Abstract
Cannabis is a major substance of abuse, and the gene encoding for the central cannabinoid receptor (CNR1) is a logical candidate gene for vulnerability toward developing symptoms of cannabis dependence. We studied four single-nucleotide polymorphisms (SNPs) in the CNR1 gene for association with having one or more symptoms of cannabis dependence in 541 adolescent subjects who had all tried cannabis five or more times. Cases (327) were defined as those who had tried marijuana and developed one or more symptoms, and controls (214) as those who had tried marijuana but developed no dependence symptoms. Cannabis dependence symptoms were assessed in these youth when they were 17 or older with the Composite International Diagnostic Interview- Substance Abuse Module. Univariate (single-marker) association tests demonstrated that SNP rs806380, located in intron 2 of the CNR1 gene, was significantly associated with developing one or more cannabis dependence symptoms, with the G allele having a protective effect (p < 0.02). This was consistent with the results of the global haplotype test (p < 0.01). One of the common haplotypes examined (present in 21% of the subjects) was significantly associated with a lower rate of having one or more cannabis dependence symptoms. Our findings provide evidence suggesting that a common CNR1 haplotype is associated with developing fewer cannabis dependence symptoms among adolescents who have experimented with cannabis.
Keywords: Cannabis, Adolescence, Genetics, CNR1
Introduction
Cannabis is the most commonly used illicit substance among adolescents and young adults. In 2004, 46% of 12th graders reported having tried cannabis at some point in their lifetime, 34% reported having used within the past month, and 5.6% reported having smoked cannabis daily (Johnston et al. 2005). Initiation into cannabis use typically begins in adolescence, as youths aged 12 to 17 constitute about two thirds of the new cannabis users (SAMHSA 2002). Approximately 14% of adolescent-onset cannabis users develop cannabis dependence, a rate roughly twice that reported for adult-onset user (Chen et al. 1997; Chen and Anthony 2003). Cannabis dependence is defined in the Diagnostic and Statistical Manual of Mental Disorder, 4th edition, text revision (DSM –IVTR) as having at least three out of seven symptoms within one year (APA, 2000). Symptoms of cannabis dependence include physiological symptoms, such as tolerance or withdrawal, cognitive symptoms such as being preoccupied with obtaining or using cannabis, and symptoms of psychosocial impairment such as continuing to use despite impairment in school or relationship functioning.
Twin studies have demonstrated genetic influences on the liability to develop cannabis dependence in both adults and adolescents (Kendler and Prescott 1998; Tsuang et al. 1998; Maes et al. 1999; Kendler et al. 2000; Miles et al. 2001; Lynskey et al. 2002; Rhee et al. 2003). A logical candidate gene that could influence the liability to develop cannabis dependence symptoms is CNR1, which codes for the cannabinoid receptor. The cannabinoid receptor is the principal site of action of delta-9-tetrahydrocannabinol (THC), the principal psychoactive ingredient in cannabis (as reviewed by Childers and Breivogel (1998) and Onaivi et al. (2002)). THC mimics the actions of endogenous cannabinoids, such as anandamide. The CNR1 receptor is found throughout the brain and is expressed at high levels (Childers and Breivogel 1998). Converging evidence from animal studies suggests that the endocannabinoid system is involved in the processing of rewarding stimuli (reviewed by Onaivi et al. (2002)) and substantial evidence now shows interactions with the opioid (Navarro et al. 2001) and dopaminergic systems (Chen et al. 1993).
