Abstract
Although considerable progress has been made in dissecting the signaling pathways involved in the innate immune response, it is now apparent that this response can no longer be productively thought of in terms of simple linear pathways. InnateDB (www.innatedb.ca) has been developed to facilitate systems-level analyses that will provide better insight into the complex networks of pathways and interactions that govern the innate immune response. InnateDB is a publicly available, manually curated, integrative biology database of the human and mouse molecules, experimentally verified interactions and pathways involved in innate immunity, along with centralized annotation on the broader human and mouse interactomes. To date, more than 3500 innate immunity-relevant interactions have been contextually annotated through the review of 1000 plus publications. Integrated into InnateDB are novel bioinformatics resources, including network visualization software, pathway analysis, orthologous interaction network construction and the ability to overlay user-supplied gene expression data in an intuitively displayed molecular interaction network and pathway context, which will enable biologists without a computational background to explore their data in a more systems-oriented manner.
Keywords: database, gene expression, innate immunity, interaction network, pathway visualization
Introduction
Humans and other mammals are constantly exposed to a deluge of microorganisms, although they usually suffer little or no detrimental effects largely because microbes are efficiently dealt with in most cases by the host's immune system. Traditionally, the immune response has been divided into two different branches, the adaptive immune response and the innate immune response. In recent years, there has been an explosion of interest in the innate immune response. It is now appreciated that most pathogens to which we are exposed are eliminated through the innate immune response without necessarily requiring the activation of adaptive immunity. Furthermore, the importance of the innate immune response is being recognized in the initiation of and interplay with the adaptive immune response (MacLeod and Wetzler, 2007), as well as the mechanism by which vaccine adjuvants operate in boosting immunity (Kwissa et al, 2007). The innate immune response, however, can also be a double-edged sword. If not tightly regulated, an overwhelming immune response can lead to what is sometimes called a cytokine storm. One such out-of-control response, sepsis, results in more than 200 000 deaths a year in the United States alone (Angus et al, 2001).
Over the course of the last decade, significant progress has been made in understanding the innate immune response, including the detailed dissection of some of the critical signaling pathways involved (Lang and Mansell, 2007; Matsukawa, 2007) and the discovery of several important pathogen recognition receptor families, such as the Toll-like receptors (TLRs) (Akira, 2006), the nucleotide binding and oligomerization domain (NOD)-like receptors (NLRs) (Inohara and Nunez, 2001; Kanneganti et al, 2007), and the retinoic acid-inducible gene 1 (RIG-1)-like receptors (RLRs) (Yoneyama et al, 2004; Thompson and Locarnini, 2007). Despite these efforts, many questions remain unanswered including how the innate immune system initiates distinct responses toward particular pathogens. It is becoming increasingly clear that the innate immune response does not involve simple linear pathways but rather complex networks of pathways and interactions, positive and negative feedback loops and multifaceted transcriptional responses (Tegner et al, 2006; Lee and Kim, 2007).
To better understand the complexities of the innate immune response and the cross-talk between its components, complementary systems-level analyses and more focused follow-up experimental approaches are now needed. Recently, researchers have started to apply systems biology approaches to the study of the immune system (Gilchrist et al, 2006; Oda and Kitano, 2006; Tegner et al, 2006; Andersen et al, 2008) and bioinformatics resources are now emerging to aid these types of analyses. So far, these resources have tended to focus on particular aspects of genomics research; the Reference Database of Immune Cells (RefDIC), for example, provides transcriptomic and proteomic data from immune-relevant cells (Hijikata et al, 2007), whereas others, such as the Innate Immunity Database, contain transcription profiles and computationally predicted transcription factor-binding sites for 2000 mouse immune genes (Korb et al, 2008). Others still have been established primarily for a specific group of researchers, such as ImmPort (http://www.immport.org), which has been created for researchers funded through the National Institute of Allergy and Infectious Diseases.
None of these resources provide detailed molecular interaction or pathway information and nor do they provide the capability to integrate disparate types of data to enable systems-level investigation of the immune response. Furthermore, despite the enormous efforts of the major publicly available interaction and pathway databases to provide as wide-ranging cover as possible (Salwinski et al, 2004; Alfarano et al, 2005; Joshi-Tope et al, 2005; Breitkreutz et al, 2007; Chatr-aryamontri et al, 2007; Kanehisa et al, 2007; Kerrien et al, 2007), it was quickly apparent to us that currently available bioinformatics resources provided poor coverage and detail of the molecular interactions and pathways relevant to innate immunity, information which is essential for the systems-orientated interpretation of large-scale genomics data. For example, TLR4, despite its status as one of the most important molecules in the innate immune response, has relatively few molecular interactions annotated in the major publicly available interaction databases. Five of these interaction databases combined contained annotated molecular interactions between TLR4 and just 11 other proteins. Through a review of the literature we have curated, in detail, a further 16 unique interactions, and provided annotation of nearly 60 different lines of evidence supporting these interactions.
To overcome these problems and to provide a resource that will enable biologists without a computational background to explore their data in a more systems-oriented manner, we have developed InnateDB. InnateDB (www.innatedb.ca) is a publicly available database and analysis platform for the genes, proteins, experimentally verified interactions and pathways involved in the human and murine innate immune responses. InnateDB captures improved coverage of the innate immunity interactome by integrating more than 100 000 known interactions and approximately 2500 pathways from major public databases together with in-house manually curated data into a centralized resource. To date, the InnateDB curation team has reviewed more than 1000 publications and curated more than 3500 innate immunity-relevant interactions, richly annotating them in terms of the experimental evidence and the context in which they occur. Building on this data, several integrated bioinformatics and novel network visualization tools are provided to facilitate systems-level investigations of the innate immune response.
