was performed using EMLD; D′ is shown in the upper left, r2 in the lower right. Non‐informative markers were removed prior to calculations. Grey bars indicate the position of the three haplotype blocks originally defined by Tagit using Caucasian data from www.hapmap.org.
Discussion
We and others have recently identified mutations within the gene LRRK2, encoding the protein dardarin, as the cause of PARK8 linked PD.1,2 Mutations in this gene are the most common genetic cause of PD identified to date.8,9,10 In the current study, we have performed a rigorous case‐control association study of genetic variation within LRRK2 as a risk factor for PD. We have identified three main haplotype blocks across the gene and defined SNPs that capture 95% of the common variability within LRRK2 in Caucasian populations of northern/western European descent.
Analysis of all SNPs in the Human Diversity Series, representing 1051 patients from 51 different world populations,16 showed considerable inter‐population variability in allele frequencies and LD across the region. These data and analysis of HapMap data for Yoruban, Han, and Japanese populations (data not shown) suggest that markedly different tSNPs will be required to tag LRRK2 in non‐European populations. The heat maps of the Greek and Finnish populations varied considerably from that seen in the European population in the diversity series. This divergence most likely represents the relative heterogeneity within this European population which consists of Orcadian, Adygei, Russian, Basque, French, Italian, Sardinian, and Tuscan populations.
We found no convincing evidence for association between LRRK2 SNPs or haplotypes and risk for PD. While there was an association between disease and SNPs rs10878258 and rs2723264 in the Greek series, this was not evident in the Finnish cohort. As there was no disease association in the haplotype block extending over this region in either population, it seems unlikely that there is a risk SNP in cis with the associated alleles of rs10878258 and rs2723264 common to both populations. It is possible that there is a risk variant in LD with these SNPs solely in the Greek series. However, the most plausible explanation for the marginal associations noted is that these are both type I errors. This seems particularly likely when one considers that p values described are uncorrected for multiple testing; after the most conservative Bonferroni correction is applied, neither of these p values remains significant.
The current study has sufficient power to detect an effect of OR greater than 1.5 (1.7 in the Finnish cohort). This study cannot, however, exclude the possibility that low frequency alleles within LRRK2 may exert an effect on lifetime risk for PD. The efficiency of the tagging methodology used here decreases dramatically at low allele frequencies and neither of these cohorts is sufficiently powered to detect moderate effects of alleles with a frequency of less than 0.05.
While these data suggest polymorphism in LRRK2 does not influence lifetime risk for disease, common variability in this gene may modulate disease presentation in LRRK2 mutation carriers. These data cannot rule out the presence of a genetic association between LRRK2 and PD in other populations. However, given that we have assessed two independent populations, this would seem unlikely.
Electronic‐database information
The International HapMap Project web page can be found at www.hapmap.org; the EMLD program is available from https://epi.mdanderson.org/˜qhuang/Software/pub.htm; the raw genotypes produced for the HGDP‐CEPH Human Genome Diversity Cell Line Panel are available at http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm; the HGDP‐CEPH Human Genome Diversity Cell Line Panel can be found at http://www.cephb.fr/HGDP‐CEPH‐Panel/; and information on GERON Genotyping can be found at http://neurogenetics.nia.nih.gov/genotyping/.
Acknowledgements
We thank the patients and their families for taking part in this research. Thanks to Dr Howard Cann for providing the HGDP‐CEPH Human Genome Diversity Cell Line Panel.
Abbreviations
LD - linkage disequilibrium
PD - Parkinson's disease
SNP - single nucleotide polymorphism
tSNP - tagging SNP
Footnotes
This research was supported in part by the Helsinki University Central Hospital, the Finnish Cultural Foundation, the Finnish Medical Foundation, and the Finnish National Graduate School of Clinical Investigation. It was also supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Competing interests: none declared
The International HapMap Project web page can be found at www.hapmap.org; the EMLD program is available from https://epi.mdanderson.org/˜qhuang/Software/pub.htm; the raw genotypes produced for the HGDP‐CEPH Human Genome Diversity Cell Line Panel are available at http://research.marshfieldclinic.org/genetics/Freq/FreqInfo.htm; the HGDP‐CEPH Human Genome Diversity Cell Line Panel can be found at http://www.cephb.fr/HGDP‐CEPH‐Panel/; and information on GERON Genotyping can be found at http://neurogenetics.nia.nih.gov/genotyping/.