CNR1 has been examined for association with a range of substance abuse phenotypes, however, to our knowledge none have focused primarily on cannabis dependence symptoms. Opioid dependence (Li et al. 2000), drug dependence (Covault et al. 2001), extreme response to cannabis use (cannabis-induced psychosis) (Hoehe et al. 2000), alcohol withdrawal delirium (Schmidt et al. 2002), intravenous drug use (Comings et al. 1997), and P300 evoked potential have been examined for their association with CNR1. Covault et al. (2001) reported no association between a CNR1 microsatellite polymorphism and drug or alcohol dependence. Li et al. (2000) reported that in a sample of 375 Han Chinese, there was no evidence of an association between alleles of the same microsatellite repeat in the CNR1 gene and heroin dependence. Hoehe et al. (2000) reported no difference in allele frequencies of two silent mutations in the coding region of CNR1 between individuals exhibiting an extreme response to cannabis and a control group. Schmidt et al. (2002) reported a positive association between alcohol withdrawal delirium and a silent mutation in the CNR1 receptor (1359 G/A). Comings et al. (1997) reported a positive association between a CNR1 microsatellite polymorphism and intravenous drug use as well as cocaine, amphetamine, and cannabis dependence. Johnson et al. (1997) also showed an association between the polymorphism and lower P300 evoked related potential, which in turn has been associated with alcohol and drug dependence. Recently, Zhang et al. (2004) reported on the structure of the CNR1 gene, including findings of three novel exons, and an association between a CNR1 haplotype and polysubstance abuse. Of the reviewed studies, only two (Comings et al. 1997; Hoehe et al. 2000) report an association between CNR1 polymorphisms and cannabis-related phenotypes. Both studies examined only one or two polymorphisms in adults. The purpose of this study was to examine the association between Single Nucleotide Polymorphisms (SNPs) within the CNR1 gene and developing cannabis dependence symptoms in adolescents using both single-marker and haplotype analyses. We are not aware of any previous studies that have examined this association in adolescents.
Methods
Subjects
Subjects were participants in ongoing studies of the genetics of adolescent and young adult substance abuse in Colorado, funded by the National Institute of Drug Abuse. Youth who participated in these studies were recruited from treatment settings for youth with substance use disorders, criminal justice settings, and community-based twin, adoption, and family studies of adolescent substance use disorders. The consents for the original studies asked whether the information gathered could be used for analyses of “genetic tests related to substance use disorders.” Only those subjects whose parents consented and themselves assented to this statement were included in these analyses.
The entire pool of potential subjects encompassed over 5000 youth and we selected for inclusion in this study those who met the following criteria: a) at least age 17 or older, b) had endorsed using marijuana at least 5 times, and c) at least one year between age of first marijuana use and age at testing. Additionally, we limited subject selection to one youth per family. Five hundred forty-one youth met the inclusion criteria and were selected for the present study.
Assessments
All youth were assessed with the Composite International Diagnostic Interview – Substance Abuse Module (CIDI-SAM). This instrument diagnoses DSM-IV Abuse and Dependence (APA 1994) for ten drug classes and assesses individual symptoms for each drug class. It primarily consists of a 30-to-60-minute interview that is designed for administration by trained, lay interviewers. It is a descendent of the NIMH Diagnostic Interview Schedule. The CIDI's reliability and validity (Cottler et al. 1989) made it the main assessment for DSM-IV Substance Field Trials. Its validity and reliability has been established in substance-dependent adolescents (Crowley et al. 2001).
Sample Description
Cases for this study were defined as those who had one or more dependence symptoms (i.e., endorsed at least one DSM-IV dependence symptom for cannabis), and controls as those who experimented with marijuana at least five times, but had no dependence symptoms. Table 1 describes the ethnic, age, and gender distribution of the cases and controls, as well as the percent meeting criteria for lifetime abuse or dependence on 7 classes of substances. We define “problem cannabis use” as meeting the criteria of “caseness” (i.e. one or more dependence symptoms).
Table I.
Sample characteristics of cases and controls
| % Cases
(N = 327) |
% Controls
(N = 214) |
|
|---|---|---|
| Race/Ethnicity | ||
|
| ||
| Caucasian | 67.9 | 72.9 |
|
| ||
| Hispanic | 19.3 | 18.7 |
|
| ||
| African American | 6.4 | 5.1 |
|
| ||
| Other | 6.4 | 3.3 |
|
| ||
| Gender | ||
|
| ||
| Male | 74.3 | 49.5 |
|
| ||
| Female | 25.7 | 50.5 |
|
| ||
| Age | 18.1+1.4 | 18.2+1.5 |