In particular, InnateDB facilitates the efficient analysis of large gene expression data sets in an interaction network and pathway context, allowing users to upload and integrate their own data that can then be interactively investigated. As a proof of concept, we have investigated the gene expression profiles of lipopolysaccharide (LPS)-stimulated THP-1 human monocytic cells at several time points. Using InnateDB, the expected significant involvement of the TNF-α and NF-κB pathways in the LPS response was identified, but InnateDB also provided deeper insight by probing the component molecular interactions involved.
By combining existing publicly available data on the general human and mouse interactomes with the manual curation of innate immune-relevant biomolecules and pathways, and by providing a user-friendly, accessible analysis environment, InnateDB provides a centralized integrative biology resource for researchers in immunology as well as the broader biomedical community.
Results
Contextually curating innate immunity-relevant interactions
One of the primary goals of InnateDB is to provide a manually curated centralized resource for experimentally verified human and mouse protein, gene and RNA molecular interactions involved in the innate immune system. To do this, a dedicated full-time team of curators and several other part-time curators have been assembled to review the relevant biomedical literature and to submit detailed annotation on these interactions and pathways to InnateDB through the customized submission system software of the database. To date, more than 3500 innate immunity-relevant interactions, involving around 1000 genes, have been manually curated through the review of approximately 1000 publications. The curation team has focused on reviewing interactions that are not annotated in other interaction databases, and have ensured that only interactions with published experimental evidence of a direct physical or biochemical interaction are submitted to InnateDB. Other types of evidence such as colocalization, overexpression, microarray or other inferences, are not deemed acceptable. A detailed guide to curation process is available from www.innatedb.ca/help.
The importance of manual curation is clear, as we are often able to double the number of interactions for a given gene or protein compared to the number currently present in the other interaction databases combined. Furthermore, this detailed manual curation permitted us to richly annotate these interactions and to place them in their relevant context. This contextual annotation includes details of the supporting publication; participant molecules; the species; the interaction detection method; the host system; the interaction type; the cell, cell-line and tissue types and several other fields.
Defining what is an innate immunity-relevant interaction is a difficult task due to the increasing understanding of the complexity of the response and the difficulty in drawing the line between innate and adaptive immunity. To date, InnateDB manual curation has prioritized molecules that are well-described members of key innate immunity signaling pathways, including the TLR pathways, the NF-κB pathway, MAPK signaling pathway, JNK signaling pathway, NOD-like receptor pathway and the RIG-I antiviral pathway (Kanneganti et al, 2007; Lee and Kim, 2007; Thompson and Locarnini, 2007). We have then curated experimentally verified interactions between these molecules and any other molecule, regardless of whether the interacting molecule has any known role in innate immunity. In this way, InnateDB quickly expands upon the simple linear view of innate immunity pathways into a more comprehensive interaction network perspective.
The InnateDB submission system software has been designed to allow submission of interaction annotation using Open Biomedical Ontology (OBO) controlled terms and in a manner that is compliant with the recently proposed ‘minimum information required for reporting a molecular interaction experiment' (MIMIx) guidelines (Orchard et al, 2007; Smith et al, 2007). Interaction data in InnateDB are also curated, stored and downloadable in the Proteomics Standards Initiative Molecular Interaction (PSI-MI) 2.5-compliant XML format (Hermjakob et al, 2004).
Going beyond innate immunity: a centralized resource for interactions and pathways
Aside from the well-known signaling pathways (e.g. TLR signaling), a range of other disparate processes, including apoptosis (Mayadas and Cullere, 2005), ubiquitination (Carmody et al, 2007), endocytosis (Tateno et al, 2007) and cell activation and recruitment (Coelho et al, 2007), are all required to mount an effective innate immune response. Adding to this complexity is the fact that the borders between the innate and adaptive immune responses are becoming increasingly blurred. Furthermore, if one hopes to identify new networks or pathways involved in innate immunity, analyses must include genes and proteins that are, as yet, not known to play specific roles in the innate immune response.
To address these issues, in addition to the detailed manual curation of the genes, proteins and their interactions and pathways that are specifically known to have a role in the innate immune response, InnateDB also incorporates data on the entire human and mouse interactomes. To do this, annotation on more than 100 000 human and mouse interactions was integrated from several of the major publicly available interaction databases into InnateDB (Figure 1). Where available, all interaction annotation was extracted from these databases as suggested by the MIMIx guidelines (Orchard et al, 2007), similar to our manually curated data. This downloaded interaction data may also be reviewed as part of the manual curation process, which systemically reviews biomedical literature for innate immunity-relevant interactions. If a curator finds, upon reading of the relevant publications, that an interaction from an external database is not supported, or is inaccurate in some way, for example specifying the incorrect species, this interaction is removed and an accurate entry is submitted to InnateDB. Curator software, developed in-house, allows the curation team to modify existing annotations and to remove externally sourced interactions that are deemed not supported upon review of the evidence. All interaction data in InnateDB, including manually curated data, are available to download in PSI-MI 2.5 format from the download page. InnateDB currently consists of 98 760 and 9018 human and mouse interactions, respectively, and 2792 hybrid interactions. The InnateDB statistics page provides weekly updated statistics on interaction data in InnateDB, broken down by species, curation, interaction type and molecule type.
Figure 1.
