Abstract
Trichome patterning in Arabidopsis serves as a model system for de novo pattern formation in plants. It is thought to typify the theoretical activator–inhibitor mechanism, although this hypothesis has never been challenged by a combined experimental and theoretical approach. By integrating the key genetic and molecular data of the trichome patterning system, we developed a new theoretical model that allows the direct testing of the effect of experimental interventions and in the prediction of patterning phenotypes. We show experimentally that the trichome inhibitor TRIPTYCHON is transcriptionally activated by the known positive regulators GLABRA1 and GLABRA3. Further, we demonstrate by particle bombardment of protein fusions with GFP that TRIPTYCHON and CAPRICE but not GLABRA1 and GLABRA3 can move between cells. Finally, theoretical considerations suggest promoter swapping and basal overexpression experiments by means of which we are able to discriminate three biologically meaningful variants of the trichome patterning model. Our study demonstrates that the mutual interplay between theory and experiment can reveal a new level of understanding of how biochemical mechanisms can drive biological patterning processes.
Keywords: Arabidopsis, competitive complex formation, pattern formation, trichomes
Introduction
Most theoretical models in biology aiming to explain de novo creation of regular spacing patterns are based on either of two principles: activator–inhibitor or substrate depletion (Gierer and Meinhardt, 1972). In both cases, pattern formation relies on a dynamic instability first suggested by Turing (1952) in the context of morphogenesis. The initiation of Arabidopsis trichomes on the leaf blade serves as an excellent model system to study the molecular mechanism underlying de novo patterning. Trichomes are leaf hairs derived from epidermal cells that are formed in a regular spacing pattern in a rapidly growing cell layer at the leaf base. New trichomes are formed at a minimal distance of three or four cells from already existing ones and their position is not correlated with any other recognizable positional landmark (Hulskamp et al, 1994). A mechanism by which the spacing pattern is achieved by a conserved cell division pattern was excluded by clonal analysis (Larkin et al, 1996; Schnittger et al, 1999). Thus, all data indicate that a de novo patterning mechanism is operating. Several independent mutational screens have identified genes that appear to act as positive or negative regulators of trichome initiation. The corresponding mutants of the positive regulators have fewer or no trichomes. They include the R2R3 MYB transcription factors GLABRA1 (GL1) and MYB23 (Oppenheimer et al, 1991; Kirik et al, 2001, 2005), the bHLH factors GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) (Payne et al, 2000; Zhang et al, 2003; Bernhardt et al, 2005) and the WD40-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) (Galway et al, 1994; Walker et al, 1999a). Yeast two-hybrid data suggest the formation of a trichome-promoting trimeric complex, due to binding of one R2R3 MYB factor and TTG1 to a bHLH factor (Payne et al, 2000). The negative regulators TRIPTYCHON (TRY) and CAPRICE (CPC) were initially identified by mutants showing trichome clusters (Schellmann et al, 2002) and a higher trichome density (Wada et al, 1997a), respectively. Both genes encode homologous single-repeat MYB-related transcription factors (Wada et al, 1997a; Schellmann et al, 2002). Later, four further homologues were found that act in a partially redundant manner, namely ENHANCER OF TRY AND CPC1 (ETC1) (Kirik et al, 2004a), ETC2 (Kirik et al, 2004b), TRICHOMELESS1 (Wang et al, 2007) and CAPRICE-LIKE MYB3 (CPL3) (Tominaga et al, 2008). These inhibitors can compete with the R2R3 MYB factor for binding to GL3/EGL3 in yeast three-hybrid assays (Esch et al, 2003). Therefore, current models assume that these inhibitors counteract the trimeric active complex by this competition mechanism under the assumption that the single-repeat MYB factor-containing complex is inactive (Larkin et al, 1996; Scheres, 2002; Marks and Esch, 2003; Pesch and Hulskamp, 2004; Ishida et al, 2008). It is assumed that the generation of the actual spacing pattern is mediated by the movement of the inhibitors. One downstream target of this machinery is the homeobox transcription factor GLABRA2 (GL2), which is thought to trigger the actual process of trichome formation (Rerie et al, 1994; Cristina et al, 1996).
Virtually the same gene cassette is operating during epidermal root hair determination. Root hairs are arranged in cell files and only epidermal cells overlying a cleft between two cortex cells develop into root hairs (Dolan et al, 1994; Berger et al, 1998). Although this suggests that cell fate is determined by position; root hair fate depends largely on the R2R3 MYB factor WEREWOLF, TTG1, GL3/EGL3 and the inhibitors CPC, TRY and ETC1. Taken together, these genes activate the expression of GL2 in non-root hair cells, where GL2 represses root hair development (Larkin et al, 1996; Scheres, 2002; Marks and Esch, 2003; Pesch and Hulskamp, 2004; Ishida et al, 2008). Thus, in contrast to trichome development on leaves, the default fate in the root epidermis is the formation of root hairs.
In this study, we create a theoretical model based on the current knowledge of the trichome patterning system. We verify experimentally the previous assumption that the expression of the inhibitor TRY is induced by the positive regulators. Further, we find (Payne et al, 2000; Zhang et al, 2003) that the inhibitors TRY and CPC are mobile, whereas GL1 and GL3 are cell autonomous. Protein interaction assays suggest three alternative scenarios by which the positive regulators are inhibited. A combination of overexpression experiments and theoretical modelling allows in the identification of the most relevant inhibition scenario for trichome patterning.