|
| ||
| Lifetime DSM-IV Substance Abuse or Dependence Diagnoses | ||
|
| ||
| Cannabis Abuse | 35.2 | 14.0 |
| Cannabis Dependence | 44.6 | 0 |
|
| ||
| Tobacco Dependence | 57.0 | 25.7 |
|
| ||
| Alcohol Abuse | 39.1 | 27.1 |
| Alcohol Dependence | 28.4 | 3.3 |
|
| ||
| Amphetamine Abuse | 4.0 | 1.0 |
| Amphetamine Dependence | 11.6 | 0.5 |
|
| ||
| Cocaine Abuse | 4.9 | 1.4 |
| Cocaine Dependence | 9.2 | 0.5 |
|
| ||
| Hallucinogen Abuse | 10.1 | 2.3 |
| Hallucingogen Dependence | 13.2 | 0.5 |
|
| ||
| Opioid Abuse | 3.7 | 1.0 |
| Opioid Dependence | 1.8 | 0 |
Selection of SNPs and genotyping
Using the Celera Discovery System, we completed preliminary bioinformatics work examining the human CNR1 gene. The gene is located on chromosome 6q14 spanning 26085 nucleotides. There have been six possible transcripts predicted by Zhang et al. (2004), with the largest consisting of four exons which produces an mRNA of 5795 nucleotides. Forty-three different SNPs have been identified and are fairly evenly distributed throughout the gene. Of these, five were selected to be genotyped using Applied Biosystems TaqMan Assays-on-Demand™ (described below). These five SNPs (rs2273512, rs6454674, rs806380, rs806377, rs104935) were selected based on information available at the time. Criteria for selection included validation status of the SNP based on the public dbSNP database and from the Celera Discovery System, minor allele frequencies (MAF) greater than 0.10 (if known), and location in the gene such that the SNPs would be approximately evenly distributed throughout the gene. There are few SNPs located within the coding regions of the gene and none that predicted amino acid changes. Genomic DNA was isolated from buccal cell swabs and preamplified using the method of Zheng et al. (2001). Data obtained using this DNA are high-quality; these methods have been shown to be reliable for genotyping (Anchordoquy et al. 2003). TaqMan® assays for allelic discrimination (Applied Biosystems) were used to determine SNP genotypes, per instructions of the manufacturer under standard conditions using ABI PRISM® 7000 and 7900 instruments. We genotyped 541 subjects for four of the SNPs (rs6454674, rs806380, rs806377, rs1049353). SNP rs2273512, located in the first intron, was genotyped in 100 subjects and we did not find it to be polymorphic. The recently reported estimated minor allele frequency for rs2273512 in dbSNP is 0.017, so it is a rare SNP. In addition, and according to the protocols of the ongoing studies, we genotyped at least one parent of 106 subjects in order to improve haplotype determination. These data are included in the analyses of the full sample.
Analytic Methods
Linkage Disequilibrium Calculations and Haplotype Estimates
Pairwise linkage disequilibrium (D′) was calculated using GOLD (Abecasis and Cookson 2000 and Haploview (Barrett et al. 2005). A logistic regression-based test of association predicting case/control status from the individual SNPs was conducted using WHAP (http://www.broad.mit.edu/personal/shaun/whap/). WHAP uses a weighted logistic regression based method based on estimated haplotypes to test for association with a phenotype. The haplotypes are estimated using SNPHAP (http://www-gene.cimr.cam.ac.uk/clayton/software/), which assigns weighted haplotypes to each individual (i.e., haplotypes are not known with certainty but must be estimated from the data). In order to confirm the accuracy of the haplotype assignments, we compared the haplotypes estimated by WHAP (SNPHAP) to those estimated by PHASE, v.2.0.2, which incorporates relative position and distance between SNPs in its algorithms (Stephens and Donnelly 2003; Stephens et al. 2001). Less than 1% of the haplotype assignments were different between PHASE and SNPHAP, and differing haplotype assignments occurred for those individuals who had multiple possible haplotype assignments. Therefore, all analyses were conducted using WHAP, because WHAP allows inclusion of the few parental genotypes we were able to obtain, which provided better estimates of subject haplotypes.
Test for association
A similar regression-based test of association for haplotypes of the four SNPs was conducted using WHAP to compare the cases and controls. The omnibus test can be used as an initial test of association with the overall gene locus. Each estimated haplotype is included in the model and regression weights (β coefficients) are calculated to provide the relative contribution of each haplotype. The effect of the most common haplotype is fixed to 0 and the effects all others are estimated relative to it. All haplotypes with frequencies less than 1% are excluded. In subsequent analyses, the haplotype specific option (“–hs”) in WHAP can be used to test the effect of each haplotype individually against all other haplotypes by constraining all other haplotypes to have equal β weights. All analyses were first conducted on the entire sample. A secondary analysis was then conducted separately for the ethnic groups Hispanic and Caucasian.
