Abstract
The photoactivatable sterol probe [3α-3H]6-Azi-5α-cholestan-3β-ol ([3H]Azicholesterol) was used to identify domains in the Torpedo californica nicotinic acetylcholine receptor (nAChR) that interact with cholesterol. [3H]Azicholesterol partitioned into nAChR-enriched membranes very efficiently (>98%), photoincorporated into nAChR subunits on an equal molar basis, and neither the pattern nor the extent of labeling was affected by the presence of the agonist carbamylcholine, consistent with photoincorporation at the nAChR lipid-protein interface. Sites of [3H]Azicholesterol incorporation in each nAChR subunit were initially mapped by S. aureus V8 protease digestion to two relatively large homologous fragments that contain either the transmembrane segments M1-M2-M3 (e.g. αV8-20) or M4 (e.g. αV8-10). The distribution of [3H]Azicholesterol labeling between these two fragments (e.g. αV8-20, 29%; αV8-10, 71%), suggests that the M4 segment has the greatest interaction with membrane cholesterol. Photolabeled amino acid residues in each M4 segment were identified by Edman degradation of isolated tryptic fragments and generally correspond to acidic residues located at either end of each transmembrane helix (e.g. αAsp-407). [3H]Azicholesterol labeling was also mapped to peptides that contain either the M3 or M1 segment of each nAChR subunit. These results establish that cholesterol likely interacts with the M4, M3, and M1 segments of each subunit and therefore the cholesterol binding domain fully overlaps the lipid-protein interface of the nAChR.
The Torpedo nicotinic acetylcholine receptor (nAChR) is the most extensively studied member of a superfamily of ligand-gated ion channels that includes muscle and neuronal nAChRs, serotonin 5-HT3 receptors, gamma-amino butyric acid type A receptors (GABAA), and glycine receptors (1,2). Each Torpedo nAChR is a pentamer comprised of four homologous transmembrane subunits with a stoichiometry of 2α:β:γ:δ that are arranged pseudosymmetrically around a central cation-selective ion channel. Conformational transitions, from the closed (nonconducting), to agonist-induced open (ion conducting), to desensitized (nonconducting) states, are critical for functioning of the nAChR (2). The ability of the nAChR to undergo these transitions is profoundly influenced by the lipid composition of the bilayer (3,4). The presence of both anionic lipid (e.g. phosphatidylserine) and neutral lipid (e.g. cholesterol) are required for the receptor to undergo agonist-induced state transitions (3,5). A requirement for cholesterol is consistent with its rich abundance in Torpedo nAChR-enriched membranes (∼35% on a molar basis; 6) and its preferential interaction with the nAChR (7,8). The molecular interactions which underlie the effects of cholesterol on nAChR functionality are not understood. Cholesterol has no detectable effect on the overall secondary structure of the receptor as revealed by FTIR spectroscopy (9). Yet nAChR state transitions are preserved in the presence of a single shell of lipids (∼45 lipids) surrounding the receptor protein (10), and amino acid substitutions at residues that contact lipid have significant effects on channel gating (11,12). These results suggest that direct interactions between cholesterol and the nAChR lipid-protein interface exert subtle yet critical structural alterations that are necessary for receptor functionality.
The structure of the nAChR lipid-protein interface has been reasonably well defined through studies employing small lipophilic photoreactive probes (12,13) and by the cryo-electron microscopy derived structure of the nAChR (4Å structure, 14,15). Each nAChR subunit is comprised of a bundle of four transmembrane helices (M1-M4). The five M2 helices are arranged about a central axis orthogonal to the membrane forming the channel lumen. Each M2 helix is shielded from the lipid bilayer by the group of helices M1, M3, and M4. The M4 helix is located on the outside of the four helical bundle, exposing a rather broad helical face to lipid, while the M1 and M3 helices have more limited surfaces that are in contact with lipid (see Fig. 4, in Ref.14).
Figure 4. Tryptic digestion of [3H]Azicholesterol labeled V8 protease fragments αV8-10, βV8-12, γV8-14, and δV8-11.