Integration of publicly available resources as a foundation for InnateDB. More than 100 000 interactions were integrated into InnateDB from the Molecular Interaction (MINT) database (Chatr-aryamontri et al, 2007); the IntAct database (Kerrien et al, 2007); the Database of Interacting Proteins (DIP) (Salwinski et al, 2004); the General Repository for Interaction Datasets (BioGRID) (Breitkreutz et al, 2007) and the Biomolecular Interaction Network Database (BIND) (Alfarano et al, 2005). Cross-references to more than 2500 pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al, 2007), the NCI-Nature Pathway Interaction Database (PID) (http://pid.nci.nih.gov), Integrating Network Objects with Hierarchies (INOH) pathway database (http://www.inoh.org/), NetPath (http://www.netpath.org) and the Reactome database (Joshi-Tope et al, 2005) were also incorporated. Up-to-date versions of Ensembl (www.ensembl.org) provide details of human and mouse genes, transcripts and proteins along with rich protein and gene annotation from the Universal Protein Resource (UniProt) (The UniProt Consortium, 2007), Gene Ontology (Ashburner et al, 2000) and Entrez Gene (http://www.ncbi.nlm.nih.gov/).
To enable the investigation of genes, proteins and their molecular interactions that are relevant to particular pathways, InnateDB also includes cross-references of genes not only to innate immunity-relevant pathways but also to more than 2500 pathways from several of the major publicly available pathway databases (Figure 1). Detailed gene and protein annotation has also been extracted from a variety of other data sources (Figure 1). As annotation in the publicly available databases that are integrated into InnateDB is being constantly updated, we have developed several scripts to enable the automatic and regular update of this information in InnateDB.
Robust orthology and gene order predictions: facilitating comparative analysis
The majority of mammalian interaction data available in InnateDB and other interaction databases primarily refers to human genes and proteins. To facilitate comparative network-based analysis of the human, mouse and bovine interactomes, detailed orthology predictions have been integrated into InnateDB. Orthologs are genes that are evolutionarily related through a speciation event and are essentially the ‘same' gene in different species. In comparison, paralogs are genes that are derived through a gene duplication event, and although evolutionarily related, are not the ‘same' gene, and are unlikely to have all of the same functions in different species. To more accurately infer the function of a gene in one species and extrapolate that to another, orthologous genes must be compared in the two species. Orthology predictions in InnateDB were generated using an in-house method, Ortholuge, which provides accurate predictions of orthology using a phylogenetic distance-based approach (Fulton et al, 2006). Orthology predictions are further supported through the development of a human and mouse gene order and synteny browser, which allows the visualization of orthologs in their genomic context and the investigation of whether these orthologs are in a region of conserved gene order.
Orthology information in InnateDB can be used to construct orthologous interaction networks in human, mouse and cow to identify common and alternative regulation networks across species. Orthologous interaction networks may be used, for example, to highlight potential differences in the signaling pathways of different species in response to particular infections.
User-friendly yet powerful searching of InnateDB
A flexible web-based interface at www.innatedb.ca allows both simple and more advanced searching of InnateDB. This interface has been developed in close collaboration with our experimental biologists to ensure that the interface is intuitive and easy to use for the biologist end user. More complex search and analysis capabilities were also included to enable more powerful systems-level analyses of the innate immune response. A guided video tutorial to using the InnateDB interface can be found at http://www.innatedb.ca/video/.
From the InnateDB search page, one has the option of searching ‘molecules', ‘interactions' or ‘pathways', or users can upload and investigate their own data in the ‘data analysis' section. Molecule searches allow one to search for particular genes or proteins of interest, either by gene or protein name or through external identifiers from a variety of other databases. Genes and proteins are frequently known, or have previously been known, by several different names or symbols, which are called synonyms. InnateDB stores lists of these synonyms from the Entrez Gene and Ensembl databases. Gene or protein name searches of InnateDB automatically search possible alternative names for the query rather than restricting searches to particular names or symbols from a specific database. InnateDB preferentially displays the HUGO Gene Nomenclature Committee (HGNC) name for human genes (Bruford et al, 2008) and Mouse Genomic Nomenclature Committee (MGNC) name for mouse genes (Blake et al, 1999). Boolean operators allow the construction of more complex queries. From the molecule search results page, rich annotation related to the genes or proteins of interest can be obtained, including details of the interactions and pathways that they are involved in, Gene Ontology annotation (Ashburner et al, 2000), orthology and gene order predictions and links to a range of external databases.
The interaction search page provides enormous flexibility in searching for molecular interactions of interest, including by the species; the molecule type and name; the interaction detection method; the interaction type (e.g. phosphorylation); the cell line, cell or tissue type that the interaction was detected in and several other parameters. One can choose to return only the molecules that directly interact with the molecules of interest (primary interactions) or the secondary interaction partners as well (i.e. molecules that are interactors of the primary interactors). Because InnateDB includes interactions from several data sources, a grouping algorithm is used to combine interactions with the same participants and interaction type to reduce redundancy. From the interaction search results (Figure 2), details of each interaction and the evidence supporting it, including links to the original supporting publication, can be obtained.
Figure 2.
A screenshot of the InnateDB interaction search results page. Each interaction is shown alongside the interaction type (e.g. phosphorylation), the supporting publications column, which shows the number of publications in the database that support the interaction, and links to the interaction details page. This page provides further information on each interaction including annotation on the experiment, the cell type, the tissue type and links to the original publication describing the interaction. Interactions can be downloaded in a variety of formats or one can click on the Cerebral button to interactively visualize the interactions (see Figure 4).
