Abstract
Recent studies have shown that the lipidation and assembly state of apolipoprotein E (apoE) determine receptor recognition and amyloid-β peptide (Aβ) binding. We previously demonstrated that apoE secreted by HEK cells stably expressing apoE3 or apoE4 (HEK-apoE) binds Aβ, and inhibits Aβ-induced neurotoxicity by an isoform-specific process that requires apoE receptors. Here we characterized the structure of HEK-apoE assemblies and determined their receptor binding specificity. By chromatography, HEK-apoE elutes in high molecular weight fractions and is the size of plasma HDL, consistent with a multi-protein assembly. No lipid was associated with these apoE assemblies. Several methods for analyzing receptor binding indicate that HEK-apoE is a ligand for low-density lipoprotein (LDL) receptor-related protein (LRP) but not the LDL receptor. This suggests that self-assembly of apoE may induce a functional conformation necessary for binding to LRP. Our results indicate that, in addition to lipid content, the assembly state of apoE influences Aβ binding and receptor recognition.
INTRODUCTION
Apolipoprotein E (apoE) is a 34-kDa glycoprotein that is a surface component of various plasma lipoprotein particles including chlyomicron remnants, β-migrating VLDL (β-VLDL), LDL, and a subclass of HDL (reviewed in 1). ApoE mediates high-affinity binding of apoE-containing lipoproteins to cell surface endocytic receptors during the transport and metabolism of plasma cholesterol (Chol) and triglycerides (TG) (1, 2). In the central nervous system (CNS), apoE is the primary apolipoprotein in the HDL-like lipoprotein particles in cerebral spinal fluid (CSF) and in the discoidal particles secreted by astrocytes in vitro (reviewed in 3). In humans, apoE exists in three isoforms that differ by a single amino acid at two polymorphic sites (E2=Cys112, Cys158; E3=Cys112, Arg158; and E4=Arg112, Arg158). The ε4 allele of apoE is a major risk factor for Alzheimer's disease (AD) (4, 5), suggesting that regulation of apoE expression and/or metabolism in the CNS may contribute to the pathogenesis of AD. While the mechanism by which apoE4 functions as an AD risk factor is not known, several hypotheses have been proposed. One possibility is an apoE isoform-specific response to brain injury (6, 7) and neuronal regeneration, a hypothesis supported by the observation that apoE3- but not apoE4-containing lipoproteins stimulate neurite outgrowth (8-11). Alternatively, apoE-containing lipoproteins may interact with amyloid β-peptide (Aβ) at several mechanistic levels. First, apoE may modulate the clearance of Aβ by forming a complex with the peptide that is then cleared by one or more of the numerous apoE receptors present on neurons or glial cells (12-15). Indeed, apoE3 has been shown to have a higher affinity for the peptide than apoE4, particularly when measured by the formation of an SDS-stable complex (16-18). Second, apoE affects the deposition of amyloid. AD patients with 1 or 2 alleles of ε4 exhibit a dose-dependent increase in total amyloid plaque burden compared to AD patients lacking ε4, an observation reproduced by crossing apoE null and human apoE transgenic mice with APP transgenic mice (19-21).
ApoE-containing lipoproteins bind members of the LDL receptor (LDLR) gene family. ApoE associated with LDL (2, 22) and DMPC vesicles (23) acts as ligands for the LDLR. In contrast, only apoE-enriched β-VLDL and recombinant apoE act as ligands for LDLR-related protein (LRP) (24-27). LRP is a large multifunctional receptor that binds and endocytosis over 30 structurally and functionally distinct ligands (26, 28-30), and is expressed abundantly in the liver and CNS. In addition to LRP, apoE-enriched lipoproteins have been shown to be ligands for other members of the LDLR gene family such as megalin/LRP2 (31), the VLDL receptor (32), and apoE receptor-2 (apoER2) (33, 34). The ability of apoE-containing lipoproteins to bind multiple members of the LDLR gene family suggests that apoE recognizes a common structural motif that is present within the ligand-binding regions of these receptors. However, apoE-containing lipoproteins also exhibit a differential affinity for members of the LDLR family, suggesting that the conformation of apoE affects its receptor-binding specificity.
We previously demonstrated that apoE particles secreted by HEK-293 cells stably transfected with human apoE3 or apoE4 cDNA (HEK-apoE) differentially form an SDS-stable complex with Aβ, a property HEK-apoE shares with apoE associated with plasma lipoproteins (16, 17). Specifically, HEK-apoE3 and plasma VLDL-containing apoE3 bind Aβ with a higher affinity than apoE4 (16, 17). However, apoE3 and E4 purified from HEK-apoE and plasma VLDL by a process that includes delipidation and denaturation, or purchased from a commercial vendor exhibit a comparable, lower affinity for Aβ (17, 18, 35). It was assumed in our earlier publications that small amounts of lipid associated with these HEK-apoE “particles” facilitated the adoption of a favorable conformation for isoform-specific interaction with Aβ. To further investigate the molecular determinates that dictate the function of apoE-containing particles, we analyzed the structure, composition, and apoE receptor binding affinity of HEK-apoE. We found that while HEK-apoE was secreted as a high molecular weight assembly, was the size of plasma HDL and measured 12 nm in size by EM analysis, HEK-apoE is virtually lipid-free. We demonstrate that these apoE assemblies are ligands for LRP but not LDLR. These results suggest that the conformation of apoE-enriched lipoproteins, and the conformation of apoE induced by its own self-assembly, both result in recognition by LRP. We further infer that the in vitro secretory process of HEK cells favors the adoption by apoE of a conformation that allows for the isoform-specific interaction with Aβ and selective affinity for apoE cell surface receptors.
EXPERIMENTAL PROCEDURES
Materials
Receptor-associated protein (RAP) was isolated and purified from a glutathione S-transferase fusion protein expressed in E. coli, as described previously (26). N-succinimidyl 3-(4-hydroxy-5-[125I] iodophenyl) propionate (Bolton and Hunter reagent) was purchased from Amersham (Oakville, Canada). Cytochrome C, TCA and PBS were from Sigma Chemical Co. (St. Louis, MO). BSA (fraction V) and Pronase were from Calbiochem-Novabiochem Corp. (La Jolla, CA). PD-10 Sephadex columns and Sepharose 6 columns were from Pharmacia Biotech (Uppsala, Sweden). Tissue culture media was from GibcoBRL (Grand Island, NY).
Preparation of HEK ApoE Particles
As previously described, HEK-293 cells were stably transfected with cDNAs encoding human apoE3 or apoE4. Serum-free conditioned media were harvested, concentrated to 500μg/ml, and dialyzed (10-kDa cutoff membrane) in PBS (HEK-apoE) (16, 36).
SDS-PAGE
Five μg HEK-apoE was added to LDS sample buffer under non-reducing conditions, loaded on a 4-12% BIS-TRIS NuPAGE gel (Invitrogen), and electrophoresed at 160 volts for 60-minutes. Gels were stained with Simply Blue coomassie stain (Invitrogen) according to manufacture's protocol. Molecular weight values determined using protein standards (BenchMark, Invitrogen).