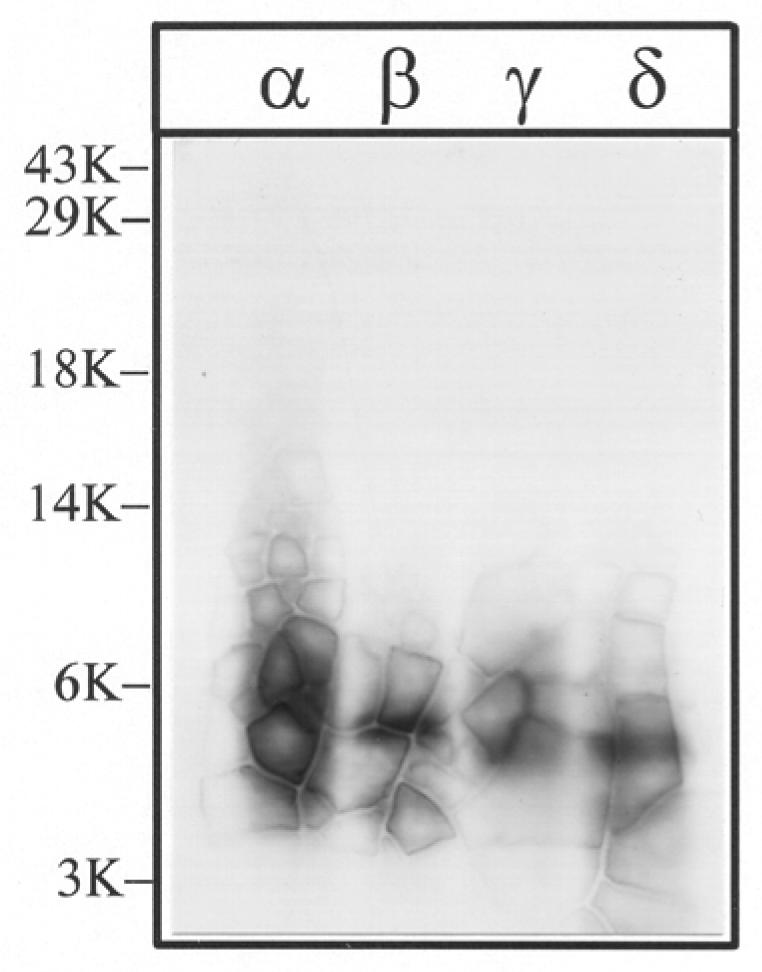
The V8 protease fragments αV8-10, βV8-12, γV8-14, and δV8-11, isolated from nAChRs labeled with 1.25 μM [3H]Azicholesterol in the absence of agonist, were exhaustively digested with trypsin (200% w/w) for four days. Aliquots of the total digests (∼5%) were fractionated by Tricine SDS-PAGE and then subjected to fluorography for 6 weeks. The migration of prestained molecular weight standards are indicated on the left. The principal band of 3H evident in each digest migrates with an apparent molecular mass of ∼ 5-kDa.
Previous labeling studies using photoactivatable cholesterol analogs have either restricted the characterization of labeling to the intact subunit level (16,17) and/or employed photoactivatable sterols that are not likely functional cholesterol substitutes (18,19). [3α-3H]6-Azi-5α-cholestan-3β-ol ([3H]Azicholesterol) is a functional cholesterol substitute (20) in which a photoactivatable diazirine functionality has been introduced into the six position (Fig. 1). Irradiation of other aliphatic diazirines with long-wavelength ultraviolet light (365 nm), that is unlikely to directly activate amino acid side chains, produces reactive intermediates which react efficiently with acidic amino acids and tyrosine, as well as with glutamine, cysteine, serine and histidine, forming adducts stable under conditons of Edman degradation (21,22).
Figure 1.
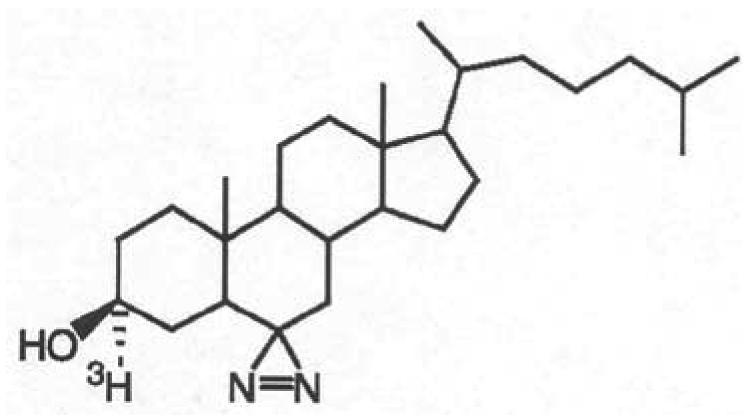
Chemical structure of [3H]Azicholesterol.