The pathway search page allows users to search for genes, proteins and interactions that are components of a pathway of interest. Defining which genes are members of a particular pathway is determined through a combination of InnateDB manual curation and through cross-references to one of the 2500 pathways that are incorporated into InnateDB (Figure 1). One can choose to return only genes that are specifically annotated as being part of a pathway (core genes) or can opt for a more comprehensive view that includes all the genes or proteins that interact with the core genes. The results of interaction and pathway searches can be interactively visualized in our novel network visualization software at the click of a button (see below) or can be downloaded in several convenient formats, including tab-delimited, comma-separated value, Simple Interaction Format (SIF) and the PSI-MI 2.5 XML format (Hermjakob et al, 2004).
Browsing capability is also provided to allow those who may be unfamiliar with the contents of the database to quickly scan for interactions or pathways of interest and to explore the types of information stored within InnateDB. One can browse InnateDB for interactions by interaction type or by relevant pathways, and can also browse annotated immune gene lists from a variety of internal and external sources (Bouwmeester et al, 2004; Calvano et al, 2005; Kelley et al, 2005; Ortutay and Vihinen, 2006).
Exploring gene/protein expression data in an interaction network/pathway context
InnateDB has been designed to enable the integration of user-supplied gene or protein expression data with network and pathway data, as this contextual view of quantitative data is useful for providing more comprehensive insights into large data sets. Traditionally, gene expression data sets have been analyzed using statistical techniques such as clustering (D'Haeseleer, 2005) or ontology over-representation analysis (Al-Shahrour et al, 2004). Although these types of analyses often reveal underlying information about potential co-regulation within a set of genes and the processes that are differentially regulated in response to a particular stimulus, additional insight into the nature of the mechanisms governing these responses may be gained through interpreting the data in a network context. Through the integration of gene expression and interaction network data, one can begin to investigate how differentially expressed genes are actually interconnected and identify the key networks or pathways and their central components that are likely regulating gene expression. These types of analyses may also enable investigators to identify important regulators of these networks that exert their regulation through protein modification or another non-transcriptional mechanism, which may not be highlighted as significant in more traditional gene expression analyses alone.
To carry out such analyses, a list of genes or protein identifiers (from the Ensembl, Entrez Gene, RefSeq and UniProt databases), along with associated gene expression values and P-values from up to 10 different experimental conditions or time points, can be uploaded. These identifiers are then cross-referenced to identifiers in InnateDB and the molecular interactions for each gene are returned, along with the uploaded expression values. There is considerable flexibility in choosing which data are returned, for example, one can limit the interactions returned to only those associated with a particular pathway. Batch interaction searches are currently limited to searching for interactions associated with a maximum of 5000 uploaded genes at one time. When one attempts to search for the interactions involving more than 5000 genes, tens of thousands of interactions in the database are usually returned and these data are difficult to analyze or visualize online. If one wishes to obtain all interactions in InnateDB, these are available in PSI-MI format from the InnateDB download page.
Batch searches can also be carried out to return detailed gene annotation or a list of pathways with which the uploaded list of genes is associated (Figure 3). These searches do not have the 5000 gene limit that is used for batch interaction searches. This integrated data can be downloaded or analyzed with our pathway or gene ontology over-representation analysis tools to identify particular pathways or ontology terms that are enriched in differentially expressed gene data sets. The over-representation analysis tools provide a number of statistical methods for pathway and gene ontology analysis, including the Fisher exact test, the hypergeometrical distribution and the χ2 test. P-values are automatically corrected using the Benjamini and Hochberg correction for the false discovery rate (Benjamini and Hochberg, 1995), although one can also opt to use the more conservative Bonferroni correction. As InnateDB uses the proportion of differentially expressed genes for each condition on the entire array to more accurately calculate which pathways are significant, it is recommended that all data from the microarray, and not just differentially expressed probes, are uploaded and analyzed using the pathway over-representation analysis tool. InnateDB also provides the option of pathway analysis of a subset of genes.
Figure 3.
A screenshot of the InnateDB pathway batch search results page. Each gene in a user-uploaded list is linked to its associated pathways. If fold changes in gene expression values are included in the uploaded file, they are shown alongside each gene and can subsequently be used in the pathway over-representation analysis (by clicking the red Pathway ORA button at the top of the page) to determine which pathways are significantly associated with differentially regulated genes.
Network/pathway visualization using Cerebral and Cytoscape
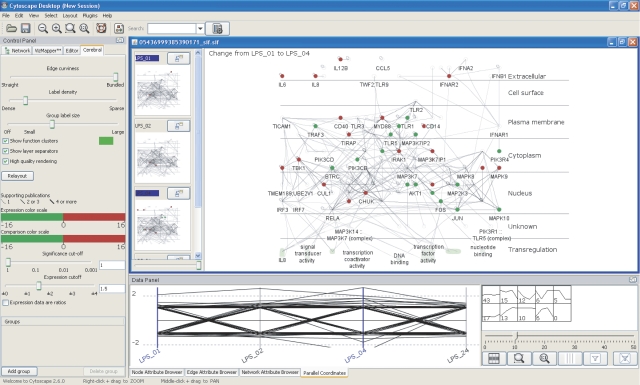
All molecular interaction and pathway data in InnateDB can be interactively investigated using a new version of our network visualization tool, Cerebral (Barsky et al, 2007). Cerebral is a Java plugin for the Cytoscape biomolecular interaction viewer (Shannon et al, 2003) that automatically generates more biologically intuitive pathway-like layouts of a network using subcellular localization information. The latest version of Cerebral allows one to visualize and overlay data from multiple quantitative experimental conditions or time points on a network. This version of Cerebral has been fully integrated into InnateDB. Upon retrieving a set of interactions or a pathway, one can launch a Java Web Start instance of Cytoscape with the Cerebral plugin loaded, such that the user does not have to install any software locally. Using information automatically extracted from InnateDB, a localization-based layout of the network is loaded (Figure 4). Nodes are placed in a layer according to the localization inferred from Gene Ontology (Ashburner et al, 2000). Users can then manipulate the network using native Cytoscape functions, and can interactively link from a particular node (molecule) or edge (interaction) of interest to a detailed page of annotation in InnateDB.