Results
Mathematical modelling and simulation of trichome patterning
We develop our initial model from the following assumptions, most of which are based on already published data in the context of trichome or root hair patterning: (1) the GL1 GL3 complex is considered to be the transcriptionally active complex (Morohashi et al, 2007); (2) similar to GL3 and WEREWOLF in the root hair system, it is assumed that GL1 GL3 activate TRY (Bernhardt et al, 2005); (3) TRY counteracts the GL1 GL3 activity by competing with GL1 for binding to GL3 (Esch et al, 2003); (4) similar to that shown for CPC in the root, it is assumed that TRY protein can move between cells (Kurata et al, 2005). Further, we consider GL3 to be mobile because GL3 can travel between cells in the root (Bernhardt et al, 2005) and GL1 to be immobile (Hulskamp et al, 1994); (5) as observed for the GL3 homologue TT8 and the GL1 homologue TT2, it is assumed that the active complex, GL1 GL3, activates the expression of GL3 (Baudry et al, 2006); (6) TTG1 is not taken into account as it is not essential for trichome formation as indicated by the fact that GL1 and GL3 overexpression or GL1 overexpression in try mutants rescues the ttg1 phenotype (Schnittger et al, 1998); (7) GL2 is considered as a downstream target gene of this machinery (Rerie et al, 1994; Cristina et al, 1996); (8) GL1 and GL3 are expressed ubiquitously in the patterning zone (Larkin et al, 1993; Zhang et al, 2003); (9) in addition, it is assumed that GL1 is activated by the active complex. This is included as a prerequisite for the model; (10) it is further assumed that all proteins are linearly degraded. The resulting interactions are shown in Figure 1 and the corresponding mathematical model is presented in the Materials and methods section.
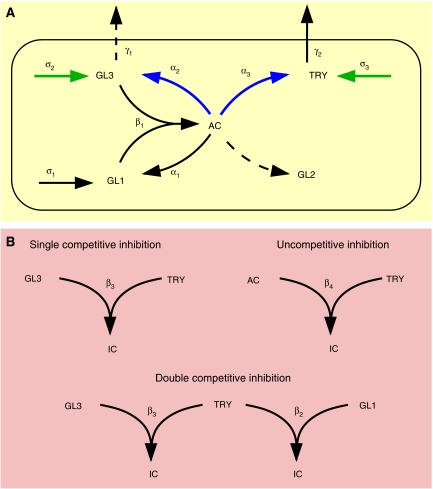
Figure 1.
Mathematical modelling. (A) Activation part of the trichome patterning model. Solid lines indicate processes that are contained in the final model, whereas dashed lines indicate hypotheses that are rejected during the analysis. Greek letters denote the corresponding rate constants. The active complex (AC) induces the expression of the patterning genes GLABRA1 (GL1), GLABRA2 (GL2), GLABRA3 (GL3) and TRIPTYCHON (TRY). GL1 and GL3 form the active complex by dimerization. GL1, GL3 and TRY are basally expressed, and GL3 and TRY are non-cell autonomous. Basal and AC-regulated expression (green and blue arrows) denote processes that are manipulated in the simulations and experiments. (B) Inhibition part of the trichome patterning model. The three inhibition scenarios characterize how TRY may inhibit the positive feedback described in (A). In the cases of single competitive inhibition, TRY prevents the formation of the active complex by binding to free GL3, whereas in the double competitive inhibition TRY binds additionally to free GL1. In case of uncompetitive inhibition, TRY directly binds to the existing active complex. In all scenarios, the resulting inactive complex is denoted by IC. The full model comprises the interactions shown in (A) and one of the inhibitions given in (B).
Simulations of the corresponding differential equations reveal a regular spacing pattern for biologically reasonable parameter ranges. To further validate our model, we directly tested several of our key assumptions that have so far only been based on indirect genetic experiments or made by analogy to the root hair system.
TRY is transcriptionally activated by the active complex and suppresses its own transcription
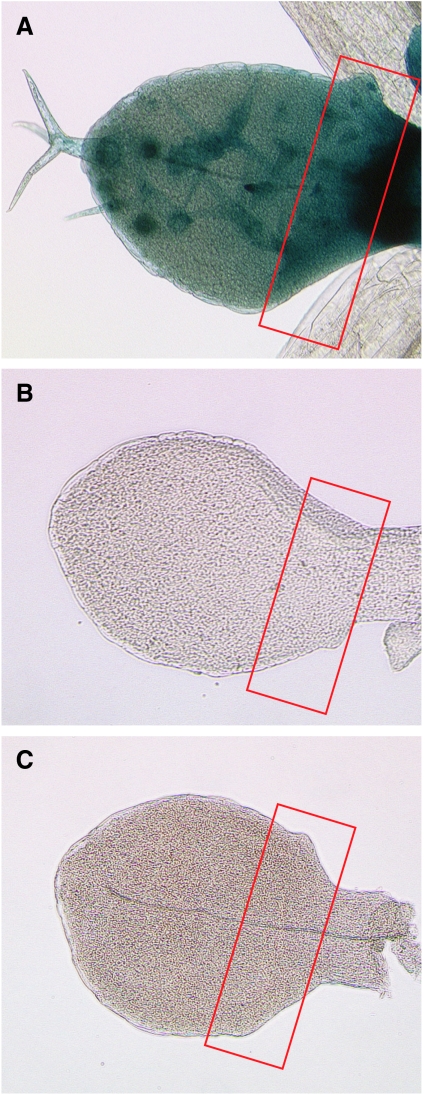
The transcriptional regulation of TRY was analysed using a TRY:GUS construct containing a promoter previously shown to truly reflect the endogenous expression (Schellmann et al, 2002). Trichome initiation takes place only at the base of young leaves (Figure 2, indicated by a red square). In this region, TRY is expressed in all epidermal cells. Expression in trichomes is stronger than in the surrounding epidermis (Schellmann et al, 2002). To corroborate the assumption that the active complex promotes the inhibitor, only those aspects of TRY expression are relevant that occur before or at the time of trichome initiation, i.e. the ubiquitous epidermal expression. This ubiquitous expression is absent in gl1, gl3 and gl3 egl3 mutants, indicating that TRY is transcriptionally induced by the active complex (Figure 2). According to our model, the inhibitors should repress the active complex and therefore indirectly themselves. This is confirmed in plants overexpressing TRY or CPC. No TRY expression is observed in a 35S:TRY or 35S:CPC background (data not shown).