HEK-apoE Molecular Weight Determination by Calibrated Size Exclusion Chromatography (SEC)
Chromatography was performed using a low dead volume HPLC equipped with bio-compatible solvent delivery (Dynamax, Rainin Instruments) and UV detection (UV-1, Rainin Instruments). A single Superose6 column was equilibrated in PBS and 100 μl samples were injected and chromatographed at a flow rate of 0.4 ml/min. Column parameters used for calculation of Kav values were obtained from the manufacturer except column exclusion limit (void volume) that was measured using blue dextran (Pharmacia) and inclusion volume which was measured using acetone diluted 1:100 in PBS.
Molecular weight and Stokes radius (size) values were calculated based on comparison of the HEK-apoE elution volume:
Ve = elution volume for the protein
Vo = Superose6 void volume
Vt = total Superose6 bed volume
Kav values were determined for a panel of eight proteins (Sigma) with known molecular weight values between 11 - 443 kDa.
| Apoferritin | 443,000 Da |
| Beta amylase | 200,000 Da |
| Alcohol dehydrogenase | 150,000 Da |
| Bovine serum albumin | 66,000 Da |
| Ovalbumin | 42,7000 Da |
| Carbonic anhydrase | 28,980 Da |
| Myoglobin | 17,000 Da |
| Cytochrome C | 11,393 Da |
A standard curve was generated plotting the log molecular weight vs. Kav. HEK-apoE molecular weight was calculated using the following equation representing the exponential fit of the data. y = 2.3133e07 × e(−10.56x) (R2 = 0.99391)
Gel Filtration Chromatography
As previously described, 1.0 ml of concentrated HEK-apoE was isolated by SEC using fast protein liquid chromatography (FPLC) with tandem Superose 6 columns (Amersham Pharmacia), in 0.02M sodium phosphate, pH 7.4, with 50mM NaCl, 0.03% EDTA, and 0.02% sodium azide (flow rate 0.4 ml/min) (37). Eighty 400μl samples were collected for analysis. Fractions 35-45 were concentrated and pooled for further analysis.
Western Blots
2X non-reducing Laemmli buffer (4% SDS, no β-mercaptoethanol) was added to SEC fractions, boiled for 5 min, and either: (1) spot blotted and probed with apoE antiserum (obtained by immunizing rabbits with apoE purified from human serum) or (2) selected fractions were electrophoresed on 10-20% SDS/Tricine gels, transferred to Immobilon-P membranes (Millipore Corp.), and probed with apoE antiserum.
Electron Microscopy (EM)
Pooled fractions 35-45 of HEK-apoE isolated by SEC were examined by using the Philips CM10 electron microscope at the Electron Microscopy Core Facility of the University of Chicago as described previously (37, 38). The diameters of 100 intact particles from an enlarged photomicrograph were measured by using a micrometer lens and expressed as mean ± standard error of the mean (SEM).
Ultracentrifugation
As previously described, 1.0ml of concentrated HEK-apoE was separated on 3-20% sodium bromide single-spin gradients by centrifugation at 38,000 rpm in an SW41 Ti rotor for 66 hours at 15°C. Thirty 400 μl fractions were collected and dialyzed against Tris-buffered saline before analysis (37, 39). In addition, the HEK-apoE media was adjusted to density 1.25 g/ml with NaBromide to isolate total lipoproteins.
Lipid Analysis
Total cholesterol (Chol), triglyceride (TG), and phospholipids (PL) was measured enzymatically using commercial kits (Boehringer Mannheim for TC and TG; Waco, Richmond, VA for PL). The sensitivity limit of these kits is <1ng/ml.
Cell Culture
Wild type mouse embryonic fibroblasts (MEF-1 cells), fibroblasts homozygous for disruption of the LRP gene (MEF-2 cells), fibroblasts homozygous for disruption of the LDLR gene (MEF-3 cells), and fibroblasts homozygous for disruption of both LRP and the LDLR genes (MEF-4 cells), were kindly provided by Joachim Herz (University of Texas Southwestern Medical Center at Dallas). MEF cells were cultured at 37°C in a humidified incubator with 5% CO2 in DMEM supplemented with 10% fetal calf serum.
LDLR Assay
The LDLR binding assays were performed essentially as described (23). Human LDL was isolated from the plasma of normal fasting subjects and radiolabeled by the iodine monochloride method. Recombinant apoE3 or apoE4 were mixed with dimyristal phosphatydolcholine (DMPC, Sigma) at a ratio of 1:3.75 (w/w). Normal human fibroblasts were cultured for one week prior to the binding assays.
Protein Iodination
HEK-apoE was iodinated using the Bolton and Hunter reagent (Amersham) according to manufacturer's instructions. Briefly, 20μg of SEC-isolated HEK-apoE3 or apoE4 was radiolabelled with 500μCi of N2-dried N-succinimidyl 3-(4-hydroxy-5-[125I]iodophenyl) propionate and kept at 4°C for 90 minutes with occasional manual mixing. The reaction was completed by quenching with 0.4M glycine buffer and passed through a PD-10 Sephadex column to separate the labeled peptide from unincorporated isotope. Specific radioactivities were 5-10μCi/μg protein.
Ligand Degradation Assays
Assay buffer for 125I-apoE binding and degradation was DMEM containing 6mg/ml BSA and 5mM CaCl2. Cellular degradation assays were performed by incubating cells in assay buffer containing 125I-apoE (5nM) in the absence or presence of unlabelled RAP (1μM). This concentration of RAP is sufficient to inhibit ligand-binding to both LRP and the LDLR. After incubation at 37°C for 4 hours, the overlying media was removed and precipitated with TCA. Degradation of ligand was defined as the appearance of TCA-soluble radioactivity in the overlying media. Non-cellular degradation of 125I-apoE was determined in parallel dishes that did not contain cells and was subtracted from each point. All experiments were repeated at least 3 times. Data are presented as mean +/− SEM.
Single Cycle Endocytosis
Single cycle endocytosis assay was performed essentially as described previously (40). Briefly, initial binding was performed with 125I-apoE3 (5nM) at 4°C for 1.5 hours. Cells were then washed three times with DMEM containing 6mg/ml BSA and 5mM CaCl2, followed by incubation at 37°C in the presence of 1μM RAP. At selected intervals, buffer overlying each monolayer was removed and subjected to TCA precipitation. Dishes were then quickly chilled on ice and cell monolayers were treated with 0.25% (w/v) Pronase in PBS for 30 minutes at 4°C to remove residual surface-bound ligand. This treatment also detached cells from culture wells. The detached cells were then separated from buffer by centrifugation. Radioactivity associated with cell pellets represents internalized protease-resistant ligand, whereas radioactivity in the supernatant fraction represents surface protease-sensitive ligand. Degraded ligand was defined as TCA-soluble radioactivity in the overlying buffer. Each determination was performed in triplicate.