Inclusion of covariates
Covariates were included in WHAP to control for other variables that may influence marijuana dependence. This is accomplished by adding the covariate in the .dat file, as described in the instructions for WHAP (http://www.broad.mit.edu/personal/shaun/whap/), and including the “—cov” option, which provides a joint test of haplotype effects while adjusting for the covariate. Age, sex, and group (i.e. ascertained from community or clinical settings) were included in the analyses as covariates in the separate data sets of Caucasians or Hispanics.
Results
Analysis of Individual SNPs
There was evidence for an association by likelihood ratio test (LRT) with SNP rs806380 (LRT=4.48, p=0.03). The results for the other SNPs did not approach statistical significance (shown in Table II). The estimated proportions of cases for each rs806380 SNP genotype are presented in Figure 2 with error bars showing 95% confidence intervals. For example, approximately 60% of individuals with the AA or AG genotype met our criteria for caseness, while only 40% of individuals with the GG genotype were cases.
Table II.
WHAP analyses for Association with CNR1 SNPs and Cannabis Dependence Symptoms. Full Sample N=541 young adults,
| SNP | Variant | MAF | LRT | p |
|---|---|---|---|---|
| rs6454674 | T/G | 0.31 | 1.793 | 0.181 |
| (hCV11418433) | ||||
|
| ||||
| rs806380 | A/G | 0.31 | 4.482 | 0.034* |
| (hCV1652583) | ||||
|
| ||||
| rs806377 | C/T | 0.49 | 0.072 | 0.789 |
| (hCV1652585) | ||||
|
| ||||
| rs1049353 | C/T | 0.24 | 0.002 | 0.962 |
| (hCV1652590) | ||||
Note. MAF-Minor Allele Frequency, LRT-Likelihood Ratio Test, p-Statistical significance
Analysis of Haplotypes
Pairwise linkage disequilibrium (LD) estimates (r2) obtained from Haploview (Barrett et al. 2005) are shown in Figure 3. One haplotype block consisting of “strong LD” between SNPs rs806380 and rs806377 was determined using the Haploview default algorithm based on Gabriel et al. (2002), using the 95% confidence D' rule. The estimated r2 between these two markers was a modest 0.42. There was also noteworthy LD between the first (rs6454674) and second (rs806380) markers (r2=0.63) and between the first (rs6454674) and third (rs806377) markers (r2=0.40). The fourth marker (rs1049353) exhibited the lowest LD with the others.
Using the omnibus test in WHAP, which tests each estimated haplotype while controlling for all others, provided evidence for an association between problem cannabis use (cases) and the CNR1 gene locus (LRT=16.47, p=0.021, shown in Table III). The regression weights (β coefficients) indicate the relative contribution of each haplotype; the effect of the most common haplotype is fixed to 0 and the effects of all others are estimated relative to it. By default, all haplotypes with frequencies less than 1% are excluded. The “–hs” option in WHAP was used to test the effect of each haplotype individually against all other haplotypes (i.e. constraining all other haplotypes to have equal β weights). Three haplotypes (GGCC, TACC, and GACC) were significantly associated when tested individually against all others. GGCC (LRT=5.984, p=0.0144) was associated with a protective effect (β= -0.366), while haplotypes TACC (LRT=5.847, p=0.0156) and GACC (LRT=6.475, p=0.0109) were associated with increased risk for dependence symptoms (β=0.585 and β=0.951, respectively). The proportion of cases for the GGCC haplotype are presented in Figure 4.
Table III.
WHAP Haplotype Analyses Omnibus Test for Association
| Full Sample N=541 young adults, 106 with at least one parent | ||
|---|---|---|
| Haplotype | Frequency | β |
| TATC | 0.380 | 0.000 |
|
| ||
| GGCC | 0.210 | -0.259 |
|
| ||
| TATT | 0.115 | 0.027 |
|
| ||
| TACC | 0.087 | 0.571 |
|
| ||
| TACT | 0.068 | -0.105 |
|
| ||
| GGCT | 0.053 | -0.076 |
|
| ||
| TGCC | 0.051 | 0.127 |
|
| ||
| GACC | 0.037 | 0.941 |
|
| ||
| LRT | 16.474 | |
| p | 0.0211 | |
| Empirical p | 0.0339 | |
Note. Frequency column shows the frequency of each estimated haplotype in the sample. β column provides the β coefficient estimate derived from the logistic regression test for association. LRT – Likelihood Ratio Test, p – statistical significance, Empirical p-determined after permutation testing
Secondary Analyses by Ethnicity
Because there is some ethnic diversity within the sample, we conducted post-hoc analyses to examine two ethnic groups Caucasians and Hispanics separately. There were significant differences in allele frequency for three of the four SNPs between Caucasians and Hispanics (rs806380, χ2=10.25, p=0.006; rs806377, χ2=7.84, p=0.02; rs1049353, χ2=12.14, p=0.002), so this was an important consideration.