Figure 2.
Expression of TRY:GUS in wild type and mutants. TRY:GUS expression is shown in young leaves: (A) wild type; (B) gl3 and (C) gl1-1. Note that the ubiquitous expression at the leaf base is absent in all single mutants.
Subcellular localization and ability for intercellular movement of TRY, CPC, GL1 and GL3
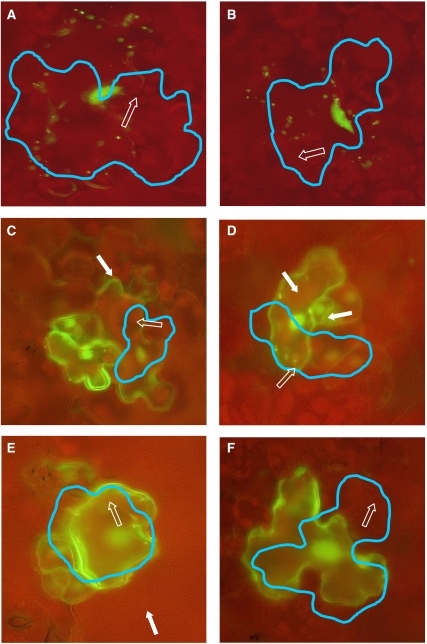
In our model, we assume that TRY and GL3 can move into neighbouring cells similar to that shown for CPC (Kurata et al, 2005) and GL3 (Bernhardt et al, 2005) in the root hair system and that GL1 acts cell autonomously (Hulskamp et al, 1994). To test this in leaves, we fused the coding sequence of TRY, CPC, GL1 and GL3 to GFP and placed them under the control of the constitutive CaMV 35S promoter. 35S:GFP and 35S:GFP:YFP constructs served as controls and a 35S:YFP:peroxisome marker was used to label the targeted cells. The functionality of the TRY and CPC fusion proteins was demonstrated by showing that their overexpression results in a loss of trichomes. The GL1 and GL3 fusion proteins have previously been shown to be functional by Esch et al (2003). Arabidopsis cotyledon and leaf epidermal cells were transformed by micro-projectile bombardment and analysed after 6–10 h. The TRY and CPC fusion proteins were localized in the targeted cell and in approximately one-third of the neighbouring cells. This demonstrates that these fusion proteins can move from the originally transformed cell to its neighbours (Figure 3; Tables I and II). GFP-GL1 and surprisingly also GFP-GL3 proteins did not move in this assay (Figure 3; Table I). The finding that GL3 is cell autonomous is incorporated into our model.
Figure 3.
Intercellular mobility of proteins involved in trichome patterning. Translational fusions of GL1, GL3, TRY and CPC under the control of the 35S promoter (35S:GFP:GL1, 35S:GFP:GL3, 35S:GFP:TRY and 35S:GFP:CPC) are co-bombarded with 35S:YFP:peroxisome into Arabidopsis cotyledons and rosette leaves by the micro-projectile bombardment method and analysed after 6–10 h. Cells expressing fluorescent-labelled peroxisomes are highlighted by a blue line. One peroxisome in the initially transformed cell is indicated by an open arrow. A cell showing fluorescence in the immediate neighbourhood is marked with a white arrow. (A) GFP:GL1 and (B) GFP:GL3 fluorescence are seen only in the transformed cell. (C) GFP:TRY and (D) GFP:CPC fluorescence are found in neighbouring cells. (E) GFP alone is also found in neighbouring cells. (F) GFP:YFP is not mobile.
Table 1.
Protein movement in the leaf epidermis
| Fusion protein | Protein size (kDa) | Cotyledons |
Rosette leaves |
||||
|---|---|---|---|---|---|---|---|
| Total | Movement | % | Total | Movement | % | ||
| GFP-TRY | 41.5 | 190 | 65 | 34 | 111 | 37 | 33 |
| GFP-CPC | 39.9 | 132 | 49 | 37 | 188 | 65 | 34 |
| GFP-GL1 | 54.9 | 101 | 0 | 0 | 68 | 0 | 0 |
| GFP-GL2 | 111.7 | 72 | 0 | 0 | 62 | 0 | 0 |
| GFP-GL3 | 99.1 | 52 | 0 | 0 | 57 | 0 | 0 |
| GFP | 28.55 | 206 | 54 | 26 | 100 | 31 | 31 |
| GFP-YFP | 55.1 | 147 | 0 | 0 | 103 | 4 | 3 |
Table 2.
Number of trichome neigbouring cell showing fluorescence
| Fusion protein | % of cells surrounding the transformed one |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| GFP-TRY | 47.5 | 32.5 | 7.5 | 8.75 | 1.25 | 1.25 | 1.25 |
| GFP-CPC | 42.37 | 35.59 | 18.64 | 3.39 | 0 | 0 | 0 |
| GFP | 31.25 | 16.25 | 16.25 | 8.75 | 8.75 | 8.75 | 10 |
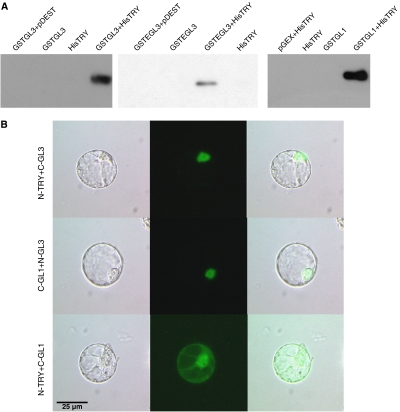
TRY physically interacts with GL1 and GL3
By assessing the interaction of TRY with other patterning genes using the bimolecular fluorescence complementation (BiFC) system and pull-down experiments, we confirmed the interaction between TRY and GL3 and EGL3 (Kirik et al, 2004b; Bernhardt et al, 2005) previously shown using yeast two-hybrid experiments (Figure 4). To our surprise, we also found an interaction of TRY with GL1 in both assays. This suggests an additional inhibitory interaction where TRY competes with GL3 for binding to GL1.