RESULTS
HEK-ApoE Can Be Isolated As a Pure ∼34 kDa Protein
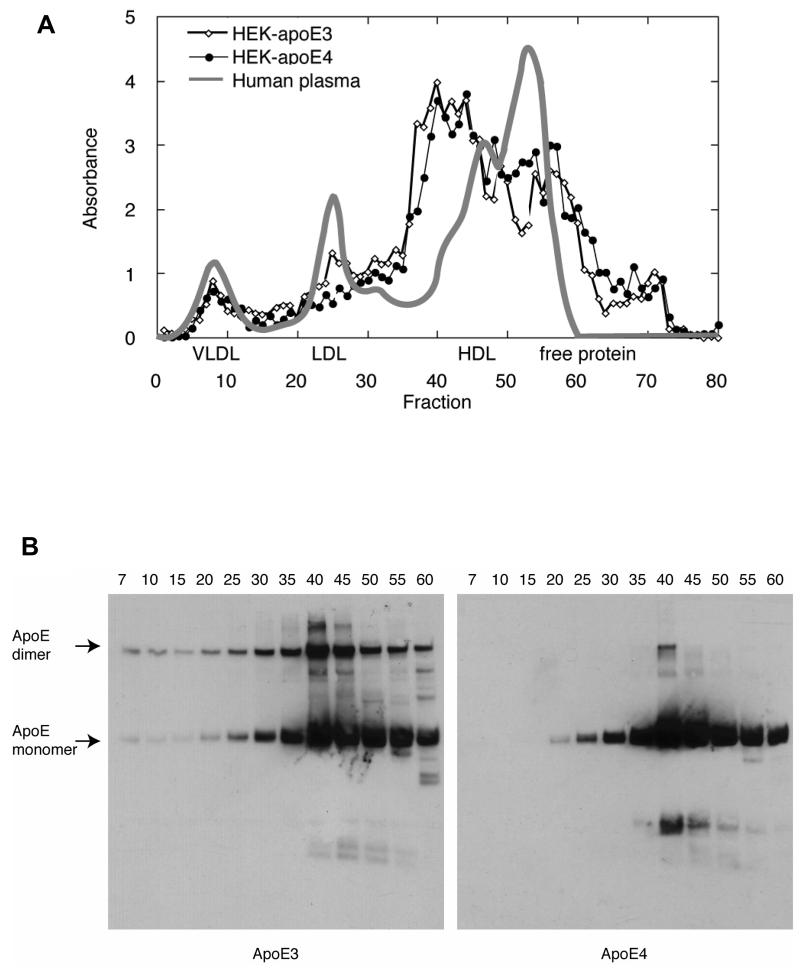
Isolation of HEK-apoE3 and E4 assemblies was performed using tandem Superose 6 columns equilibrated in PBS. A representative trace (Figure 1A) demonstrates that HEK-apoE eluted as a single peak with Gaussian distribution. Analysis of the peak fractions by SDS-PAGE demonstrated that the majority of protein migrated as a single species with a molecular weight of ∼34 kDa (Figure 1B). The observed value is slightly greater than 34 kDa predicted by apoE sequence and may be due to post-translational modifications resulting from expression in eukaryotic HEK cells. Under non-reducing conditions dimeric HEK-apoE3 was detected (Figure 1B, arrow). As predicted, no dimer was detected for HEK-apoE4 lacking the Cys112 residue necessary for intermolecular disulfide bond formation.
FIGURE 1.

Isolation of HEK-apoE. (A) Representative Superose-6 elution profile of concentrated media from HEK cells stably transfected with the cDNA encoding human apoE isolated in PBS. Cell culture media containing secreted apoE was concentrated 50-fold and injected on Superose 6 columns at a flow rate of 0.4 ml/min. Fractions across the primary peak, detected by absorbance at A280, were pooled (HEK-apoE arrows) and protein concentration determined spectrophotometrically. (B) Coomassie stained SDS-PAGE analysis of HEK-apoE3 (E3) and HEK-apoE4 (E4). 5 μg of protein was loaded under non-reducing conditions for each isoform of HEK-apoE. Molecular weight values standards determined by protein standards (BenchMark, Invitrogen). ∼37 kDa HEK-apoE monomer and ∼80 kDa HEK-apoE3 dimer indicated with arrows.
HEK-ApoE Migrates As a 360–390 kDa High-Molecular Weight Complex As Determined by Calibrated SEC Analysis
To compare the hydrodynamic properties of HEK-apoE3 and HEK-apoE4, samples were subjected to calibrated SEC analysis. The size of the HEK-apoE assemblies that eluted as a single peak from the Superose 6 column (Figure 1A) was estimated by calibrating the Superose 6 column using a panel of eight known proteins (Figure 2). An exponential fit to the data yielded good predictive value (R2 = 0.9939). Based on the equation for the exponential fit, HEK-apoE3 assemblies had a calculated molecular weight of 360 kDa, and HEK-apoE4 had a calculated molecular weight of 390 kDa (Figure 2, open boxes). These results suggest a possible assembly of 10-12 apoE monomers.
FIGURE 2.

Calibrated SEC analysis of HEK-apoE. Calibration of a single Superose 6 gel filtration column. Eight proteins that ranged in size from 11-440 kDa were used to generate a standard curve for the gel filtration column. The molecular weight was plotted against the partition coefficient Kav (calculations described in methods). The solid line represents the best exponential fit for the data. Calculated values for HEK-apoE3 and HEK-apoE4 (open boxes) were generated using the equation representing the standard curve.
HEK-ApoE3 and ApoE4 Assemblies Are 8-15nm in Size and Lipid-Poor
Using SEC with tandem Superose-6 columns (37, 38), the size distribution of protein assemblies in conditioned media from HEK cells expressing apoE3 and E4 was compared to human plasma lipoproteins (Figure 3A). The major portion of the apoE in the media from HEK-apoE3 and apoE4 cells eluted in fractions 35-45, indicating an approximate size of 8-15 nm, comparable in size to plasma HDL-2 particles. This localization of apoE is confirmed by Western blot analysis of selected FPLC fractions via non-reducing SDS-PAGE (Figure 3B) with apoE3 appearing as monomer and dimer and apoE4 primarily as monomer. The small amount of apoE4 dimer likely resulted from non-covalent dimerization of apoE and the bands smaller than the apoE monomeric size likely represent products of apoE proteolytic processing (50). The secondary apoE peak at fractions 50-60 is likely to be monomeric apoE as free proteins, such as albumin, elute in these fractions. EM confirms the presence of apoE assemblies with an average diameter of 12 nm ± 1.87 (Figure 4). Interestingly, the size and shapes of cell-secreted apoE assemblies are also similar to those of lipoproteins isolated from CSF (37) and γ-migrating, lipid-poor apoE-containing particles in plasma (12-16nm) (41). The number of apoE monomers that make-up the HEK-apoE assemblies has not yet been determined as SEC and EM analyses suggest an approximate size and morphology but not a molecular weight.
FIGURE 3.
SEC profiles of HEK-apoE3 and E4 and human plasma. (A) 1 ml of concentrated conditioned media from HEK cells expressing human apoE3 or E4 (HEK-apoE) was fractionated by size exclusion chromatography (SEC) using tandem Superose-6 columns and detected by absorbance at A280, with the profile of human plasma included for comparison (37). ApoE immunoreactivity was determined by dot blot. Graphs are an average of n = 4 separate experiments. (B) Representative Western blots of SDS-PAGE of selected HEK-apoE3 and E4 SEC fractions probed with apoE antisera.
FIGURE 4.

Electron microscopy analysis of HEK-apoE. Pooled fractions 35-45 of HEK-apoE isolated by SEC were concentrated and an aliquot placed on a carbon-coated electron microscopy grid and negatively stained with 2% phosphotungstic acid. Using a Philips CM10 electron microscope, the diameters of 100 intact particles from an enlarged photomicrograph were measured by using a micrometer lens. Representative particles are shown below, with a mean diameter of 12nm ± 1.87 standard deviations. Results shown are for HEK-apoE3. Comparable results were observed using HEK-apoE4 (data not shown).