Secondary analysis of the individual SNPs by ethnic group are shown in Table IV. For Caucasians (N=386), there was a significant association between rs806380 and problem cannabis use (p < 0.04), as we found in the full sample. Although there was no evidence for significant association with this SNP and problem cannabis use in the Hispanic sample, this may be the result of reduced power, as the size of the Hispanic sample is low (N = 104). The pattern of estimated proportions of cases by SNP rs806380 genotype within each ethnic group is similar to that of the full sample. In the Caucasians, approximately 60% of subjects with genotypes AA and AG are estimated to be cases, while only 40% with the GG genotype are. In Hispanics, the effect is more modest; approximately 60% of AA and AG individuals are cases and approximately 50% of GG subjects are cases.
Table IV.
Secondary WHAP analyses for Association with CNR1 SNPs and Problem Marijuana Use in Caucasian and Hispanic Sub-Groups
| Caucasian sample N=386 young adults | Hispanic Sample N=104 young adults | |||||
|---|---|---|---|---|---|---|
| SNP Variant | MAF | LRT | p | MAF | LRT | p |
| rs6454674 T/G | 0.32 | 2.406 | 0.121 | 0.26 | 0.871 | 0.351 (hCV11418433) |
| rs806380 A/G | 0.34 | 4.262 | 0.039* | 0.24 | 0.257 | 0.612 (hCV1652583) |
| rs806377 C/T | 0.46 | 0.033 | 0.857 | 0.42 | 0.018 | 0.894 (hCV1652585) |
| rs1049353 C/T | 0.29 | 0.052 | 0.820 | 0.17 | 3.971 | 0.046 (hCV1652590) |
Note. MAF-Minor Allele Frequency, LRT-Likelihood Ratio Test, p-Statistical significance
When the sample is divided into ethnic groups for the haplotype analyses, the omnibus test is no longer significant in either group (Table V). However, within both groups, the overall directions of the β coefficients are largely comparable in direction and strength, particularly for the three significant haplotypes observed in the full sample when using the “—largest” option. In Caucasians, the GGCC haplotype was statistically significant (β=-0.402, LRT=5.230, p=0.0222), and results for the TACC and GACC haplotypes were suggestive (β=0.563, LRT=3.078, p=0.0794 and β=1.121, LRT=3.943, p=0.0471). In Hispanics, the β coefficient is -0.374 (LRT=1.173, p=0.279) for the GGCC haplotype, -0.004 (LRT=0.000, p=0.995) for the TACC haplotype, and 0.289 (LRT=0.179, p=0.672) for the GACC haplotype. To further illustrate this point, the proportion of cases for the GGCC haplotype are presented in Figure 4 for the combined sample.
Table V.
WHAP haplotype analyses Omnibus Test for Association in Caucasian and Hispanic Sub-Groups
| Caucasian sample
N=386 young adults |
Hispanic Sample
N=104 young adults |
|||
|---|---|---|---|---|
| Haplotype | Frequency | β | Frequency | β |
| TATC | 0.331 | 0.000 | 0.488 | 0.000 |
|
| ||||
| GGCC | 0.236 | -0.384 | 0.176 | -0.116 |
|
| ||||
| TATT | 0.130 | -0.139 | 0.096 | 1.137 |
|
| ||||
| TACC | 0.078 | 0.449 | 0.077 | 0.152 |
|
| ||||
| TACT | 0.095 | -0.162 | 0.041 | 0.610 |
|
| ||||
| GGCT | 0.064 | -0.127 | 0.020 | -0.048 |
|
| ||||
| TGCC | 0.046 | 0.106 | 0.033 | 0.874 |
|
| ||||
| GACC | 0.020 | 1.001 | 0.057 | 0.409 |
|
| ||||
| GATC | n.o. | n.o. | 0.012 | -0.259 |
|
| ||||
| LRT | 11.239 | 5.268 | ||
| p | 0.129 | 0.729 | ||
| Empirical p | 0.179 | 0.778 | ||
Note. Frequency column shows the frequency of each estimated haplotype in the sample. β column provides the β coefficient estimate derived from the logistic regression test for association. LRT – Likelihood Ratio Test, p – statistical significance, Empirical p-determined after permutation testing. N.o. – not observed.