Figure 4.
Molecular interactions of TRY with GL3, EGL3 and GL1. (A) The corresponding single protein fusions or combinations were purified by a GST pull down and detected on a western blot using an anti-His antibody. (B) BiFC was used to detect the interaction between TRY and GL1 or GL3 in protoplasts. Left lane shows a light micrograph, middle lane shows the BiFC fluorescence and the right micrograph shows the overlay.
Challenging the system by overexpression experiments
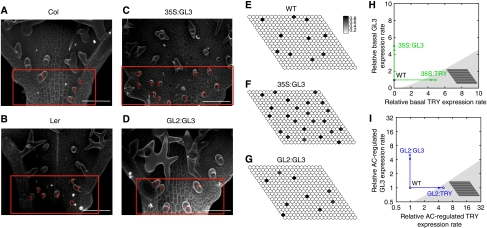
The finding that GL1 can bind to TRY as well as the fact that TRY can bind to GL3 raises the possibility for three different inhibition scenarios (Figure 1B). (1) Single competitive inhibition: TRY binds to free GL3 and prevents GL1 GL3 dimerization; (2) Double competitive inhibition: TRY binds to free GL3 or GL1, thereby preventing the interaction between GL1 and GL3; (3) Uncompetitive inhibition: TRY binds to the GL1 GL3 dimer and represses its function. Although the double competitive model contains the single competitive model as an extreme case, the uncompetitive model is independent of the other two inhibition scenarios. In all three cases, we found regular spacing patterns for biologically meaningful parameters, although with different parameter ranges and sensitivities (Supplementary Figure 4). This suggests manipulation of the system experimentally to discriminate the biological relevance of the three scenarios. We decided to consider overexpression of TRY and GL3 either constitutively under the 35S promoter or under the common downstream promoter of GL2. We created the respective plant lines to investigate the results of these experimental interventions. All GL2:TRY plants are completely glabrous in two independent lines. 35S:GL3 lines show a higher trichome density in the patterning zone (compare Figure 5B and C). In the most basal part of the leaf spanning the region containing only unbranched trichomes, a significant difference is found between wild-type Ler (9.2±2.7, n=30) and 35S:GL3 (14.7±4.3, n=30) by the Student's t-test (P<0.01). However, in GL2:GL3 lines, we observe no significant difference in trichome density in the patterning zone (wild type: 6.3±1.7, n=30; GL2:GL3: 7±2.77, n=30; compare Figure 5A and D). It is noteworthy that in both GL3 overexpression lines, the mature part of the leaf displays additional young trichomes regularly scattered between fully mature trichomes. This situation is rarely found in wild-type Columbia (7.3%±7.3, n=20) and Ler (7.8%±8.4, n=20). In GL2:GL3 and 35S:GL3, they represent a significant fraction of all trichomes (47%±7.3, n=21) and (44.1%±11.5, n=22), respectively. This indicates that these young trichomes are formed later in leaf development, suggesting that the time window allowing trichome formation is expanded in GL2:GL3 and 35S:GL3 lines.
Figure 5.
Results of the experimental and simulated GL3 and TRY overexpressions. (A–D) Scanning electron microscopy. The initiation zone is highlighted by a red box and red T's denote developing trichomes. The scale bar corresponds to 50 mm. (A) Columbia (Col) wild type. (B) Landsberg erecta (Ler) wild type. (C) 35S:GL3 overexpression results in a higher trichome density compared with the corresponding wild type ecotype Columbia. (D) GL2:GL3 overexpression results in a similar trichome density compared with the corresponding wild-type ecotype Landsberg erecta. (E–G) Simulation results for the single competitive inhibition scenario. The relative level of the active complex AC is given in grey scale. High level indicates a future trichome. The parameter values are given in Materials and methods. (E) Wild type (WT). (F) 35S:GL3 overexpression. (G) GL2:GL3 overexpression. The change of the trichome density observed in the experiments is reflected in the simulations. (H, I) Two-dimensional sections of the parameter space. White area denotes the Turing space. The wild-type parameter set is indicated by the black circle and the corresponding simulated pattern is shown in (E). (H) Effect of 35S:GL3 and 35S:TRY overexpression in the single competitive scenario. The overexpression is simulated by increasing the rescaled basal expression rates k4 and k9 of GL3 and TRY, respectively. The 35S:TRY overexpression (right arrow) leads to a loss of trichome patterning (shaded area). Conversely, a five-fold 35S:GL3 overexpression relative to wild type level (top arrow) preserves the ability to form patterns (white area). The corresponding pattern is shown in (F). (I) Effect of GL2:GL3 and GL2:TRY overexpression in the single competitive scenario. The overexpression is simulated by five-fold increased AC-regulated expression rates k5 and k10 of GL3 and TRY, respectively. The corresponding pattern of five-fold GL2:GL3 overexpression is shown in (G).
We simulated the corresponding overexpression experiments of TRY and GL3 using the three inhibition scenarios. Because the model parameters are unknown, we cannot directly test the accordance of the different simulation results with our experimental findings. However, we can confine the model parameters to biologically meaningful ranges and evaluate the accordance within these ranges (see Table III). We randomly sampled parameters inside the reduced parameter space and tested for each sample whether the following five criteria are fulfilled: (1) pattern formation from homogeneous initial conditions is possible, i.e. the parameter set is inside the Turing space; (2) 35S:TRY overexpression leads to a loss of patterning; (3) GL2:TRY overexpression leads to a loss of patterning; (4) 35S:GL3 overexpression yields a higher trichome density compared to wild type; (5) GL2:GL3 overexpression results in a similar trichome density compared to wild type. For details of the simulations, see Materials and methods.