Several methods of analysis were used to determine if the HEK-apoE assemblies contain lipid. First, we analyzed their density via density gradient centrifugation (37). We found that HEK-apoE distributed in the fractions corresponding to free protein, indicating an absence of lipid (Figure 5). This absence of lipid was confirmed by enzymatic analysis of the apoE-containing density gradient and SEC fractions for Chol, PL, or TG, (sensitivity limit <1ng/ml). Based on the concentration of apoE in the HEK apoE-containing SEC fractions, we anticipated being able to determine the lipid profile for SEC fractions of HEK-apoE as we had for CSF and astrocyte-secreted particles (37, 38). However, no lipid was detected. Lipid was also not detected in concentrated, pooled SEC HEK-apoE fractions 35-45 nor in the d<1.25 g/ml fraction from total HEK-apoE media. Thus, some eukaryotic cells may secrete apoE3 and apoE4 assemblies with little or no lipid, unlike both CSF and astrocyte-secreted lipoproteins that contain both PL and Chol, as measured by the same enzymatic assays used for HEK-apoE (37, 38).
FIGURE 5.
Equilibrium ultracentrifugation profiles of HEK-apoE3 and E4. 1 ml of concentrated HEK-apoE3 or E4 conditioned media was fractionated via single-spin equilibrium ultracentrifugation and the positions of various human plasma lipoproteins are indicated for comparison. ApoE immunoreactivity was determined by dot blot. Graphs are average of n = 2 separate experiments.
HEK-ApoE3 and ApoE4 Assemblies Are Ligands for LRP But Not LDLR
Previous studies have shown that apoE-containing plasma lipoproteins are ligands for the LDLR, whereas apoE-enriched β-VLDL, remnant particles and recombinant apoE are ligands for LRP (2, 25, 42). To examine whether cell-secreted apoE3 and apoE4 assemblies are ligands for LDLR and/or LRP, we performed receptor-mediated ligand uptake and degradation assay using a series of genetically generated mouse embryonic fibroblast cell lines (MEF). These cell lines include 1) wild type (MEF-1, expressing both LRP and LDLR), 2) LRP-deficient (MEF-2), 3) LDLR-deficient (MEF-3), or 4) LRP and LDLR double deficient (MEF-4) cells. In addition, we utilized RAP in these assays to ensure the specificity for LRP and LDLR. RAP functions normally within the early secretory pathway as a molecular chaperone to assist the folding and prevent premature ligand binding to the receptors during trafficking (43). The recombinant form of RAP has been used extensively as an antagonist to inhibit all known ligand interactions with members of the LDLR family (43). We chose to perform ligand uptake and degradation assays instead of saturation binding analysis because previous studies have shown that heparan sulfate proteoglycan (HSPG) is involved in the initial binding of apoE/lipoproteins, whereas apoE receptors (e.g. LRP) mediate subsequent uptake (44, 45). Thus, MEF-1, MEF-2, MEF-3, and MEF-4 cells were incubated with 125I-apoE3 or 125I-apoE4 (5nM) in the absence or presence of 1μM RAP at 37°C for 4 hours. Cellular degradation of 125I-apoE was assessed by TCA precipitation of overlying media after incubation with cells. We found that MEF-1 cells exhibited significant RAP-inhibitable degradation of 125I-apoE3 (Figure 6A). The portion of 125I-apoE3 degradation that was not inhibited by RAP likely reflects extracellular or cell surface degradation by secreted proteases, or intracellular degradation following HSPG-mediated cellular uptake (44, 45). Interestingly, when compared to MEF-1 cells, RAP-inhibitable degradation of 125I-apoE3 was comparable in LDLR-deficient MEF-3 cells, but significantly less in LRP-deficient MEF-2 cells. The RAP-inhibitable degradation of 125I-apoE3 was minimal in MEF-4 cells, lacking both LDLR and LRP. Similar results were seen when 125I-apoE4 was used as the ligand (Figure 6B). Together these results suggest that lipid-poor, cell-secreted apoE assemblies prefer LRP to the LDLR as the endocytic receptor.
FIGURE 6.

Degradation of cell-secreted HEK-125I-apoE3 and E4 in MEF cells. MEF-1 (wild type), MEF-2 (LRP-deficient), MEF-3 (LDLR-deficient), and MEF-4 (LRP- and LDLR-double deficient) cells were incubated with 125I-HEK apoE3 (5nM) (A) or 125I-HEK apoE4 (5nM) (B) in the absence or presence of 1μM RAP for 4 hours at 37°C. The degradation of 125I-apoE under each condition was analyzed via TCA-precipitation of the overlying media. Symbols represent the average of triplicate determinations +/− SEM shown from one of three independent experiments. Non-specific degradation of 125I-apoE in parallel dishes that did not contain cells was subtracted from each assay. *, p<0.05 when compared to the column without RAP competitor.
As a confirmation that HEK-apoE is not a high affinity ligand for the LDLR, the ability of HEK-apoE3 to compete for binding of 125I-LDL to fibroblasts was compared to LDL and DMPC-apoE3 and DMPC-apoE4 (Figure 7). Competition curves comparing LDLR binding by LDL (Figure 7A) and dimyristoyl phosphatidylcholine (DMPC) vesicles (Figure 7B) indicate that both LDL and DMPC vesicles exhibit a clear dose response, with 50% binding inhibition at 3.44 μg/ml for LDL, and 0.039 and 0.036 μg/ml for E3-DMPC and E4-DMPC, respectively. In contrast, HEK-apoE is a poor competitor for bound 125I-LDL and does not reach 50% binding inhibition. These results confirm that HEK-apoE is not a ligand for LDLR, as suggested by the results in Fig. 6.
FIGURE 7.
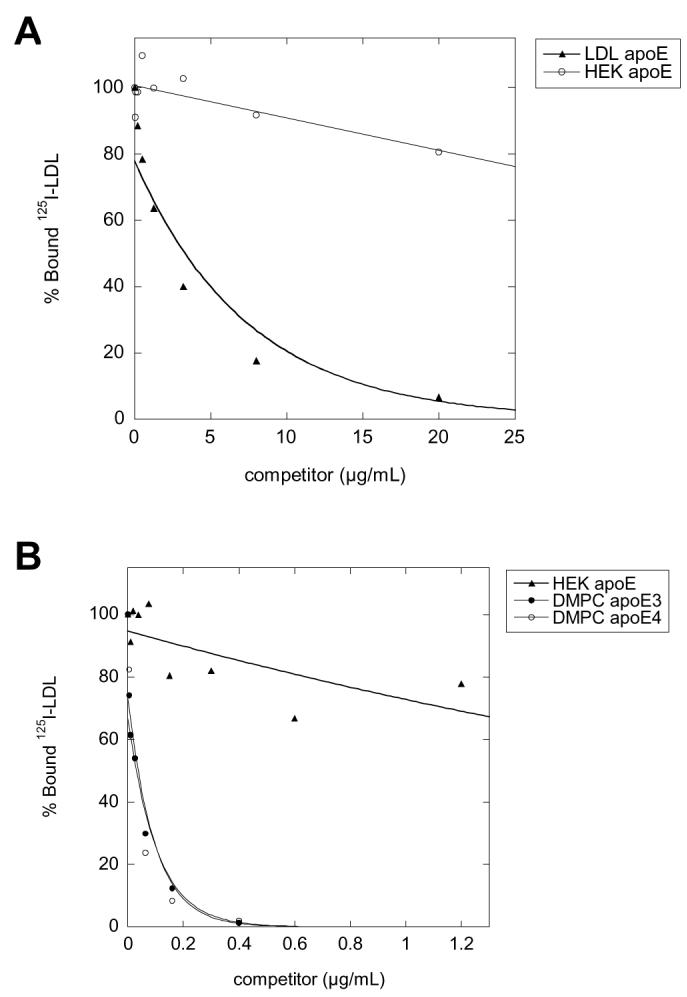
Ability of various apoE assemblies to compete with 125I-LDL for binding to LDLR on normal human fibroblasts. Normal human fibroblasts were incubated at 4°C for 2 h in a medium containing 2 μg/ml 125I-LDL and various concentrations of unlabeled LDL (A) or DMPC-apoE3 and DMPC-apoE4 (B). The ability of HEK-apoE3 to compete with 125I-LDL binding was compared to those of LDL (A) and the DMPC-apoE (B) at identical concentrations. Data were average from triplicate determinations.