Figure 4.
Cerebral enables the overlay of quantitative data in a molecular interaction network context. To facilitate the side-by-side comparison of specific experimental conditions, Cerebral uses a series of small, linked views to visualize quantitative data (gene expression values, for example) across multiple conditions simultaneously, alongside a larger central window, which permits more detailed investigation of particular conditions or regions of the network. Nodes (i.e. molecules) are colored according to user-defined thresholds of fold change in gene expression (red, significantly upregulated; green, significantly downregulated). The small multiple windows and the larger overview window are linked––if one zooms in or out, pans, mouses over or selects a node in one of the views, the same action will be perpetuated across all views. By selecting one of the small multiple windows, the expression values for that condition will be promoted to the larger overview window. If two small multiple windows are selected simultaneously, the difference in gene expression in the two conditions is computed and displayed in the main window. In this view, the nodes in the main window are colored according to the magnitude of the change between the two conditions. InnateDB uses the number of pieces of evidence supporting an interaction, usually separate publications or experiments, as a measure of confidence in the interaction. Cerebral uses weighted lines in its display to represent these confidence scores (heavier weighted lines=higher confidence). By right clicking on a node or edge, one may interactively link to the relevant pages in InnateDB for more detailed annotation regarding the gene, protein or interaction of interest.
If expression data have also been provided, for example through the ‘data analysis' function of InnateDB, this data will be integrated with the network and displayed in a series of small windows, each providing an overview of the experimental conditions that were submitted (Figure 4). The uploaded quantitative data associated with each experiment are used to color the nodes in the network, allowing one to interactively visualize how a particular network changes, for example, in its gene expression, over several conditions.
Two further features have been integrated into Cerebral to facilitate a user's exploration of their expression data. First, the parallel coordinates window is a line graph showing the expression profile for genes of interest, providing a profile-based view of a gene's behavior across all conditions. Second, k-means clustering has been implemented to group genes with similar expression profiles. The number of clusters can be adjusted using a slider, and the recalculation occurs so quickly as to not be noticeable. A thumbnail showing the average profile of a specific cluster and the number of genes with that profile is displayed. By selecting a thumbnail, the genes within that cluster alone will be displayed. The k-means clustering algorithm is not limited to working on only the 10 conditions or time points that can be uploaded to InnateDB at one time. Should one wish to analyze more than 10 conditions, one may install a local version of Cytoscape with the Cerebral plugin installed.
As mentioned, Cerebral is a Java plugin that is loaded within a Web Start instance of the Cytoscape network visualization and analysis tool. The Cerebral layout may be deleted by the user and alternative Cytoscape layouts of the network can be created. One can then use any of the other Cytoscape plugins for additional analysis. The results of any interaction search in InnateDB are also provided in a PSI-MI 2.5 XML format. These data can readily be uploaded to several online network analysis tools such as Hubba (http://hub.iis.sinica.edu.tw/Hubba/), which accepts data in this format for network analysis (Lin et al, 2008).
Investigating the LPS-induced inflammatory response in a network and pathway context
We have previously reported a microarray investigation of differential gene expression responses of LPS-stimulated human THP-1 monocytic cells at 1, 2, 4 and 24 h, in the presence or absence of the host defense peptide LL-37 (Mookherjee et al, 2006). As a case example of how InnateDB can facilitate the analysis of such studies, we have re-analyzed this data (Hokamp et al, 2004) and used InnateDB to investigate the pathways and interactions that are significantly involved in the LPS response in this cell type. Re-analysis of the LPS-stimulated human THP-1 data set in the absence of LL-37 revealed that there were 454 differentially expressed genes in total (Supplementary Table 1).
In the original paper, the pathways involved in these responses were identified through the manual assignment of differentially expressed genes to pathways based on the biomedical literature (Mookherjee et al, 2006). In contrast, InnateDB enabled the comparative pathway analysis of each of the time points simultaneously in less than a few minutes. Using the ‘data analysis' feature of InnateDB, this entire data set was uploaded to the database, the pathways associated with each gene were returned (Figure 3), and the pathway over-representation analysis tool in InnateDB was used to very rapidly determine which of the more than 2500 pathways, sourced from several of the publicly available pathway databases, were significantly associated with differentially regulated genes.
At the 1, 2 and 4 h time points following LPS stimulation, InnateDB identified the TNF-α signaling pathway and the NF-κB pathway as being significantly associated with upregulated genes, confirming the well-known involvement of these pathways in this response (Lin and Yeh, 2005). At the 4 h time point, which was associated with the largest number of differentially expressed genes, a range of other pathway terms were also identified as being significantly upregulated, including ‘TLR signaling pathway'; ‘IL-1 signaling'; ‘IL-23-mediated signaling events'; ‘IL-10 anti-inflammatory signaling pathway'; ‘keratinocyte differentiation'; ‘p75 (NTR)-mediated signaling' and ‘small cell lung cancer'. Several other pathways had a number of component genes that were upregulated in response to LPS, including the MAPK signaling pathway, the JAK-STAT pathway, the EGFR1 signaling pathway and the TGF-β pathway, although these were not significant after correction for multiple testing.