Table 3.
Parameters of the mathematical model
| Dimensionless parameter | Functional relation to dimensional parameters | Biologically reasonable parameter range | SCI: median (IQR) single competition scenario (median (IQR)) | DCI: median (IQR) double competitive scenario (Median (IQR)) |
|---|---|---|---|---|
| k1 | σ1β1/ρ12 | 0.01–100 | 8.37 (25.71) | 4.07 (11.90) |
| k2 | σ1/ρ1 | 0–100 | 4.02 (7.12) | 2.47 (2.92) |
| k3 | β2/β1 | 0.01–100 | 0 | 0.57 (8.68) |
| k4 | σ2β1/ρ12 | 0.01–100 | 9.49 (32.93) | 13.97 (39.89) |
| k5 | σ2/ρ1 | 0–100 | 2.40 (2.98) | 2.89 (3.79) |
| k6 | ρ2/ρ1 | 0.1–10 | 1.27 (3.71) | 1.29 (3.61) |
| k7 | β3/β1 | 0.01–100 | 13.18 (31.54) | 0.91 (12.77) |
| k8 | γ1/ρ2 | 0 | 0 | 0 |
| k9 | σ3β1/ρ12 | 0 | 0 | 0 |
| k10 | α3/β1 | 0.1–1 | 0.46 (0.44) | 0.46 (0.43) |
| k11 | ρ3/ρ1 | 0.1–10 | 0.83 (1.91) | 1.60 (3.75) |
| k12 | β4/β1 | 0.01–100 | 0 | 0 |
| k13 | γ2/ρ3 | 0.1–100 | 35.16 (44.28) | 44.51 (47.97) |
| k14 | ρ4/ρ1 | 0.1–10 | 0.82 (1.20) | 0.72 (0.97) |
The last two columns give the median and IQR of all parameter samples that meet the overexpression criteria as given in the text. Single competitive inhibition (SCI), double competitive inhibition (DCI) and interquartile range (IQR).
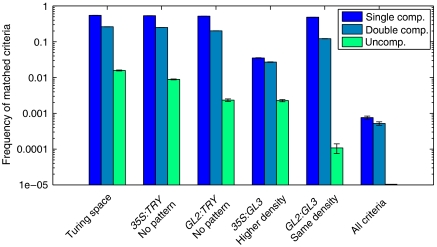
The frequency of matched criteria for the three competition scenarios is presented on a logarithmic scale in Figure 6. The single competitive inhibition scenario meets each criterion most often, followed by the double competitive scenario. The uncompetitive scenario matches each criterion at least one order of magnitude less compared to the two other cases. In particular, the frequency of randomly hitting the Turing space is only 2%, whereas it is more than 30% in the two other scenarios. Interestingly, we found parameter samples that match all criteria simultaneously only for the single and double competitive scenarios but not for the uncompetitive case. As we could not reproduce all the experimental overexpression phenotypes simultaneously with the uncompetitive inhibition scenario, we conclude that it is not consistent with our data.
Figure 6.
Frequency of matched criteria for the three inhibition scenarios. The frequency of each criterion is determined from 106 random samples of the parameter space. The criteria reflect the agreement between the results of a model simulation and the experimentally observed phenotype in wild type and the four overexpression situations. Note the logarithmic scale of the y axis. Although the single and double competition scenarios fulfil all criteria within the same order of magnitude, the frequency of matches for the uncompetitive scenario is one order of magnitude lower than both other cases. Only the single and the double competition scenario, respectively, can match all criteria simultaneously. Mean and standard deviation of each criterion are determined from 10 blocks of 105 samples. For details of the criteria, see Materials and methods.
Single competitive inhibition is a submodel of double competitive inhibition as it contains only the GL3 TRY interaction, whereas the latter additionally contains the GL1 TRY interaction. Hence, we investigate the relation between the parameter samples matching all criteria of these two models simultaneously. In particular, we ask whether the double competitive inhibition is only able to meet all criteria in the extreme case of single competitive inhibition. We calculate the pairwise correlations between all parameters of the double competitive inhibition model and find a strong negative correlation (ρk3, k7= −0.55, P<0.01) between the binding rates of TRY to GL1 and GL3. Parameter sets with comparable binding affinities are found far less frequent than these two extreme cases (see Supplementary Figure 3). Thus, the double competitive inhibition explains in most cases all experimental data either by a strong binding affinity of TRY to GL3 and a weak binding affinity to GL1 or vice versa. Therefore, we conclude that single competitive inhibition is sufficient to explain all experimental findings and double competitive inhibition is not necessary to describe trichome patterning in the context of our data.
A qualitative comparison between the simulated and experimental overexpression of GL3 in the single competitive inhibition scenario is presented in Figure 5C–G. Note that the irregularities of the simulated trichome pattern are due to the fact that only TRY acts non-cell autonomously and can also be observed experimentally. In Figure 5H and I, the effect of the GL3 and TRY overexpression is illustrated by a two-dimensional section of the parameter space. Parameter regions that give rise to a Turing instability, as determined from a linear stability analysis, are indicated in white, whereas regions outside the Turing space are given in grey. For details of the analysis, see the Supplementary information. This local visualization of the Turing space provides an explanation why the overexpression of GL3 and TRY yield different phenotypes. Because of the local convexity of the Turing space, an increased TRY expression yields a parameter set outside the Turing space and consequently leads to a loss of pattern. In contrast, an increased GL3 expression preserves the pattern-forming ability of the model. Note that we only consider the relative increase in expression rates, as the absolute values of the expression rates in wild type and mutant are unknown. The increased sensitivity towards a manipulation of the TRY level in comparison to the GL3 level holds for constitutive as well as AC-driven overexpression. It is noteworthy that this increased sensitivity is not a local feature of the Turing space at this particular parameter set given in Figure 5, but hold for very different parameter sets as indicated by the results presented in Figure 6 and Supplementary Figure 2.