To confirm that cell-secreted apoE3 is internalized by LRP via a receptor-mediated endocytic pathway, we performed single cycle endocytosis analyses. 125I-apoE3 (5nM) was incubated with MEF-1 cells for 1.5 hours at 4°C to allow cell surface binding, followed by incubation at 37°C for selected intervals to allow ligand uptake and degradation in the presence of 1μM RAP. After each interval, media was removed and subjected to TCA precipitation, whereas cell monolayers were quickly cooled to prevent further ligand internalization. Cells were then treated with Pronase at 4°C to remove residual surface ligand. The partitioning of ligand after each interval was assessed as described in the Experimental Procedures. As shown in Figure 8A, more than 70% of surface bound ligand disappeared within the first 10 minutes. Concomitantly, ligand was internalized rapidly and reached a peak level at 15-20 minutes before subsequently declining. Ligand appeared in the media simultaneously with the disappearance of cell-surface ligand. TCA-soluble radioactivity (representing degraded ligand) was detected only after a delay of 15-20 min, consistent with an endocytic trafficking and lysosomal degradation. Thus, the kinetic distribution pattern of 125I-apoE3 during a single cycle of endocytosis is typical of that of other ligands degraded in a receptor-mediated fashion (46). When 125I-apoE4 was used in the single-cycle endocytosis assay, similar kinetics were observed (Figure 8B). It is interesting to note that when compared to other ligands of LRP (e.g. tissue-type plasminogen activator, or tPA (40), a large portion of intracellular apoE remained undegraded even after 2.5 hours of incubation, suggesting that there may be an intracellular retention mechanism that is specific for apoE.
FIGURE 8.

Distribution of cell-secreted HEK-125I-apoE3 and E4 during a single cycle endocytosis in MEF-1 cells. Binding of 125I-HEK apoE3 (5nM) (A) or 125I-HEK apoE4 (5nM) (B) to MEF-1 cells was performed at 4°C for 1.5 hours. After washing, cells were incubated at 37°C for selected intervals in the presence of 500nM RAP. Overlying buffer was then removed and subjected to TCA-precipitation, whereas cell monolayers were chilled and treated with Pronase. Cell-surface ligand (Pronase-sensitive, ⨯) and internalized ligand (Pronase-resistant, ▲) were quantified for cells following Pronase treatment. Dissociated ligand (TCA-precipitable, ■), as well as degraded ligand (TCA-soluble, ◆), associated with media was also determined. Symbols represent means of triplicate determinations.
DISCUSSION
This paper describes the structure and apoE receptor binding affinity of apoE assemblies secreted by eukaryotic cells stably expressing human apoE3 or E4 (HEK-apoE). Cell-secreted HEK-apoE isoforms were isolated to homogeneity by SEC and subsequent analysis using a column calibrated for molecular weight confirmed a retention time consistent with 360–390 kDa assembly. These measured values are larger than previous reports of ∼225 kDa using SEC analysis for purified recombinant and plasma apoE in aqueous solutions (47-49). However, subsequent sedimentation equilibrium analysis by analytical ultracentrifugation (AUC) of recombinant apoE demonstrated an apparent molecular weight of ∼ 134 kDa suggesting a tetrameric assembly, the presumed form of purified apoE (50). Although concentrating the conditioned medium may force some apoE complexes into higher order structures than would usually be obtained at in vivo concentrations, this possibility is unlikely as the concentration of apoE even after concentrating is still well below those that can result in significant aggregation under our buffer conditions (50). We have also carefully compared the elution profiles of unconcentrated and concentrated media and observed identical results. In general the higher molecular weight values predicted by SEC analysis for apoE may be the result of non-globular behavior during chromatographic separation. In the absence of lipid, several apoE assemblies including tetramers, octamers, and higher order apoE oligomers in dynamic equilibrium have been detected by both AUC (51) and fluorescence anisotropy (52). However, HEK-apoE migrates as a single gaussian peak and does not exhibit the broad chromatographic distribution observed with recombinant apoE, suggesting that HEK-apoE is a stable, homogenous multimeric assembly. As the conformation of apoE varies depending on its assembly state (50, 53), it is likely that the functional properties of these different apoE structural assemblies will also vary.
Although HEK-apoE is in the approximate size range of HDL when compared to plasma lipoproteins, these assemblies are virtually lipid-free. In vivo, apoE is detected in different lipoprotein particles. First, the most common form of apoE is as a component of several classes of plasma lipoproteins, including chylomicron remnants, LDL and a sub-set of HDL. Second, γ-migrating, lipid-poor apoE-containing particles in plasma have been described by Huang and coworkers (41). This particle and HEK-apoE are lipid poor/free and appear to be the same size by SEC. Third, in the CNS, HDL-like spherical apoE-containing lipoproteins are present in the CSF and apoE-containing discoidal lipoproteins are secreted primarily by astrocytes in vitro (reviewed in 3).
HEK-apoE acts as a ligand for LRP but not LDLR. Members of the LDLR gene family bind apoE (26), although the conformation of apoE affects it's affinity for the individual receptors. In the extreme, purification of apoE and the removal of lipid abolishes its ability to bind LDLR (54), while reconstitution with lipid restores receptor-binding affinity (55). ApoE can adopt several conformations that influence its affinity for specific apoE receptors, including association with lipoproteins and assembly state. ApoE-containing LDL and DMPC vesicles bind LDLR, but not LRP (2, 22, 23). Only apoE-enriched particles (24-26), recombinant apoE (42) and now HEK-apoE act as ligands for LRP. The common feature of these apoE assemblies that are ligands for LRP is that they are all rich in apoE. The most interesting example of receptor-specificity is the β-VLDL. Native apoE-containing β-VLDL is a ligand for LDLR but not LRP. However, apoE enrichment, with either recombinant apoE or apoE extracted from native β-VLDL, converts these particles to a high affinity ligand for LRP (24, 56). Therefore, the inability of apoE on the native β-VLDL to bind LRP is not due to any permanent modifications of apoE but rather to a lack of an apoE assembly that can be generated in the presence of higher concentrations of apoE (56). Kowal et al. have suggested that the apoE on the β-VLDL can be “activated” in vivo to become an LRP ligand when the β-VLDL acquires additional apoE while trapped in the space of Disse. Supporting this in vivo relevance, studies have shown that injection of apoE into Watanabe heritable hyperlipidemic rabbits that lack functional LDLR resulted in a significant and rapid reduction of plasma cholesterol levels (44). These results suggest that apoE enrichment of lipoprotein particles occurs in vivo, allowing apoE to facilitate cholesterol clearance via an LRP-mediated pathway. Together with our current study, it appears that the conformation of apoE induced by its enrichment on lipoprotein particles or its own self-assembly is required for recognition by LRP.