In the present work, [3H]Azicholesterol has been used as a photoaffinity probe to localize the regions of the nAChR that interact with membrane cholesterol. [3H]Azicholesterol partitions into nAChR-enriched membranes with high efficiency (>98%) and photolabels each nAChR subunit on an equal molar basis. Neither the pattern nor the extent of labeling is affected by the inclusion of the agonist carbamylcholine, consistent with incorporation at the lipid-protein interface. Nearly three-quarters of the total [3H]Azicholesterol incorporation into each nAChR subunit is localized to S. aureus protease fragments that contain the M4 segment (e.g. αV8-10), suggesting that the transmembrane M4 helix has the greatest interaction with membrane cholesterol. In addition, [3H]Azicholesterol labeling was mapped to peptides that contain either the M1 or M3 segments of each receptor subunit, which suggests that the cholesterol binding domain (M1, M3, and M4 segments) fully overlaps the lipid-protein interface of the nAChR.
EXPERIMENTAL PROCEDURES
Materials
nAChR-enriched membranes were isolated from frozen Torpedo californica electric organs obtained from Aquatic Research Consultants (San Pedro, CA) by differential and sucrose density gradient centrifugation, as described previously (23). The final membrane suspensions in ∼38% sucrose were stored at −80°C. [3H]Azicholesterol ([3α-3H]6-Azi-5α-cholestan-3β-ol; 20 Ci/mmol)) was obtained from American Radiolabeled Chemicals (St. Louis, MO) and was stored in ethanol at −20°C. Carbamylcholine chloride was purchased from Sigma-Aldrich (St. Louis, MO); Staphylococcus aureus glutamyl endopeptidase (V8 protease) from MP Biochemicals (Irvine, CA); Trypsin (TPCK-treated) from Worthington (Lakewood, NJ); Trifluoroacetic acid (TFA) from Pierce (Rockford, IL); and Genapol C-100 from Calbiochem (San Diego, CA). Prestained low range molecular weight standards were purchased from Life Technologies, Inc (Gaithersburg, MD).
Photolabeling of nAChR-enriched membranes with [3H]Azicholesterol
Freshly thawed nAChR-enriched Torpedo membranes (in ∼38% sucrose) were diluted four-fold with vesicle dialysis buffer (VDB, 100mM NaCl, 0.1mM EDTA, 0.02% NaN3, 10mM MOPS, pH 7.5) and pelleted by centrifugation (18,000 rpm for 1 h using a JA-20 rotor in a Beckman J2-HS centrifuge). Pelleted membranes were then resuspended in VDB to achieve a protein concentration of 1.25 mg/mL (∼1 μM nAChR). For photolabeling experiments, membrane aliquots in glass test tubes or vials (1 mg analytical scale; 25 mg preparative scale) were incubated with [3H]Azicholesterol (from an ethanolic stock solution; final concentrations are 1.25μM and <1% ethanol) either in the presence or absence of 400μM carbamylcholine (carb). After 2 h in the dark at room temperature, samples were irradiated at 365nm (Spectroline EN-280L) for 10 minutes at a distance of < 1 cm. Photolyzed membrane suspensions were then pelleted, solubilized in electrophoresis sample buffer, and subjected to SDS-PAGE.
SDS-Polyacrylamide Gel Electrophoresis
SDS-PAGE was performed according to the method of Laemmli (24) with analytical (1.0 mm thick) and preparative (1.5 mm) separating gels comprised of 8% polyacrylamide/0.33% bis-acrylamide. Following electrophoresis, nAChR subunits were visualized by staining with Coomassie Blue R-250 (0.25% (w/v) in 45% methanol, 10% acetic acid, 45% dH2O) and destaining (25% methanol, 10% acetic acid, 65% dH2O). For fluorography, stained gels were impregnated with fluor (Amplify; GE Biosciences) for 30 min, dried and exposed at −80 °C to Eastman Kodak X-Omat LS film for various times (3-6 weeks). 3H incorporation into individual polypeptides was also quantified by liquid scintillation counting of excised gel slices (25). For either analytical or preparative [3H]Azicholesterol photolabeling experiments, individual nAChR subunits were fragmented by limited ‘in gel’ digestion with S. aureus V8 protease (26,27). Following electrophoresis, stained 8% acrylamide gels were soaked in distilled water overnight and nAChR subunit bands (α, β, γ, δ) from each labeling condition (−/+ carb) were excised and transferred directly to the well(s) of either a 1.0 mm (analytical) or 1.5 mm (preparative) thick mapping gel, composed of a 5-cm-long 4.5% acrylamide