References
- 1.Paisán‐Ruiz C, Jain S, Evans E W, Gilks W P, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil A M, Khan N, Johnson J, Martinez J R, Nicholl D, Carrera I M, Pena A S, de Silva R, Lees A, Marti‐Masso J F, Perez‐Tur J, Wood N W, Singleton A B. Cloning of the gene containing mutations that cause PARK8 linked Parkinson disease. Neuron 200444(4)595–600. [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti R J, Calne D B, Stoessl A J, Pfeiffer R F, Patenge N, Carbajal I C, Vieregge P, Asmus F, Muller‐Myhsok B, Dickson D W, Meitinger T, Strom T M, Wszolek Z K, Gasser T. Mutations in LRRK2 cause autosomal‐dominant parkinsonism with pleomorphic pathology. Neuron 200444(4)601–607. [DOI] [PubMed] [Google Scholar]
- 3.Paisan‐Ruiz C, Lang A E, Kawarai T, Sato C, Salehi‐Rad S, Fisman G K, Al‐Khairallah T, St George‐Hyslop P, Singleton A, Rogaeva E. LRRK2 gene in Parkinson disease. Neurology. 2005 Jun 15; [Epub ahead of print]. [DOI] [PubMed]
- 4.Deng H, Le W, Guo Y.et al Genetic and clinical identification of Parkinson's disease patients with LRRK2 G2019S mutation. Ann Neurol 200557(6)933–934. [DOI] [PubMed] [Google Scholar]
- 5.Aasly J O, Toft M, Fernandez‐Mata I, Kachergus J, Hulihan M, White L R, Farrer M. Clinical features of LRRK2‐associated Parkinson's disease in central Norway. Ann Neurol 200557(5)762–765. [DOI] [PubMed] [Google Scholar]
- 6.Kachergus J, Mata I F, Hulihan M, Taylor J P, Lincoln S, Aasly J, Gibson J M, Ross O A, Lynch T, Wiley J, Payami H, Nutt J, Maraganore D M, Czyzewski K, Styczynska M, Wszolek Z K, Farrer M J, Toft M. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet 200576(4)672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bras J M, Guerreiro R J, Ribeiro M H, Januario C, Morgadinho A, Oliveira C R, Cunha L, Hardy J, Singleton A. G2019S dardarin substitution is a common cause of Parkinson's disease in a Portuguese cohort. Mov Disord 200520(12)1653–1655. [DOI] [PubMed] [Google Scholar]
- 8.Di Fonzo A, Rohe C F, Ferreira J, Chien H F, Vacca L, Stocchi F, Guedes L, Fabrizio E, Manfredi M, Vanacore N, Goldwurm S, Breedveld G, Sampaio C, Meco G, Barbosa E, Oostra B A, Bonifati V, Italian Parkinson Genetics Network A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson's disease. Lancet 2005365(9457)412–415. [DOI] [PubMed] [Google Scholar]
- 9.Gilks W P, Abou‐Sleiman P M, Gandhi S, Jain S, Singleton A, Lees A J, Shaw K, Bhatia K P, Bonifati V, Quinn N P, Lynch J, Healy D G, Holton J L, Revesz T, Wood N W. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet 2005365(9457)415–416. [DOI] [PubMed] [Google Scholar]
- 10.Nichols W C, Pankratz N, Hernandez D, Paisan‐Ruiz C, Jain S, Halter C A, Michaels V E, Reed T, Rudolph A, Shults C W, Singleton A, Foroud T, ; Parkinson Study Group‐PROGENI investigators Genetic screening for a single common LRRK2 mutation in familial Parkinson's disease. Lancet 2005365(9457)410–412. [DOI] [PubMed] [Google Scholar]
- 11.Eerola J, Hernandez D, Launes J, Hellstrom O, Hague S, Gulick C, Johnson J, Peuralinna T, Hardy J, Tienari P J, Singleton A B. Assessment of a DJ‐1 (PARK7) polymorphism in Finnish PD. Neurology 200361(7)1000–1002. [DOI] [PubMed] [Google Scholar]
- 12.Hughes A J, Daniel S E, Kilford L, Lees A J. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico‐pathological study of 100 cases. J Neurol Neurosurg Psychiatry 199255(3)181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weale M E, Depondt C, Macdonald S J, Smith A, Lai P S, Shorvon S D, Wood N W, Goldstein D B. Selection and evaluation of tagging SNPs in the neuronal‐sodium‐channel gene SCN1A: implications for linkage‐disequilibrium gene mapping. Am J Hum Genet 200373(3)551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J H, Lissarrague S, Essioux L, Sham P C. GENECOUNTING: haplotype analysis with missing genotypes. Bioinformatics 200218(12)1694–1695. [DOI] [PubMed] [Google Scholar]
- 15.Abecasis G R, Cookson W O. GOLD‐‐graphical overview of linkage disequilibrium. Bioinformatics 200016(2)182–183. [DOI] [PubMed] [Google Scholar]
- 16.Cann H M, de Toma C, Cazes L, Legrand M F, Morel V, Piouffre L, Bodmer J, Bodmer W F, Bonne‐Tamir B, Cambon‐Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara G B, Friedlaender J S, Groot H, Gurwitz D, Jenkins T, Herrera R J, Huang X, Kidd J, Kidd K K, Langaney A, Lin A A, Mehdi S Q, Parham P, Piazza A, Pistillo M P, Qian Y, Shu Q, Xu J, Zhu S, Weber J L, Greely H T, Feldman M W, Thomas G, Dausset J, Cavalli‐Sforza L L. A human genome diversity cell line panel. Science 2002296(5566)261–262. [DOI] [PubMed] [Google Scholar]