InnateDB also enabled the rapid identification of which genes in which pathways were differentially expressed. By automatically integrating the uploaded gene expression data with molecular interaction network information, InnateDB readily enabled the visualization and interactive interrogation of the genes and proteins with which they might interact (Figure 4). In this way, InnateDB not only enabled high-level pathway over-representation analysis, but allowed more sophisticated analysis of the regulatory networks involved in this gene expression data set. Investigating the data in this manner allowed us to identify potential functional relationships between groups of genes/proteins that otherwise might not have been so evident. For example, transcription factor-binding site over-representation analysis of differentially expressed genes, using the oPOSSUM program (Ho Sui et al, 2005), identified the enrichment of both NF-κB and CCAAT/enhancer-binding protein (C/EBP)-binding site motifs, in genes differentially expressed at 1, 2 and 4 h. Enrichment of binding sites for two other transcription factors, hepatic leukemia factor (HLF) and nuclear factor, interleukin 3 regulated (NFIL3) were also identified at these time points. C/EBP is a family of transcription factors that are well known to cooperate with NF-κB in transcriptional regulation (Xia et al, 1997; Xiao et al, 2004), and both HLF and NFIL3 were annotated in InnateDB as interacting with C/EBP proteins. This allowed us to quickly construct the hypothesis that these HLF and NFIL3 may play an important role along with C/EBP and NF-κB in the regulation of the LPS response. Further discussion of this analysis is available in the Supplementary Information.
Discussion
Systems biology approaches to investigating the innate immune system are in their infancy (Smith and Bolouri, 2005; Tegner et al, 2006). InnateDB provides the first publicly available consolidated effort to facilitate such approaches for innate immunity research and to enable this and other research communities to investigate their data in a more systems-orientated manner. InnateDB provides detailed manually curated annotation for innate immunity-relevant molecular interactions and pathways, as well as computationally integrated data on the wider human and mouse interactomes from several major publicly available databases. Specific interactions, pathways and genes or proteins of interest can be interactively searched for in InnateDB through the flexible web-based search interface of the database, providing a knowledge base for the community, whereas the bioinformatics and network visualization tools incorporated into InnateDB elevate the system from database to robust analysis platform.
InnateDB allows one to integrate quantitative data (such as differential gene expression) into a molecular interaction network and pathway context, enabling the interrogation of such data in novel and insightful ways. Investigating differentially expressed molecular interaction networks may identify subnetworks or as-yet unidentified pathways as being significantly involved in the response to a particular stimulus. By incorporating Cytoscape into InnateDB, investigators are able to take a closer look at the interactions involved in these pathways or subnetworks, potentially identifying cross-talk between key pathways, and highlighting the molecules that are the hubs of these networks. The Cerebral plugin allows one to further extend this experience, visually interrogating data across multiple conditions.
Integrated pathway over-representation analysis can identify those pathways that are significantly associated with differentially regulated genes. Through such pathway analysis, it is possible to identify common pathways that are involved in the innate immune response to particular infections, and to identify the common central regulators of these pathways as attractive targets for immune modulation. The current pathway analysis tool integrated into InnateDB is expected to be more powerful than many of the existing tools that tend to use only one or two sources of pathway annotation for analysis (Goffard and Weiller, 2007), whereas InnateDB has collected data from several major pathway databases (Figure 1).
InnateDB manual curation has already resulted in the contextual annotation of more than 3500 innate immunity-relevant molecular interactions, through the review of approximately 1000 journal articles. Continued manual curation of innate immunity-relevant molecular interactions and the annotation of interactions in their relevant biological pathways will be an important component to improve the accuracy of systems-level investigations of the innate immune system. Additionally, annotating the context in which an interaction occurs, as is done in InnateDB, will be essential if investigators are to truly adopt systems-level approaches. Whether or not an interaction occurs may depend on a number of factors including the subcellular localization, cell type, tissue type and stimulus, all of which must be considered for a more accurate systems perspective. Regarding curation of molecular interactions, much remains to be done by the community. It is estimated that only about 15% of all interactions likely to exist have been reported in the literature to date (Bader et al, 2008), and of those already published many are not available in a consolidated standardized format in the publicly available interaction databases. Our own manual curation efforts indicate that the number of interactions for a given gene can often be doubled through literature review compared to the number of interactions currently present in all of the interaction databases combined. Importantly, more complete annotation of pathways will lead to more accurate pathway analysis. We have already noted cases where a particular pathway was significant in an over-representation analysis only when all components of that pathway had been accurately identified and annotated.
Innate immunity is an increasingly complex field of study. To assist in increasing the coverage and accuracy of data in InnateDB, experts from the innate immunity research community could be recruited to participate in the additional review and curation of molecules and pathways of interest, adopting an approach similar to our successful Pseudomonas Community Annotation Project (Brinkman et al, 2000; Winsor et al, 2005). It now seems likely that the submission of interaction data to a database, such as InnateDB, will be a requirement prior to publication in the near future, analogous to the current procedure for many journals that require deposition of sequence and microarray data in a relevant database. InnateDB welcomes the participation of researchers in its curation project. By contacting innatedb-mail@sfu.ca, researchers will be able to submit interactions over the internet through the user-friendly InnateDB submission system software. A detailed guide to the submission process has been developed and is available at http://www.innatedb.ca/help.