stacking gel and a 11-cm-long 15% acrylamide separating gel. For analytical labelings, each gel slice was overlaid with 6 μg S. aureus V8 protease in overlay buffer (5% sucrose,125 mM Tris-HCl, 0.1% SDS, pH 6.8) and electrophoresed at 60V constant voltage for ∼ 3 h and then at ∼6 mA constant current overnight. After Coomassie Blue R-250 staining (2 h) and destaining (3-4 h), analytical gels were impregnated with fluor, dried and exposed to film (up to 16 weeks). Preparative 8% acrylamide gels (1.5 mm thick) were soaked in distilled water overnight and each nAChR subunit excised as a strip (∼15 cm). The gel strips were transferred to individual 15% acrylamide mapping gels and overlaid with 200 μg V8 protease in overlay buffer. Following electrophoresis, staining, and soaking in distilled water overnight, the following subunit proteolytic fragment bands were excised, using the nomenclature of (12): αV8-20 (αSer-173-Glu-338), αV8-10 (αAsn-339-Gly-437), βV8-22 (βIle-173-Glu383), βV8-12 (βMet-384-Ala-469), γV8-24 (γAla-167-Glu-372), γV8-14 (γLeu-373-Pro-489), δV8-20 (δIle192-Glu372),and δV8-11 (δLys436-Ala501). The excised proteolytic fragments were then isolated by passive diffusion into 25 mLs of Elution Buffer (0.1M NH4HCO3, 0.1% (w/v) SDS, 1% β-mercaptoethanol, pH 7.8) for 4 d at room temperature with gentle mixing (12,18). The gel suspensions were then filtered (Whatman No. 1 paper) and concentrated using Centriprep-10 concentrators (10 kDa cutoff, Amicon). Excess SDS was removed by acetone precipitation (>85% acetone at −20°C overnight).
Proteolytic digestions
For digestion with trypsin, acetone precipitated V8 protease subunit fragments, αV8-20, αV8-10, βV8-22, βV8-12, γV8-24, γV8-14, δV8-20 and δV8-11 were initially resuspended in a small volume (60 μL) of SDS containing buffer (0.1M NH4HCO3 , 0.1% SDS, pH 7.8). The SDS concentration was then reduced by diluting with buffer without SDS and Genapol C-100 was added, resulting in final concentrations of 0.02% (w/v) SDS, 0.5% Genapol C-100, pH 7.8 (1-2 mg/mL protein). To ensure complete proteolysis, trypsin was added at a 200% (w/w) enzyme to substrate ratio and the digestion allowed to proceed for 4 days at room temperature. Small aliquots (∼5%) of the total digest were resolved on analytical (1.0 mm thick) small pore (16.5%T/6%C) Tricine SDS-PAGE gels (28,12). Analytical Tricine gels were placed in destain for 1 h, impregnated with fluor, dried, and exposed to film (2-12 weeks). The bulk of the tryptic digests were separated by reverse-phase HPLC. For exhaustive V8 protease digestion, acetone precipitated V8 protease subunit fragments, αV8-20, βV8-22, γV8-24 and δV8-20 were resuspended in 300 μL of 0.1M NH4HCO3, 0.01% (w/v) SDS, pH 7.8, then incubated with V8-protease (400% w/w) for 6 days at room temperature. Aliquots (∼10%) of each digest were resolved on an analytical Tricine SDS-PAGE gel and processed for fluorography (12-16 week exposure). The bulk of the material from each V8 protease digest was separated on individual preparative scale (1.5 mm thick) Tricine SDS-PAGE gels. Following staining and destaining of each preparative Tricine gel, select fragments were excised based upon the results of fluorographs of analytical Tricine gels as well as previous results (12) and with the aid of low range molecular weight standards (Life Technologies, Inc). The selected fragments: αV8-8.5K, βV8-7K, γV8–7.5K and δV8-7.5K were then isolated by passive elution into 4 mL of elution buffer over 4 d at room temperature. The gel suspensions were filtered, concentrated (Centriprep-3, Amicon) and further purified by reversed-phase HPLC.
HPLC purification
Protelytic fragments of nAChR subunits were purified by reversed-phase HPLC on a Shimadzu LC-10A binary HPLC system, using a Brownlee Aquapore C4 column (100 × 2.1mm). Solvent A was 0.08% trifluoroacetic acid (TFA) in water and Solvent B was 0.05% TFA in 60% acetonitrile/40% 2- propanol. A nonlinear elution gradient at 0.2 ml/min was employed (25% to 100% Solvent B in 100 min) and fractions were collected every 2.5 min (42 fractions/run). The elution of peptides was monitored by the absorbance at 210 nm and 25 or 50 μl aliquots of each collected fraction were counted for radioactivity using liquid scintillation counting.