Discussion
The use of phenomenological descriptions in the study of development has a long tradition (Hofmeister, 1868; Thompson, 1942). Essential principles of development were formulated as mathematical models that describe the observed developmental pattern on a phenomenological basis (Gierer and Meinhardt, 1972; Cooke and Zeeman, 1976; Othmer, 1977; Mitchison, 1977; Erneux, 1978; Mitchison, 1980). The overwhelming advances in the molecular understanding of developmental processes in the last decades demand an adaptation of this successful approach to the details of current knowledge and experimental possibilities. Mechanistic in contrast to phenomenological descriptions can fill this gap. Mechanistic models can predict the effect of specific genetic or biochemical interventions and can suggest the design of specific experiments. In this study, we present a mechanistic model for trichome patterning that can be used to address several experimentally feasible interventions of the system.
Our study reveals several new properties of trichome patterning in Arabidopsis leaves. First, we show that GL1 and GL3 are actually positive regulators for the expression of TRY and CPC, which has been assumed previously. This fact is a necessary requirement of theoretical models and has not been experimentally confirmed before. Second, we demonstrate by micro-projectile bombardment that the inhibitors TRY and CPC can move into neighbouring cells. In contrast to the root system, we find that GL3 is cell autonomous. Both results are in agreement with the requirement of the activator–inhibitor model for a farther transport of the inhibitors compared to the activators. As predicted by theoretical models including only the mobility of the inhibitor, the resulting pattern is less regular than in a model including the mobility of both substances. This is also consistent with the less orderly pattern observed in the initiation zone of the young leaf. Further, we confirm the previously shown interaction of GL3 and TRY and find an additional interaction between TRY and GL1. This opens the possibility for different ways how TRY can exert its inhibitory function. We construct three alternative models that reflect different inhibition scenarios: (1) single competitive inhibition in which TRY binds to free GL3 and blocks the formation of the active complex, (2) double competitive inhibition that additionally includes the binding of TRY to free GL1 and (3) uncompetitive inhibition whereby TRY binds only to the activator complex and renders it inactive. Analysis of the three models suggests experimental interventions in the form of different overexpressions to identify the most relevant of these scenarios. We use the 35S promoter and the promoter of the downstream target GL2 to overexpress GL3 and TRY either constitutively or under the control of the active complex, respectively. For each of the three scenarios, we compare simulation results with the overexpression phenotypes. This comparison depends on the particular choice of parameter values. As these are unknown, we employ a global sampling strategy that allows us to evaluate the different scenarios and identify constraints on the parameters. Using this approach, we can predict that the interaction between TRY and GL3 is the most relevant for trichome patterning. In particular, the uncompetitive inhibition scenario cannot reproduce our experimental findings, whereas the double competition scenario mostly does so in the extreme case of single competition. Interestingly, the single competitive inhibition scenario tolerates larger parameter variations than both other scenarios while still forming the correct patterns (cf. Supplementary Figure 4). In this sense, the model can explain why the trichome patterning system is robust and can yield a similar qualitative output despite naturally occurring perturbations, e.g. in the form of genetic and environmental variability.
Although we have considered only few components of the patterning system, our combined experimental and theoretical effort reveals specific system properties far beyond any intuitive judgement. Yet the model is not sufficient to explain many additional observations, e.g. the single and double mutant phenotypes of the inhibitors. Additional experiments on the specific properties of the inhibitors are needed. With joint endeavour of theory and experiment, it should be possible to uncover the principles of trichome pattern formation.
Materials and methods
Mathematical model
From the interactions depicted in Figure 1, we derive the following system of coupled ordinary differential equations describing the time evolution of GL1, GL3, TRY and the active complex (AC).
![]()
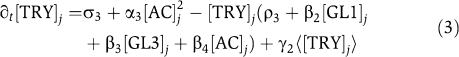 |
The model does not include an equation describing the time evolution of any inactive complex as these do not feed back into the system. Note that the square in equation (3) is needed for the stability of the system. For biological relevance, all variables and parameters have to be non-negative. The model is formulated on a hexagonal grid with coordinates j=(y, x) as depicted in Supplementary Figure 1. Here, 1⩽x⩽xmax and 1⩽y⩽ymax, and xmax and ymax are the number of cells in x and y directions, respectively. We assume periodic boundary conditions of the grid. To describe the coupling between neighbouring cells, we define the passive transport of a variable [C]i as
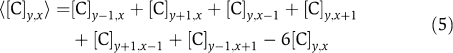 |
where C stands for GL3 or TRY. The parameters in our model are rates for basal expression (σi), regulated expression (αi), degradation (ρi), complex formation (βi) and transport (γi) of the corresponding species. Note that the basal expression of TRY is only incorporated to enable simulations of 35S:TRY overexpression experiments (σ3>0i); in all other cases, σ3=0. The complex formation rates, which together determine the type and strength of the inhibition, are adapted to reflect three different scenarios. These are single competitive (β2=0, β3>0, β4=0), double competitive (β2>0, β3>0, β4=0) and uncompetitive (β2=0, β3=0, β4>0) inhibition. As all model parameters are unknown, a rescaling of time and concentration is applied. This approach allows us to reduce the total number of model parameters and to confine the resulting dimensionless parameters to biologically reasonable ranges, while retaining a mathematically equivalent set of equations. All concentrations are rescaled by the factor β1/ρ1. Time is expressed in units of half-life of GL1, i.e. τ=ρ1t. The resulting dimensionless model has the following form:
![]()
Table III lists the dimensionless parameters together with their corresponding functional relation to the dimensional parameters. Note that due to our experimental finding that GL3 is cell autonomous, we set k8=0 in all numerical simulations.
