Abstract
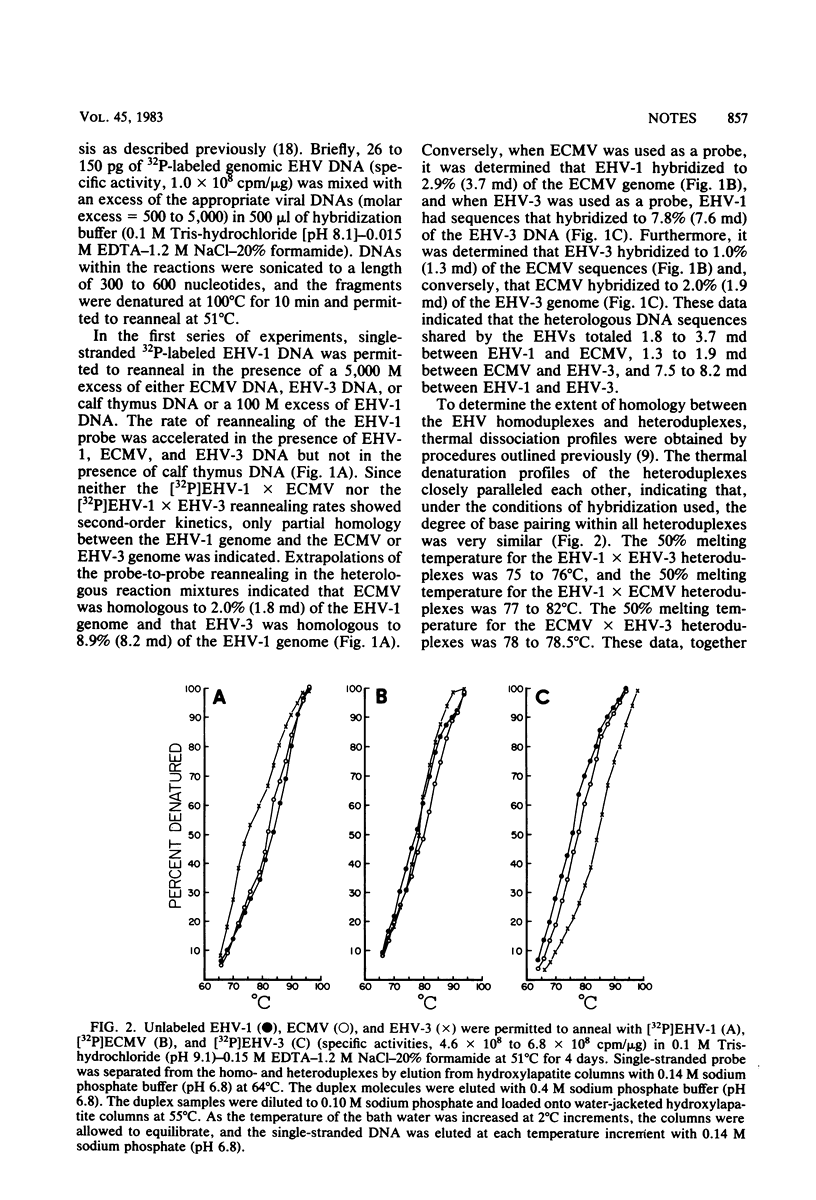
Genomic DNAs of equine herpesvirus type 1 (EHV-1), EHV-2 (equine cytomegalovirus), and EHV-3 were examined by reassociation kinetic and thermal denaturation analyses to determine the extent and degree of homology among the three viral DNAs. Results of reassociation analyses indicated a limited homology among the three EHV genomes. Homologous DNA sequences equivalent to 1.8 to 3.7 megadaltons between EHV-1 and equine cytomegalovirus, 7.6 to 8.2 megadaltons between EHV-1 and EHV-3, and 1.3 to 1.9 megadaltons between equine cytomegalovirus and EHV-3 were detected. Examination by thermal denaturation of the DNA homoduplexes and heteroduplexes formed during reassociation revealed a high degree of base pairing within the duplexes, suggesting that closely related sequences may be conserved among the genomes of EHV.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen G. P., O'Callaghan D. J., Randall C. C. Genetic relatedness of equine herpesvirus types 1 and 3. J Virol. 1977 Dec;24(3):761–767. doi: 10.1128/jvi.24.3.761-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G. P., Randall C. C. Proteins of equine herpesvirus type 3. I. Polypeptides of the purified virion. Virology. 1979 Jan 15;92(1):252–257. doi: 10.1016/0042-6822(79)90232-0. [DOI] [PubMed] [Google Scholar]
- Atherton S. S., Sullivan D. C., Dauenhauer S. A., Ruyechan W. T., O'Callaghan D. J. Properties of the genome of equine herpesvirus type 3. Virology. 1982 Jul 15;120(1):18–32. doi: 10.1016/0042-6822(82)90003-4. [DOI] [PubMed] [Google Scholar]
- Dauenhauer S. A., Robinson R. A., O'Callaghan D. J. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J Gen Virol. 1982 May;60(Pt 1):1–14. doi: 10.1099/0022-1317-60-1-1. [DOI] [PubMed] [Google Scholar]
- Heilman C. J., Jr, Zweig M., Stephenson J. R., Hampar B. Isolation of a nucleocapsid polypeptide of herpes simplex virus types 1 and 2 possessing immunologically type-specific and cross-reactive determinants. J Virol. 1979 Jan;29(1):34–42. doi: 10.1128/jvi.29.1.34-42.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- MIYAZAWA Y., THOMAS C. A., Jr NUCLEOTIDE COMPOSITION OF SHORT SEGMENTS OF DNA MOLECULES. J Mol Biol. 1965 Feb;11:223–237. doi: 10.1016/s0022-2836(65)80053-5. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan D. J., Cheevers W. P., Gentry G. A., Randall C. C. Kinetics of cellular and viral DNA synthesis in equine abortion (herpes) virus infection of L-M cells. Virology. 1968 Sep;36(1):104–114. doi: 10.1016/0042-6822(68)90120-7. [DOI] [PubMed] [Google Scholar]
- PLUMMER G. SEROLOGICAL COMPARISON OF THE HERPES VIRUSES. Br J Exp Pathol. 1964 Apr;45:135–141. [PMC free article] [PubMed] [Google Scholar]
- Perdue M. L., Cohen J. C., Kemp M. C., Randall C. C., O'Callaghan D. J. Characterization of three species of nucleocapsids of equine herpesvirus type-1 (EHV-1). Virology. 1975 Mar;64(1):187–204. doi: 10.1016/0042-6822(75)90091-4. [DOI] [PubMed] [Google Scholar]
- Perdue M. L., Kemp M. C., Randall C. C., O'Callaghan D. J. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974 May;59(1):201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Henry B. E., Duff R. G., O'Callaghan D. J. Oncogenic transformation by equine herpesviruses (EHV). I. Properties of hamster embryo cells transformed by ultraviolet-irradiated EHV-1. Virology. 1980 Mar;101(2):335–362. doi: 10.1016/0042-6822(80)90449-3. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Dauenhauer S. A., O'Callaghan D. J. Electron microscopic study of equine herpesvirus type 1 DNA. J Virol. 1982 Apr;42(1):297–300. doi: 10.1128/jvi.42.1.297-300.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOEHNER R. L., GENTRY G. A., RANDALL C. C. SOME PHYSICOCHEMICAL CHARACTERISTICS OF EQUINE ABORTION VIRUS NUCLEIC ACID. Virology. 1965 Jul;26:394–405. doi: 10.1016/0042-6822(65)90003-6. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Pettersson U., Sambrook J. Viral DNA in transformed cells. I. A study of the sequences of adenovirus 2 DNA in a line of transformed rat cells using specific fragments of the viral genome. J Mol Biol. 1974 Jul 15;86(4):709–726. doi: 10.1016/0022-2836(74)90348-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wharton J. H., Henry B. E., O'Callaghan D. J. Equine cytomegalovirus: cultural characteristics and properties of viral DNA. Virology. 1981 Feb;109(1):106–119. doi: 10.1016/0042-6822(81)90475-x. [DOI] [PubMed] [Google Scholar]