The dependence of apoE receptor recognition on the conformation of apoE is further demonstrated by two recent publications. Using reduced apoE, Ruiz and co-workers demonstrated that lipid-free apoE binds VLDLR but not LRP or LDLR (57). ApoE receptor specificity is further established by Fryer and coworkers who demonstrated that astrocyte apoE isolated in vitro binds only LDLR and not other the LDLR family members, including LRP, ER2, and VLDLR (58). Clearly, this receptor-dependent specificity for various forms of apoE requires further investigation to determine the pattern and function of this selectivity.
In addition to functioning as ligands for LRP but not LDLR, we have previously demonstrated that HEK-apoE3 and apoE4 assemblies differentially form an SDS-stable complex with Aβ, a property it shares with apoE associated with plasma lipoproteins (16, 17). Indeed, both small, lipid-poor HEK-apoE3 assemblies and large, lipid-rich plasma VLDL-containing apoE3 bind Aβ with a higher affinity than apoE4 (16, 17), while apoE3 and E4 purified from HEK-apoE and plasma VLDL exhibit a comparable, lower affinity for Aβ (17, 18, 35). That HEK-apoE assemblies exhibits functions comparable to lipoprotein-associated apoE is further suggested by the several in vitro models for the role of apoE in the brain. We have demonstrated that HEK-apoE inhibits both Aβ-induced neurotoxicity and glial-mediated inflammation and this inhibition is blocked by RAP (59, 60). Further, HEK-apoE3 but not HEK-apoE4 supports neurite sprouting in primary rat cortical neurons (Teter and LaDu, unpublished observations), as has been described for glial-apoE3 and E4 particles expressed in hippocampal slice cultures from apoE transgenic animals (10). Murine glial cells secreting apoE3- but not apoE4-containing lipoproteins also promote neurite outgrowth (11). In addition, apoE-enriched β-VLDL also exhibited this apoE isoform-specific effect on neurite outgrowth in a neuronal cell line and the effect was demonstrated to require LRP (9). Thus, HEK-apoE mimics the function of a variety of lipoprotein particles including LRP receptor binding and complex formation with Aβ, functions which may further contribute to the effect of apoE on Aβ-induced neurotoxicity and neuroinflammation, as well as the isoform-specific effect of apoE on neurite outgrowth.
One of the primary functions of apoE-containing lipoproteins in the plasma is the delivery and clearance of lipid, particularly cholesterol. However, Huang and co-workers identified a γ-migrating, 12nm lipid-poor apoE-containing “particle” in plasma that appears to serve as a cholesterol acceptor (41). A similar process may occur in the CNS. Following its secretion, apoE and/or apoE-containing CNS lipoproteins appear to exhibit a paracrine-like function distributing lipids via binding to cell surface apoE receptors on neurons and glia. It is possible that when apoE is acutely produced following injury in the peripheral or CNS (6, 61-63), it is secreted in lipid-poor or lipid-free forms. These apoE assemblies can then take-up extracellular lipid for delivery via apoE receptors to neurons to support regeneration. In addition, apoE-containing CNS particles may also accept cholesterol from neural cells via ABCA1 transporters (64, 65). Although the in vivo roles of the LDLR and LRP in CNS apoE/lipoprotein metabolism are not clear, over-expression of LRP in CNS neurons via a transgenic approach (66) results in a significant decrease in brain apoE levels (Zerbinatti and Bu, manuscript in preparation).
Taken together, these results suggest that in addition to lipid distribution in the CNS, apoE and LRP may play a role in the clearance of apoE/Aβ complexes (12, 14, 15, 18), similar to the clearance of α2-macroglobulin/Aβ complexes by LRP (14, 67). Although apoE3 and apoE4 exhibit a similar affinity for cell surface receptors, because apoE3 has a greater affinity for Aβ, apoE can potentially mediate the catabolism of Aβ in an isoform-specific manner. The fact that HEK-apoE is a ligand for LRP and exhibits isoform-specific binding to Aβ suggests that these apoE assemblies are a viable reagent for the further investigation of the role of apoE isoform and assembly state on apoE receptor-mediated clearance of Aβ, studies that may give insights into the pathogenesis of AD. In addition, these results contribute to the growing body of evidence that conformation of apoE, as induced by assembly state, lipidation or experimental conditions, significantly affects the receptor specificity of apoE (57, 58). Thus, in designing future experiments on the biological role of apoE, the form of apoE is critically important to any results that are influenced by apoE receptor specificity.
ACKNOWLEDGMENTS
The authors thank Reemy Balestra and Yvonne Newhouse at the Gladstone Institute for Cardiovascular Disease for performing the LDLR binding assay (Figure 6), John Lukens and Daniel Schneider for technical assistance at the University of Chicago, Yadong Huang and Karl Weisgraber for spirited discussions and input, and Joachim Herz for kindly providing MEF cell lines.
ABBREVIATIONS
1The abbreviations used are:
- apoE
apolipoprotein E
- Aβ
amyloid-β peptide
- AD
Alzheimer's disease
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- VLDL
very low-density lipoprotein
- DMPC
dimyristoyl phosphatidylcholine
- LDLR
LDL receptor
- LRP
LDL receptor-related protein
- HSPG
heparan sulfate proteoglycan
- RAP
receptor-associated protein
- MEF
mouse embryonic fibroblasts
- SEM
standard error of the mean
- CNS
central nervous system
- CSF
cerebral spinal fluid
- SEC
Size exclusion chromatography
- FPLC
fast protein liquid chromatography
- EM
electron microscopy
Footnotes
This work was supported by NIH grants NS37525 (GB), AG19121 (MJLD), NS520138 (GSG and CAR); American Health Assistance Foundation ADR97006 (GSG and CAR); and the Charles Walgreen Jr. Fund (MJLD). GB is an Established Investigator of the American Heart Association.