Numerical simulation
All simulations are performed with MATLAB from Math Works Inc. The ODE system (6)–(9) is integrated using the ode15s function of MATLAB, which is designed to solve stiff ODEs. As we could not obtain an analytical expression for the homogeneous steady state of equations (6), (7), (8) and (9), we determined the steady state by numerical integration of the single-cell model starting with zero initial conditions for all protein concentrations. This steady state is used with additional 1% random variation per cell as initial conditions for the simulation of the grid of cells, including spatial coupling.
Parameter scan
For each inhibition model, 106 random parameter samples are drawn from an exponential distribution to sample the orders of magnitude of each parameter uniformly. Each sample is confined to the ranges specified in Table III. For a given parameter sample, the condition for Turing instability is checked by a linear stability analysis as described in detail in the Supplementary information. Basal GL3 overexpression is simulated by a five-fold increase in the expression rate k4. As a basal expression of TRY is absent in planta, 35S:TRY overexpression is simulated by setting the basal expression rate of TRY, i.e. k9, equal to the value of k4 in the simulated 35S:GL3 overexpression. The overexpression of GL3 and TRY under the GL2 promoter is simulated by a five-fold increased rate of active complex-dependent expression, i.e. rate k5 and k10, respectively, as the active complex binds to the promoter of GL2. The effect of each of these four parameter perturbations is checked by a linear stability analysis (for details of the analysis, see Supplementary information). Additionally, numerical simulation of the 35S:GL2 and GL2:GL3 plants is used to determine the trichome density of the corresponding plants. The simulated 35S:GL3 plant is said to fulfil the experimental observation of an increased trichome density if it is at least 1.5 times higher than the simulated WT density. The density of the simulated GL2:GL3 plants DGL2:GL3 is said to match the WT density DWT if (DGL2:GL3−DWT)2⩽0.0004 holds.
Parameters used for simulation presented in Figure 5
Wild-type parameter values: k1=8.2707, k2=3.4869, k3=0, k4=15.0952, k5=1.3488, k6=0.4503, k7=7.9509, k8=0, k9=0, k10=0.4117, k11=0.9565, k12=0, k13=10 and k14=0.2703. Parameter values of the overexpressed mutants are chosen as described above.
Plant material, growth condition and genetic methods
Plants were grown at 22°C for 16 h white light a day. The Arabidopsis lines TRY:GUS, gl1-1, gl3, gl3 egl3, 35S:TRY and 35S:CPC have been described previously (Oppenheimer et al, 1991; Larkin et al, 1993; Hulskamp et al, 1994; Lloyd et al, 1994; Wada et al, 1997b; Szymanski and Marks, 1998; Walker et al, 1999b; Schellmann et al, 2002). Crosses of the TRY:GUS lines to recessive mutants were performed by using the respective GUS line as male parent. The F2 generation was screened for BASTA resistance.
For the swapping experiment, GL2:GL3 lines have been described previously (Kirik et al, 2005). 35S:GL3 was generated by recombination of full-length GL3 cDNA into pAMPAT vector. 35S:TRY and GL2:TRY were generated by cloning of TRY cDNA in pCAMBIA 1300 (Mathur et al, 2003). The constructs were transformed into the Agrobacterium strain GV3101 by electroporation (Bio-Rad gene pulser) and Landsberg erecta plants were transformed using the floral dip method (Clough and Bent, 1998). Transformants were selected in the T1 generation using MS plates containing 50 mg/l kanamycin or on soil with a 0.1% BASTA solution.
Constructs, biolistic transformation and BiFC
The constructs used for the transient assay in leaf epidermis were created by fusing GFP (Clontech) to the N terminus of the coding sequence of GL1, GL3, TRY and CPC and placed under the control of the CaMV 35S promoter. Details are available on request. Transient expression analysis was carried out by using the particle bombardment method as described previously (Mathur et al, 2003).
TRY, GL1 and GL3 were recombined into BiFC vectors (pBatTL) and transfected in Arabidopsis protoplast (Uhrig et al, 2007). The transfected cells were incubated at 23°C for 16–20 h in the dark before microscopic observation.
Histology and microscopy
GUS staining was performed as described by Malamy and Benfey (1997). Plant specimens were analysed using the LEICA-DMRE microscope equipped with a high-resolution KY-F70 3-CCD JVC camera and frame-grabbing software (DISKUS; Technisches Büro, Königswinter). Photos were edited with Adobe Photoshop 6.
Expression and purification of recombinant fusion proteins
GL1, GL3, EGL3 and TRY cDNA were recombined into pGEX2TMGW and pDEST 17 vectors by Gateway Cloning (Invitrogen) to create GST and His fusion proteins, respectively. Recombinant proteins were expressed in Escherichia coli BL21DE3RIL cells (Stratagene) by the induction of GST fusion proteins with a final IPTG concentration of 0.1 mM at 37°C for 4 h and of His fusion proteins with a final concentration of IPTG of 1 mM at 37°C for 3 h. Bacteria expressing GST fusion proteins were lysed as described by Frangioni and Neel (1993) and proteins were purified through the glutathione sepharose (GE Healthcare cat. no: 17-5279-01) by batch method as described by Sambrook and Russell (2001). The buffer of the purified proteins was changed against 50 mM Tris–HCl pH 7.8, 1 mM EDTA, 150 mM NaCl, 0.1% Nonidet P-40 and 4% BSA by the Amicon Ultra 10k. Bacteria expressing His fusion proteins were pelleted after induction, and lysis was achieved by sonicating five times using 10-s pulses in the lysis buffer (100 mM NaCl, 50 mM Tris pH 7.9, 2 mM EDTA, 1% Triton X-100). His fusion proteins were purified through Ni-NTA resin (Qiagen), according to the product instructions. Final elutions were done with phosphate-buffered saline buffer containing a final concentration of 20 mM EDTA. Purified proteins were dialysed against 50 mM Tris–HCl pH 7.8, 1 mM EDTA, 150 mM NaCl. All of the buffers used contained Roche Complete Protease Inhibitor Cocktail™.