We have shown here how InnateDB facilitates the rapid investigation of large gene expression data sets, enabling one to quickly identify the pathways that are significantly altered in their gene expression and to investigate the interactions between the molecular components of these pathways. InnateDB was used to rapidly confirm that the TNF-α and NF-κB pathways were significantly induced at the early time points in LPS-stimulated THP-1 cells and to detail and visualize how the components of these pathways interact. The types of analyses discussed here are just a sample of those possible when using gene expression data integrated into a molecular interaction network and pathway context using InnateDB. InnateDB, along with other emerging resources for bioinformatics and systems-level analysis of immunology (Kelley et al, 2005; Ortutay and Vihinen, 2006; Hijikata et al, 2007; Korb et al, 2008), will undoubtedly lead to novel and much deeper insights into the innate immune response to particular pathogens.
Materials and methods
The InnateDB website is installed on an IBM x3550 server with 16 GB RAM running the openSUSE Linux v10.2 operating system, the Apache Tomcat servlet container and the Apache HTTP web server. A separate MySQL database server, with identical hardware configuration, hosts the data, whereas front-end access to the database is through JavaServer Pages and the Apache Struts Framework. Cerebral v2 is currently launched within the webstart version of Cytoscape v2.6. Regular SQL dumps of the InnateDB database are available on request and it is anticipated that an API will be developed in the future. A figure illustrating the InnateDB schema is available as Supplementary Information.
Supplementary Material
Supplementary Figure
Supplementary Table
Supplementary Information
Acknowledgments
We thank Kathleen Wee, Eddie Yuen, Patrick Taylor, Sheena Tam, Tom Yang and other members of the Pathogenomics of Innate Immunity project for their assistance in manual curation of InnateDB. InnateDB has been funded by Genome Canada and Genome BC through the Pathogenomics of Innate Immunity (PI2) project and by the Foundation for the National Institute of Health and the Canadian Institutes of Health Research (CIHR) under the Grand Challenges in Global Health Research Initiative (Grand Challenges ID: 419). DJL and JLG hold Postdoctoral Trainee Awards from the Michael Smith Foundation for Health Research (MSFHR), and JLG also holds a Sanofi Pasteur CIHR fellowship. MDW holds a Junior Graduate Studentship Award from the MSFHR. FSLB is a CIHR New Investigator and an MSFHR Senior Scholar. REWH holds a Canada Research Chair (CRC). We also thank the various interaction, pathway and annotation databases that have been integrated into InnateDB for freely providing their data to the public.
References
- Akira S (2006) TLR signaling. Curr Top Microbiol Immunol 311: 1–16 [DOI] [PubMed] [Google Scholar]
- Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D'Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H et al. (2005) The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res 33: D418–D424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics (Oxf) 20: 578–580 [DOI] [PubMed] [Google Scholar]
- Andersen J, VanScoy S, Cheng TF, Gomez D, Reich NC (2008) IRF-3-dependent and augmented target genes during viral infection. Genes Immun 9: 168–175 [DOI] [PubMed] [Google Scholar]
- Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 29: 1303–1310 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader S, Kuhner S, Gavin AC (2008) Interaction networks for systems biology. FEBS Lett 582: 1220–1224 [DOI] [PubMed] [Google Scholar]
- Barsky A, Gardy JL, Hancock REW, Munzner T (2007) Cerebral: a Cytoscape plugin for layout of and interaction with biological networks using subcellular localization annotation. Bioinformatics 23: 1040–1042 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B 57: 289–300 [Google Scholar]
- Blake JA, Richardson JE, Davisson MT, Eppig JT (1999) The Mouse Genome Database (MGD): genetic and genomic information about the laboratory mouse. The Mouse Genome Database Group. Nucleic Acids Res 27: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, Dolinski K, Tyers M (2007) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res 36: D637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman FS, Hancock REW, Stover CK (2000) Sequencing solution: use volunteer annotators organized via internet. Nature 406: 933. [DOI] [PubMed] [Google Scholar]
- Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E (2008) The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res 36: D445–D448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF (2005) A network-based analysis of systemic inflammation in humans. Nature 437: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH (2007) Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science 317: 675–678 [DOI] [PubMed] [Google Scholar]
- Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res 35: D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho AL, Schaller MA, Benjamim CF, Orlofsky AZ, Hogaboam CM, Kunkel SL (2007) The chemokine CCL6 promotes innate immunity via immune cell activation and recruitment. J Immunol 179: 5474–5482 [DOI] [PubMed] [Google Scholar]
- D'Haeseleer P (2005) How does gene expression clustering work? Nat Biotechnol 23: 1499–1501 [DOI] [PubMed] [Google Scholar]
- Fulton DL, Li YY, Laird MR, Horsman BG, Roche FM, Brinkman FS (2006) Improving the specificity of high-throughput ortholog prediction. BMC Bioinformatics 7: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Kennedy K, Hai T, Bolouri H, Aderem A (2006) Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441: 173–178 [DOI] [PubMed] [Google Scholar]
- Goffard N, Weiller G (2007) PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res 35: W176–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, Roechert B, Poux S, Jung E, Mersch H, Kersey P, Lappe M, Li Y, Zeng R, Rana D, Nikolski M et al. (2004) The HUPO PSI's molecular interaction format––a community standard for the representation of protein interaction data. Nat Biotechnol 22: 177–183 [DOI] [PubMed] [Google Scholar]
- Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, Kanagawa O, Kawamoto H, Kawano K, Koseki H, Kubo M, Kurita-Miki A, Kurosaki T, Masuda K, Nakata M, Oboki K et al. (2007) Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics 23: 2934–2941 [DOI] [PubMed] [Google Scholar]
- Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW (2005) oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res 33: 3154–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokamp K, Roche FM, Acab M, Rousseau ME, Kuo B, Goode D, Aeschliman D, Bryan J, Babiuk LA, Hancock REW, Brinkman FS (2004) ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res 32: W457–W459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Nunez G (2001) The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene 20: 6473–6481 [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L (2005) Reactome: a knowledgebase of biological pathways. Nucleic Acids Res 33: D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2007) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G (2007) Intracellular NOD-like receptors in host defense and disease. Immunity 27: 549–559 [DOI] [PubMed] [Google Scholar]
- Kelley J, de Bono B, Trowsdale J (2005) IRIS: a database surveying known human immune system genes. Genomics 85: 503–511 [DOI] [PubMed] [Google Scholar]
- Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, Kohler C, Khadake J, Leroy C, Liban A, Lieftink C, Montecchi-Palazzi L, Orchard S, Risse J, Robbe K, Roechert B et al. (2007) IntAct—open source resource for molecular interaction data. Nucleic Acids Res 35: D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb M, Rust AG, Thorsson V, Battail C, Li B, Hwang D, Kennedy KA, Roach J, Rosenberger CM, Gilchrist M, Zak D, Johnson C, Marzolf B, Aderem A, Shmulevich I, Bolouri H (2008) The Innate Immune Database (IIDB). BMC Immunol 9: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwissa M, Kasturi SP, Pulendran B (2007) The science of adjuvants. Expert Rev Vaccines 6: 673–684 [DOI] [PubMed] [Google Scholar]
- Lang T, Mansell A (2007) The negative regulation of Toll-like receptor and associated pathways. Immunol Cell Biol 85: 425–434 [DOI] [PubMed] [Google Scholar]
- Lee MS, Kim YJ (2007) Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem 76: 447–480 [DOI] [PubMed] [Google Scholar]
- Lin CY, Chin CH, Wu HH, Chen SH, Ho CW, Ko MT (2008) Hubba: hub objects analyzer—a framework of interactome hubs identification for network biology. Nucleic Acids Res 36: W438–W443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Yeh WC (2005) Implication of Toll-like receptor and tumor necrosis factor alpha signaling in septic shock. Shock 24: 206–209 [DOI] [PubMed] [Google Scholar]
- MacLeod H, Wetzler LM (2007) T cell activation by TLRs: a role for TLRs in the adaptive immune response. Sci STKE 2007: pe48. [DOI] [PubMed] [Google Scholar]
- Matsukawa A (2007) STAT proteins in innate immunity during sepsis: lessons from gene knockout mice. Acta Med Okayama 61: 239–245 [DOI] [PubMed] [Google Scholar]
- Mayadas TN, Cullere X (2005) Neutrophil beta2 integrins: moderators of life or death decisions. Trends Immunol 26: 388–395 [DOI] [PubMed] [Google Scholar]
- Mookherjee N, Brown KL, Bowdish DM, Doria S, Falsafi R, Hokamp K, Roche FM, Mu R, Doho GH, Pistolic J, Powers JP, Bryan J, Brinkman FS, Hancock REW (2006) Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J Immunol 176: 2455–2464 [DOI] [PubMed] [Google Scholar]
- Oda K, Kitano H (2006) A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol 2: 2006.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, Salama JJ, Moore S, Wojcik J, Bader GD, Vidal M, Cusick ME, Gerstein M, Gavin AC, Superti-Furga G, Greenblatt J et al. (2007) The minimum information required for reporting a molecular interaction experiment (MIMIx). Nat Biotechnol 25: 894–898 [DOI] [PubMed] [Google Scholar]
- Ortutay C, Vihinen M (2006) Immunome: a reference set of genes and proteins for systems biology of the human immune system. Cell Immunol 244: 87–89 [DOI] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D (2004) The Database of Interacting Proteins: 2004 update. Nucleic Acids Res 32: D449–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, Leontis N, Rocca-Serra P, Ruttenberg A, Sansone SA, Scheuermann RH, Shah N, Whetzel PL, Lewis S (2007) The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25: 1251–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Bolouri H (2005) Dissecting innate immune responses with the tools of systems biology. Curr Opin Immunol 17: 49–54 [DOI] [PubMed] [Google Scholar]
- Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, Paulson JC (2007) Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol 27: 5699–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegner J, Nilsson R, Bajic VB, Bjorkegren J, Ravasi T (2006) Systems biology of innate immunity. Cell Immunol 244: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res 36: D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Locarnini SA (2007) Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol 85: 435–445 [DOI] [PubMed] [Google Scholar]
- Winsor GL, Lo R, Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock REW, Brinkman FS (2005) Pseudomonas aeruginosa Genome Database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res 33: D338–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Cheshire JK, Patel H, Woo P (1997) Cross-talk between transcription factors NF-kappa B and C/EBP in the transcriptional regulation of genes. Int J Biochem Cell Biol 29: 1525–1539 [DOI] [PubMed] [Google Scholar]
- Xiao W, Hodge DR, Wang L, Yang X, Zhang X, Farrar WL (2004) Co-operative functions between nuclear factors NFkappaB and CCAT/enhancer-binding protein-beta (C/EBP-beta) regulate the IL-6 promoter in autocrine human prostate cancer cells. Prostate 61: 354–370 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Table
Supplementary Information