REFERENCES
- 1.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 3.LaDu MJ, Reardon CA, Van Eldik LJ, Fagan AM, Bu G, Holtzman D, Getz GS. Lipoproteins in the central nervous system. Annals N.Y. Acad. Sci. 2000;903:167–175. doi: 10.1111/j.1749-6632.2000.tb06365.x. [DOI] [PubMed] [Google Scholar]
- 4.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high avidity binding to ß-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corder EH, Saunder SM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 6.Poirier J. Apolipoprotein E in animal models of CNS injury and in Alzheimer's disease. Trends Neurosci. 1994;17:525–530. doi: 10.1016/0166-2236(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 7.Horsburgh K, McCarron MO, White F, Nicoll JA. The role of apolipoprotein E in Alzheimer's disease, acute brain injury and cerebrovascular disease: evidence of common mechanisms and utility of animal models. Neurobiol. Aging. 2000;21:245–255. doi: 10.1016/s0197-4580(00)00097-x. [DOI] [PubMed] [Google Scholar]
- 8.Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 9.Holtzman DM, Pitas RE, Kilbridge J, Nathan B, Mahley RW, Bu G, Schwartz AL. Low density lipoprotein receptor-related protein mediates apolipoprotein E-dependent neurite outgrowth in a central nervous system-derived neuronal cell line. Proc. Natl. Acad. Sci. USA. 1995;92:9480–9484. doi: 10.1073/pnas.92.21.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teter B, Xu PT, Gilbert JR, Roses AD, Galasko D, Cole GM. Defective neuronal sprouting by human apolipoprotein E4 is a gain-of-negative function. J. Neurosci. Res. 2002;68:331–336. doi: 10.1002/jnr.10221. [DOI] [PubMed] [Google Scholar]
- 11.Peng D, Song C, Reardon CA, Liao S, Getz GS. Lipoproteins produced by ApoE−/− astrocytes infected with adenovirus expressing human ApoE. J. Neurochem. 2003;86:1391–1402. doi: 10.1046/j.1471-4159.2003.01950.x. [DOI] [PubMed] [Google Scholar]
- 12.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer's disease: allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 13.Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, Higgs R, Liu F, Malkani S, Bales KR, Paul SM. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat. Med. 2004;10:719–726. doi: 10.1038/nm1058. [DOI] [PubMed] [Google Scholar]
- 14.Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, Zlokovic BV. Clearance of Alzheimer's amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J. Clin. Invest. 2000;106:1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beffert U, Aumont N, Dea D, Lussier-Cacan S, Davignon J, Poirier J. Beta-amyloid peptides increase the binding and internalization of apolipoprotein E to hippocampal neurons. J. Neurochem. 1998;70:1458–1466. doi: 10.1046/j.1471-4159.1998.70041458.x. [DOI] [PubMed] [Google Scholar]
- 16.LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to ß-amyloid. J. Biol. Chem. 1994;269:23404–23406. [PubMed] [Google Scholar]
- 17.LaDu MJ, Pederson TM, Frail DE, Reardon CA, Getz GS, Falduto MT. Purification of apolipoprotein E attenuates isoform-specific binding to ß-amyloid. J. Biol. Chem. 1995;270:9030–9042. doi: 10.1074/jbc.270.16.9039. [DOI] [PubMed] [Google Scholar]
- 18.Tokuda T, Calero M, Matsubara E, Vidal R, Kumar A, Permanne B, Zlokovic B, Smith J, LaDu MJ, Rostagno A, Frangione B, Ghiso J. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer's amyloid β-peptide. Biochem. J. 2000;348:359–365. [PMC free article] [PubMed] [Google Scholar]
- 19.Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, Piccardo P, Ghetti B, Paul SM. Lack of apolipoprotein E dramatically reduces amyloid β-peptide deposition. Nat. Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 20.Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Macky B, Olney J, McKeel D, Wozniak D, Paul SM. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolev I, Michaelson DM. A nontransgenic mouse model shows inducible amyloid-beta (Abeta) peptide deposition and elucidates the role of apolipoprotein E in the amyloid cascade. Proc. Natl. Acad. Sci. USA. 2004;101:13909–13914. doi: 10.1073/pnas.0404458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradley WA, Gianturco SH. ApoE is necessary and sufficient for the binding of large triglyceride-rich lipoproteins to the LDL receptor; apoB is unnecessary. J. Lipid Res. 1986;27:40–48. [PubMed] [Google Scholar]
- 23.Dong LM, Innerarity TL, Arnold KS, Newhouse YM, Weisgraber KH. The carboxyl terminus in apolipoprotein E2 and the seven amino acid repeat in apolipoprotein E-Leiden: role in receptor-binding activity. J. Lipid Res. 1998;39:1173–1180. [PubMed] [Google Scholar]
- 24.Kowal RC, Herz J, Goldstein JL, Esser V, Brown MS. Low density lipoprotein receptor-related protein mediates uptake of cholesteryl esters derived from apoprotein E-enriched lipoproteins. Proc. Natl. Acad. Sci USA. 1989;86:5810–5814. doi: 10.1073/pnas.86.15.5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beisiegel U, Weber W, Ihrke G, Herz J, Stanley KK. The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature. 1989;341:162–164. doi: 10.1038/341162a0. [DOI] [PubMed] [Google Scholar]
- 26.Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP) Annu. Rev. Biochem. 1994;63:601–637. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- 27.Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strickland DK, Kounnas MZ, Argraves WS. LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 1995;9:890–898. doi: 10.1096/fasebj.9.10.7615159. [DOI] [PubMed] [Google Scholar]
- 29.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herz J. The LDL receptor gene family: (un)expected signal transducers in the brain. Neuron. 2001;29:571–581. doi: 10.1016/s0896-6273(01)00234-3. [DOI] [PubMed] [Google Scholar]
- 31.Willnow TE, Goldstein JL, Orth K, Brown MS, Herz J. Low density lipoprotein receptor-related protein and gp330 bind similar ligands, including plasminogen activator-inhibitor complexes and lactoferrin, an inhibitor of chylomicron remnant clearance. J. Biol. Chem. 1992;267:26172–26180. [PubMed] [Google Scholar]
- 32.Takahashi S, Kawarabayasi Y, Nakai T, Sakai J, Yamamoto T. Rabbit very low density lipoprotein receptor: a low density liporportein receptor-like protein with distinct ligand specficity. Proc. Natl. Acad. Sci. USA. 1992;89:9252–9256. doi: 10.1073/pnas.89.19.9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DH, Iijima H, Goto K, Sakai J, Ishii H, Kim HJ, Suzuki H, Kondo H, Saeki S, Yamamoto T. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J. Biol. Chem. 1996;271:8373–8380. doi: 10.1074/jbc.271.14.8373. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J. Biol. Chem. 1996;271:24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
- 35.Strittmatter WJ, Weisgraber KH, Huang DY, Dong L-Y, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid β peptide: isoform specific-effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, Akama KT, Krafft GA, Chromy BA, Van Eldik LJ. Amyloid-beta peptide activates cultured astrocytes: morphological alterations, cytokine induction and nitric oxide release. Brain Res. 1998;785:195–206. doi: 10.1016/s0006-8993(97)01318-8. [DOI] [PubMed] [Google Scholar]
- 37.LaDu MJ, Gilligan SM, Lukens SR, Cabana VG, Reardon CA, Van Eldik LJ, Holtzman DM. Nascent astrocyte particles differ from lipoproteins in CSF. J. Neurochem. 1998;70:2070–2081. doi: 10.1046/j.1471-4159.1998.70052070.x. [DOI] [PubMed] [Google Scholar]