Covariate Analyses
In the Caucasian sample, we included the covariates age, sex, and group status (clinical or community) in the omnibus test and in a test of the individual SNP rs806380. When all three covariates were included the likelihood ratio test actually improved, resulting in a suggestive p-value (LRT=13.13, p=0.069). Similarly, the association with SNP rs806380 became significant once the covariates were included in the model (LRT=7.045, p=0.0079). However, in the Hispanic sample, neither the omnibus test nor the individual SNP were significant (LRT=2.937, p=0.94 omnibus; LRT=0.602, p=0.44 SNP rs806380).
Discussion
Taken together, these data support the hypothesis that variation within the CNR1 gene may be associated with developing one or more symptoms of cannabis dependence in adolescents who have experimented with cannabis. The results support findings by Zhang et al. (2004), who reported an association between a haplotype between three SNPs (hCV1652584, hCV8943758, and hCV11600616) in intron 2 of the CNR1 gene and polysubstance abuse in adults. Of the SNPs studied here, rs806380, which was significantly associated with cannabis problem use in the current sample, is most proximal to the three SNPs examined by Zhang et al. (2004). However, given the relatively low linkage disequilibrium between the SNPs in this study, it is possible that more genetic diversity within the gene could be captured by examining additional SNPs in this sample.
The main limitation of this study was the difficulty in controlling for population stratification. Although we did not find a statistically significant association when analyzing the data separately for Caucasians and Hispanics, the sample sizes were substantially smaller. Also, the direction of effect was similar in the Caucasians and Hispanics, and in the larger ethnic group, Caucasians, we did find a significant result for the rs806380 SNP and a trend toward significance for the omnibus test. In addition, once the covariate age, sex, and group status were included in the model, the omnibus test in Caucasians approached significance and the individual SNP rs806380 association became highly significant.
Another potential limitation is that cases and controls were drawn from three groups: youth in treatment for substance dependence, adjudicated youth, and youth who were subjects of general-population twin and family studies. All of the studies of these groups used common assessment batteries and were designed to be able to compare across groups. In order to increase power, cases and controls were drawn from any of the groups if they met the criteria of having experimented with marijuana. Recently, Hartman et al. (in press) demonstrated that the patterns of endorsement of marijuana abuse and dependence were similar across all three groups.
Despite this limitation, the study has a number of strengths. First, to our knowledge, it is the first report that has specifically examined symptoms of cannabis dependence as a phenotype for association with CNR1. Second, by focusing on adolescents, we are examining the association during a time where initiation and progression toward developing dependence symptoms frequently occurs. Previous studies of CNR1 and its association with a range of drug abuse phenotypes have examined only one or two polymorphisms and have typically examined alcohol or “polysubstance abuse” phenotypes in adults. In addition, to our knowledge, it is the largest study to examine an association between CNR1 and any drug dependence phenotype.
In addition to having one or more cannabis dependence symptoms, many of the cases in this study met full criteria for cannabis dependence, as well as abuse or dependence on other substances. Thus, CNR1 polymorphisms may be associated with a broader range of substance abuse phenotypes. This would be consistent with results by Zhang et al. (2004) as well as twin studies that have reported that the genetic risk for developing marijuana dependence overlaps with the genetic risk for other substance dependence diagnoses (Kendler et al. (2003), Young et al. (in press)), although other authors (Tsuang et al. 1998) have reported that there are some marijuana –specific genetic influences.
Future work should focus on careful definition and examination of quantitative marijuana-and drug-related phenotypes to further clarify this possibility. In addition, future studies should include dense SNP mapping as such data become available through the HapMap project (Gibbs et al., 2003), combined with resequencing in select samples for SNP discovery. Bioinformatics and functional molecular approaches will be necessary to understand the possible functional roles of associated SNPs.
Figure 1.
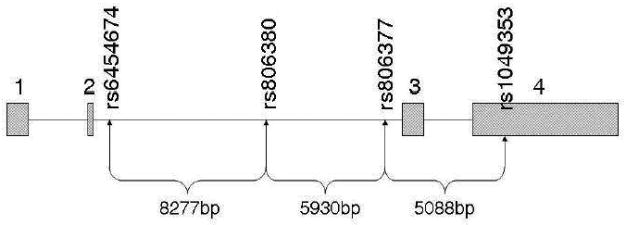
Diagram of CNR1 gene and locations of SNPs examined. Exons are shown as boxes with cross-hatched fill. The rs numbers indicate the SNPs genotyped with the base pair (bp) distances between them.