Pull down
GST and His fusion purified proteins (0.5 μg) were incubated together for 2 h at 4°C on a rocking platform. Then 50 μl of glutathione resin (Mathur et al, 2003; GE Healthcare cat. no: 17-5279-01) was added and the mixture was incubated further for 2 h at 4°C on a rocking platform. After incubation, the mixture was washed for five times with buffer (50 mM Tris–HCl pH 7.8, 1 mM EDTA, 150 mM NaCl, 0.1% Nonidet P-40). Pull-down assays and the mixture was washed either in the presence or absence of BSA. SDS gel extraction buffer was added to the samples. Samples were run on SDS–PAGE and blotted against PVDF membranes. Anti-His antibodies were used for the detection of the His-tagged pulled down proteins.
Supplementary Material
Supplementary Figures 1–4
Supplementary Information
Acknowledgments
We appreciate the support of Amanda Walker for the completion of some work in her laboratory. This study was supported by the Sonderforschungsbereich 572 of the Deutsche Forschungsgemeinschaft to MH. SD was supported by the Graduate School for Biological Sciences and BD by the European ADOPT program. BG was supported by the FP6 COSBICS Project (512060), FG by BMBF NGFN II 101 35 05 201 and CF by BMBF FRISYS 0313921. pGEX-2TM-GW vector is kindly provided by Dr Bekir Uelker and Imre Somssich, Max-Planck Institute for Plant Breeding Research, Department of Plant Microbe Interactions, Cologne, Germany.
References
- Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Berger F, Haseloff J, Schiefelbein J, Dolan L (1998) Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr Biol 8: 421–430 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC (1976) A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol 58: 455–476 [DOI] [PubMed] [Google Scholar]
- Cristina MD, Sessa G, Dolan L, Linstead P, Ruberti S, Morelli G (1996) The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J 10: 393–402 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Erneux T (1978) Turing's theory in morphogenesis. Bull Math Biol 40: 771–789 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen M, Sanders M, Hillestad M, Ndkium S, Idelkope B, Neizer J, Marks MD (2003) A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894 [DOI] [PubMed] [Google Scholar]
- Frangioni JV, Neel BG (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem 210: 179–187 [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gierer A, Meinhardt H (1972) A theory of biological pattern formation. Kybernetik 12: 30–39 [DOI] [PubMed] [Google Scholar]
- Hofmeister W (1868) Allgmeine Morphologie der Gewachse. Leipzing: Engelmann [Google Scholar]
- Hulskamp M, Misera S, Jürgens G (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer D, Schiefelbein J, Hulskamp M (2005) Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kirik V, Schnittger A, Radchuk V, Adler K, Hulskamp M, Baumlein H (2001) Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev Biol 235: 366–377 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Hulskamp M, Schiefelbein J (2004a) The ENHANCER OF TRY AND CPC1 (ETC1) gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev Biol 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M (2004b) ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol 55: 389–398 [DOI] [PubMed] [Google Scholar]
- Kurata T, Okada K, Wada T (2005) Intercellular movement of transcription factors. Curr Opin Plant Biol 8: 600–605 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD (1993) Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD (1996) The control of trichome spacing and number in Arabidopsis. Development 122: 997–1005 [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW (1994) Epidermal cell fate determination in Arabidopsis: patterns defined by steroid-inducible regulator. Science 266: 436–439 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Marks MD, Esch JJ (2003) Initiating inhibition. Control of epidermal cell patterning in plants. EMBO Rep 4: 24–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, Hulskamp M (2003) Arabidopsis CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development 130: 3137–3146 [DOI] [PubMed] [Google Scholar]
- Mitchison GJ (1977) Phyllotaxis and the Fibonacci series. Science 196: 270–275 [DOI] [PubMed] [Google Scholar]
- Mitchison GJ (1980) A model for vein formation in higher plants. Proc R Soc Lond B Biol Sci 207: 79–109 [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E (2007) Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Othmer H (1977) Current theories of pattern formation. Lect Math Life Sci 9: 57–87 [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hulskamp M (2004) Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev 14: 422–427 [DOI] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD (1994) The glabra 2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jurgens G, Hulskamp M (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B (2002) Plant patterning: TRY to inhibit your neighbors. Curr Biol 12: R804–R806 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Folkers U, Schwab B, Jürgens G, Hulskamp M (1999) Generation of a spacing pattern: the role of TRIPTYCHON in trichome patterning in Arabidopsis. Plant Cell 11: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A, Jurgens G, Hulskamp M (1998) Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125: 2283–2289 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD (1998) GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10: 2047–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA (1942) On Growth and Form. Cambridge: Cambridge University Press [Google Scholar]
- Tominaga R, Iwata M, Sano R, Inoue K, Okada K, Wada T (2008) Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 135: 1335–1345 [DOI] [PubMed] [Google Scholar]
- Turing A (1952) The chemical basis of morphogenesis. Philos Trans R Soc Lond Ser B 237: 37–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig JF, Mutondo M, Zimmermann I, Deeks MJ, Machesky LM, Thomas P, Uhrig S, Rambke C, Hussey PJ, Hulskamp M (2007) The role of Arabidopsis SCAR genes in ARP2-ARP3-dependent cell morphogenesis. Development 134: 967–977 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997a) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K (1997b) Epidermal cell differentiation in Arabidopsis determined by a myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999a) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999b) The TTG1 (transparent testa, glabra1) locus which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis encodes a WD40-repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Kwak SH, Zeng Q, Ellis BE, Chen XY, Schiefelbein J, Chen JG (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–4
Supplementary Information