- 38.Fagan AM, Holtzman DM, Munson G, Mathur T, Schneider D, Chang LK, Getz GS, Reardon CA, Lukens J, Shah JA, LaDu MJ. Unique lipoproteins secreted by primary astrocytes from wild type, apoE(−/−), and human apoE transgenic mice. J. Biol. Chem. 1999;274:30001–30007. doi: 10.1074/jbc.274.42.30001. [DOI] [PubMed] [Google Scholar]
- 39.Cabana VG, Siegel JN, Sabesin SM. Effects of the acute phase response on the concentration and density distribution of plasma lipids and apolipoproteins. J. Lipid Res. 1989;30:39–49. [PubMed] [Google Scholar]
- 40.Bu G, Morton PA, Schwartz AL. Identification and partial characterization by chemical cross-linking of a binding protein for tissue-type plasminogen activator (t-PA) on rat hepatoma cells. J. Biol. Chem. 1992;267:15595–15602. [PubMed] [Google Scholar]
- 41.Huang Y, von Eckardstein A, Wu S, Maeda N, Assmann G. A plasma lipoprotein containing only apolipoprotein E and with gamma mobility on electrophoresis releases cholesterol from cells. Proc. Natl. Acad. Sci. USA. 1994;91:1834–1838. doi: 10.1073/pnas.91.5.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narita M, Holtzman DM, Fagan AM, LaDu MJ, Yu L, Han X, Gross RW, Bu G, Schwartz AL. Cellular catabolism of lipid poor apolipoprotein E via cell surface LDL receptor-related protein. J. Biochem. 2002;132:743–749. doi: 10.1093/oxfordjournals.jbchem.a003282. [DOI] [PubMed] [Google Scholar]
- 43.Bu G, Schwartz AL. RAP, a novel type of ER chaperone. Trends Cell Biol. 1998;8:272–276. doi: 10.1016/s0962-8924(98)01283-5. [DOI] [PubMed] [Google Scholar]
- 44.Mahley RW, Weisgraber KH, Hussain MM, Greenman B, Fisher M, Vogel T, Gorecki M. Intravenous infusion of apolipoprotein E accelerates clearance of plasma lipoproteins in rabbits. J. Clin. Invest. 1989;83:2125–2130. doi: 10.1172/JCI114126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji ZS, Dichek HL, Miranda RD, Mahley RW. Heparan sulfate proteoglycans participate in hepatic lipaseand apolipoprotein E-mediated binding and uptake of plasma lipoproteins, including high density lipoproteins. J. Biol. Chem. 1997;272:31285–31292. doi: 10.1074/jbc.272.50.31285. [DOI] [PubMed] [Google Scholar]
- 46.Schwartz MW, Sipols A, Kahn SE, Lattemann DF, Taborsky GJ, Jr., Bergman RN, Woods SC, Porte D., Jr. Kinetics and specificity of insulin uptake from plasma into cerebrospinal fluid. Am. J. Physiol. 1990;259:E378–383. doi: 10.1152/ajpendo.1990.259.3.E378. [DOI] [PubMed] [Google Scholar]
- 47.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J. Biol. Chem. 1993;268:15745–15750. [PubMed] [Google Scholar]
- 48.Chan W, Fornwald J, Brawner M, Wetzel R. Native complex formation between apolipoprotein E isoforms and the Alzheimer's disease peptide Aβ. Biochemistry. 1996;35:7123–7130. doi: 10.1021/bi952852v. [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama S, Kawai Y, Tajima S, Yamamoto A. Behavior or human apolipoprotein E in aqueous solutions and at interfaces. J. Biol. Chem. 1985;260:16375–16382. [PubMed] [Google Scholar]
- 50.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv. Prot. Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 51.Perugini MA, Schuck P, Howlett GJ. Self-association of human apolipoprotein E3 and E4 in the presence and absence of phospholipid. J. Biol. Chem. 2000;275:36758–36765. doi: 10.1074/jbc.M005565200. [DOI] [PubMed] [Google Scholar]
- 52.Dergunov AD, Shuvaev VV, Yanushevskaja EV. Quaternary structure of apolipoprotein E in solution: fluorimetric, chromatographic and immunochemical studies. Biol. Chem. Hoppe. Seyler. 1992;373:323–331. doi: 10.1515/bchm3.1992.373.1.323. [DOI] [PubMed] [Google Scholar]
- 53.Lund-Katz S, Weisgraber KH, Mahley RW, Phillips MC. Conformation of apolipoprotein E in lipoproteins. J. Biol. Chem. 1993;268:23008–15. [PubMed] [Google Scholar]
- 54.Innerarity TL, Mahley RW. Enhanced binding by cultured human fibroblasts of apo-E-containing lipoproteins as compared with low density lipoproteins. Biochemistry. 1978;17:1440–1447. doi: 10.1021/bi00601a013. [DOI] [PubMed] [Google Scholar]
- 55.Innerarity T, Hui D, Bersot T, Mahley RW. Type III hyperlipoproteinemia: a focus on lipoprotein receptor-apolipoprotein E2 interactions. Adv. Exp. Med. & Biol. 1986;201:273–288. doi: 10.1007/978-1-4684-1262-8_24. [DOI] [PubMed] [Google Scholar]
- 56.Kowal RC, Herz J, Weisgraber KH, Mahley RW, Brown MS, Goldstein JL. Opposing effects of apoliporoteins E and C on lipoprotein binding to low density lipoprotein receptor-related protein. J. Biol. Chem. 1990;265:10771–10779. [PubMed] [Google Scholar]
- 57.Ruiz J, Kouiavskaia D, Migliorini M, Robinson S, Saenko EL, Gorlatova N, Li D, Lawrence D, Hyman BT, Weisgraber KH, Strickland DK. Characterization of the apoE isoform binding properties of the VLDL receptor reveal marked differences from LRP and the LDL receptor. J. Lipid Res. 2005;46:1721–1731. doi: 10.1194/jlr.M500114-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Fryer JD, Demattos RB, McCormick LM, O'Dell M,A, Spinner ML, Bales KR, Paul SM, Sullivan PM, Parsadanian M, Bu G, Holtzman DM. The low-density lipoprotein receptor regulates the level of CNS human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J. Biol. Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- 59.Jordan J, Galindo MF, Miller RJ, Reardon CA, Getz GS, LaDu MJ. Isoform-specific effect of apolipoprotein E on cell survival and beta-amyloid-induced toxicity in rat hippocampal pyramidal neuronal cultures. J. Neurosci. 1998;18:195–204. doi: 10.1523/JNEUROSCI.18-01-00195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaDu MJ, Shah JA, Reardon CA, Getz GS, Bu G, Hu J, Guo L, Van Eldik LJ. Apolipoprotein E and apolipoprotein E receptors modulate A beta-induced glial neuroinflammatory responses. Neurochem. Int. 2001;39:427–434. doi: 10.1016/s0197-0186(01)00050-x. [DOI] [PubMed] [Google Scholar]
- 61.Ignatius MJ, Gebicke-Harter PJ, Skene JHP, Schilling JW, Weisgraber KH, Mahley RW, Shooter EM. Expression of apolipoprotein E during nerve degeneration and regeneration. Proc. Natl. Acad. Sci. USA. 1986;83:1125–1129. doi: 10.1073/pnas.83.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holtzman DM, Fagan AM. Potential role of apoE in structural plasticity in the nervous system: Implications for diseases of the central nervous system. Trends Cardiovasc. Med. 1998;8:250–255. doi: 10.1016/s1050-1738(98)00017-6. [DOI] [PubMed] [Google Scholar]
- 63.Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ, Shooter EM. A role for apolipoprotein E, apolipoprotein A-I, and low density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J. Clin. Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J. Biol. Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 65.Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 Impairs Apolipoprotein E Metabolism in Brain. J. Biol. Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- 66.Zerbinatti CV, Wozniak DF, Cirrito J, Cam JA, Osaka H, Bales KR, Zhuo M, Paul SM, Holtzman DM, Bu G. Increased soluble amyloid-beta peptide and memory deficits in amyloid model mice overexpressing the low-density lipoprotein receptor-related protein. Proc. Natl. Acad. Sci. USA. 2004;101:1075–1080. doi: 10.1073/pnas.0305803101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narita M, Holtzman DM, Schwartz AL, Bu G. Alpha2-macroglobulin complexes with and mediates the endocytosis of beta-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J. Neurochem. 1997;69:1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x. [DOI] [PubMed] [Google Scholar]