Acknowledgments
Supported by DA11015 DA015522 DA012845. We acknowledge Rachel Nichols and Jennifer Yu for their assistance in the preparation of this manuscript.
References
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV) American Psychiatric Association; Washington, DC: 1994. [Google Scholar]
- Anchordoquy HC, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of three candidate genes after whole-genome preamplification of DNA collected from buccal cells. Behav Genet. 2003;33:73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Chen CY, Anthony JC. Possible age-associated bias in reporting of clinical features of drug dependence: epidemiological evidence on adolescent-onset marijuana use. Addiction. 2003;98:71–82. doi: 10.1046/j.1360-0443.2003.00237.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Marmur R, Pulles A, Paredes W, Gardner EL. Ventral tegmental microinjection of delta 9-tetrahydrocannabinol enhances ventral tegmental somatodendritic dopamine levels but not forebrain dopamine levels: evidence for local neural action by marijuana's psychoactive ingredient. Brain Res. 1993;621:65–70. doi: 10.1016/0006-8993(93)90298-2. [DOI] [PubMed] [Google Scholar]
- Chen K, Kandel DB, Davies M. Relationships between frequency and quantity of marijuana use and last year proxy dependence among adolescents and adults in the United States. Drug Alcohol Depend. 1997;46:53–67. doi: 10.1016/s0376-8716(97)00047-1. [DOI] [PubMed] [Google Scholar]
- Childers SR, Breivogel CS. Cannabis and endogenous cannabinoid systems. Drug Alcohol Depend. 1998;51:173–187. doi: 10.1016/s0376-8716(98)00075-1. [DOI] [PubMed] [Google Scholar]
- Coffey C, Carlin JB, Lynskey M, Li N, Patton GC. Adolescent precursors of cannabis dependence: findings from the Victorian Adolescent Health Cohort Study. Br J Psychiatry. 2003;182:330–336. doi: 10.1192/bjp.182.4.330. [DOI] [PubMed] [Google Scholar]
- Comings DE, Muhleman D, Gade R, Johnson P, Verde R, Saucier G, MacMurray J. Cannabinoid receptor gene (CNR1): association with i.v. drug use. Mol Psychiatry. 1997;2:161–168. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- Cottler LB, Robins LN, Helzer JE. The reliability of the CIDI-SAM: a comprehensive substance abuse interview. Br J Addict. 1989;84:801–814. doi: 10.1111/j.1360-0443.1989.tb03060.x. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Kranzler H. Association study of cannabinoid receptor gene (CNR1) alleles and drug dependence. Mol Psychiatry. 2001;6:501–502. doi: 10.1038/sj.mp.4000925. [DOI] [PubMed] [Google Scholar]
- Crowley TJ, Macdonald MJ, Whitmore EA, Mikulich SK. Cannabis dependence, withdrawal, and reinforcing effects among adolescents with conduct symptoms and substance use disorders. Drug Alcohol Depend. 1998;50:27–37. doi: 10.1016/s0376-8716(98)00003-9. [DOI] [PubMed] [Google Scholar]
- Crowley TJ, Mikulich SK, Ehlers KM, Whitmore EA, MacDonald MJ. Validity of structured clinical evaluations in adolescents with conduct and substance problems. J Am Acad Child Adolesc Psychiatry. 2001;40:265–273. doi: 10.1097/00004583-200103000-00005. [DOI] [PubMed] [Google Scholar]
- Fergusson DM, Horwood LJ, Lynskey MT, Madden PA. Early reactions to cannabis predict later dependence. Arch Gen Psychiatry. 2003;60:1033–1039. doi: 10.1001/archpsyc.60.10.1033. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Belmont JW, Hardenbol P, Willis TD, Yu F, Yang H, et al. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Hartman C, Gelhorn H, Sakai J, Stallings MC, Young S, Rhee SH, Corley R, Hewitt JK, Crowley TJ, Hopfer CJ. An Item Response Theory Analysis of DSM-IV marijuana abuse and dependence criteria in an adolescent sample. Abstract, College of Problems on Drug Dependence. 2006 In Press. [Google Scholar]
- Hoehe MR, Rinn T, Flachmeier C, Heere P, Kunert HJ, Timmermann B, Kopke K, Ehrenreich H. Comparative sequencing of the human CB1 cannabinoid receptor gene coding exon: no structural mutations in individuals exhibiting extreme responses to cannabis. Psychiatr Genet. 2000;10:173–177. doi: 10.1097/00041444-200010040-00004. [DOI] [PubMed] [Google Scholar]
- Johnson JP, Muhleman D, MacMurray J, Gade R, Verde R, Ask M, Kelley J, Comings DE. Association between the cannabinoid receptor gene (CNR1) and the P300 event-related potential. Mol Psychiatry. 1997;2:169–171. doi: 10.1038/sj.mp.4000246. [DOI] [PubMed] [Google Scholar]
- Johnston LD, O'Malley PM, Bachman JG, Schulenberg JE. Monitoring the Future national results on adolescent drug use: Overview of key findings, 2004 (NIH Publication No 05-5726) National Institute on Drug Abuse; Bethesda, MD: 2005. [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry. 2000;57:261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–95. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA. Cannabis use, abuse, and dependence in a population-based sample of female twins. Am J Psychiatry. 1998;155:1016–1022. doi: 10.1176/ajp.155.8.1016. [DOI] [PubMed] [Google Scholar]
- Li T, Lie X, Zhu ZH, Zhao J, Hu X, Ball DM, Sham PC, Collier DA. No association between (AAT)n repeats in the cannabinoid receptor gene (CNR1) and heroin abuse in a Chinese population. Mol Psychiatry. 2000;5:128–130. doi: 10.1038/sj.mp.4000670. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Heath AC, Nelson EC, Bucholz KK, Madden PA, Slutske WS, Statham DJ, Martin NG. Genetic and environmental contributions to cannabis dependence in a national young adult twin sample. Psychol Med. 2002;32:195–207. doi: 10.1017/s0033291701005062. [DOI] [PubMed] [Google Scholar]
- Maes HH, Woodard CE, Murrelle L, Meyer JM, Silberg JL, Hewitt JK, Rutter M, Simonoff E, Pickles A, Carbonneau R, Neale MC, Eaves LJ. Tobacco, alcohol and drug use in eight- to sixteen-year-old twins: the Virginia Twin Study of Adolescent Behavioral Development. J Stud Alcohol. 1999;60:293–305. doi: 10.15288/jsa.1999.60.293. [DOI] [PubMed] [Google Scholar]
- Miles DR, van den Bree MB, Gupman AE, Newlin DB, Glantz MD, Pickens RW. A twin study on sensation seeking, risk taking behavior and marijuana use. Drug Alcohol Depend. 2001;62:57–68. doi: 10.1016/s0376-8716(00)00165-4. [DOI] [PubMed] [Google Scholar]
- Navarro M, Carrera MR, Fratta W, Valverde O, Cossu G, Fattore L, Chowen JA, Gomez R, del Arco I, Villanua MA, Maldonado R, Koob GF, Rodriguez de Fonseca F. Functional interaction between opioid and cannabinoid receptors in drug self-administration. J Neurosci. 2001;21:5344–5350. doi: 10.1523/JNEUROSCI.21-14-05344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Leonard CM, Ishiguro H, Zhang PW, Lin Z, Akinshola BE, Uhl GR. Endocannabinoids and cannabinoid receptor genetics. Prog Neurobiol. 2002;66:307–344. doi: 10.1016/s0301-0082(02)00007-2. [DOI] [PubMed] [Google Scholar]
- Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- SAMHSA. Substance Abuse and Mental Health Services Administration; 2002. Marijuana Treatment Admissions Increase: 1993-1999. SAMHSA. 4. 2002. http://www.samhsa.gov/oas/2k2/MJtx.htm. [Google Scholar]
- Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, Wendel B, Rommelspacher H, Hoehe MR. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002;65:221–224. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Young SE, Rhee SHyun, Stallings MC, Corley RP, Hewitt JK. Genetic and environmental vulnerabilities underlying adolescent substance use and problem use: General or specific? Behavior Genetics. doi: 10.1007/s10519-006-9066-7. in press, Behavior Genetics. [DOI] [PubMed] [Google Scholar]
- Zhang PW, Ishiguro H, Ohtsuki T, Hess J, Carillo F, Walther D, Onaivi ES, Arinami T, Uhl GR. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry. 2004;9:916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. [PubMed] [Google Scholar]