Sequence Analysis
Amino terminal sequence analysis was performed on a Beckman Instruments (Porton) 20/20 automated protein sequencer using gas phase cycles (Texas Tech Biotechnology Core Facility). Pooled HPLC fractions were dried by vacuum centrifugation, resuspended in a small volume (20 μL) of 0.1% SDS and immobilized on chemically modified glass fiber disks (Beckman Instruments), which were used to improve the sequencing yields of hydrophobic peptides. Peptides were subjected to at least 10 sequencing cycles. Alternatively, fractions containing the α-M4, β-M4, γ-M4 and δ-M4 peptides, isolated by HPLC from tryptic digests of αV8-10, βV8-12, γV8-14 and δV8-11, were sequenced on an Applied Biosystems Procise 492 protein sequencer using liquid phase cycles. The volume of pooled HPLC fractions was reduced by vacuum centrifugation (to ∼1 mL); diluted with 2 volumes of 0.1% TFA in distilled water (to reduce organic concentration) and loaded onto PVDF disks using Prosorb sample preparation cartridges (Applied Biosystems # 401959). Approximately one-sixth of the released PTH-amino acids were analyzed for released PTH-amino acids, and the remaining five-sixths were collected for scintilation counting. During the sequence analysis of nAChR subunit fragments photolabeled by [3H]Azicholesterol, we found that the very hydrophobic photolabeled cleavage products were apparently inefficiently transfered from the cleavage cartridge and/or conversion flask by the solvents normally used for sequencing. For example, when the conversion flask was rinsed with 20% CH3CN and then methanol prior to beginning a new sequence run, depending upon the amount of 3H loaded in the previous sequence run, between 100 and 300 cpm were recovered from the flask (i.e. 6-20 times the normal background). Such residual radioactivity in the conversion flask was not seen in previous studies even with other hydrophobic probes such as [125I]TID (12) or [3H]promegestone (19). After sequencing samples labeled with [3H]Azicholesterol, several days of washing with methanol were required to reduce the residual 3H wash-through from ∼200 cpm/cycle to background levels (∼15 cpm). Because this inefficient transfer of labeled PTH amino acids is also likely to result in significant recovery of 3H in the cycles following cleavage at a labeled amino acid, as detailed in Results, we interpret 3H release profiles cautiously. Yield of PTH-derivatives was calculated from peak height compared with standards using the model 610A Data Analysis Program, version 2.1A. Initial and repetitive yields were calculated by a nonlinear least squares regression to the equation M = I0 × Rn, where M is the observedrelease, I0 is the initial yield, R is the repetitive yield,and n is the cycle number.
RESULTS
Photoincorporation of [3H]Azicholesterol into nAChR-rich membranes
Initial photolabeling experiments were designed to characterize the general pattern of [3H]Azicholesterol photoincorporation into Torpedo nAChR-enriched membranes as well as to assess the sensitivity of photoincorporation to the presence of agonist. nAChR-enriched membranes (1.25 mg/mL) were equilibrated with 1.25 μM [3H]Azicholesterol (>98% membrane partitioning) in the absence and in the presence of 400 μM carbamylcholine (carb). After irradiation (365 nm, 10 min), membrane suspensions were pelleted and resuspended in electrophoresis sample buffer, and the pattern of incorporation monitored by SDS-PAGE followed by fluorography. A representative fluorograph of an 8% polyacrylamide gel (Fig. 2) demonstrates that there is appreciable incorporation of [3H]Azicholesterol into each of the nAChR subunits. As expected for a lipid bilayer probe, [3H]Azicholesterol photoincorporates into several other membrane proteins that are also present in nAChR-enriched membranes including the Na/K-ATPase (Figure 2, αNK), the voltage-gated chloride channel CLC-0 (band just below αNK), and most prominently a 34-kDa polypeptide identified as the mitochondrial voltage-dependent anion channel (VDAC) (29). Neither the overall labeling pattern nor the relative incorporation into individual nAChR subunits (or non nAChR polypeptides) was affected by the inclusion of 400 μM carb (Fig. 2, + lanes). Based on liquid scintillation counting of excised gel bands, ∼0.05% of nAChR subunits incorporated [3H]Azicholesterol, with approximately equal molar incorporation into each subunit (2α/1β/1γ/1δ: 1:1.01: 0.92:1.22, n= 2). The presence of carb resulted in no detectable (<10%) change in the extent of [3H]Azicholesterol incorporation into any polypeptide, including nAChR subunits.
Figure 2. Photoincorporation of [3H]Azicholesterol into nAChR-enriched membranes in the absence and presence of carbamylcholine.
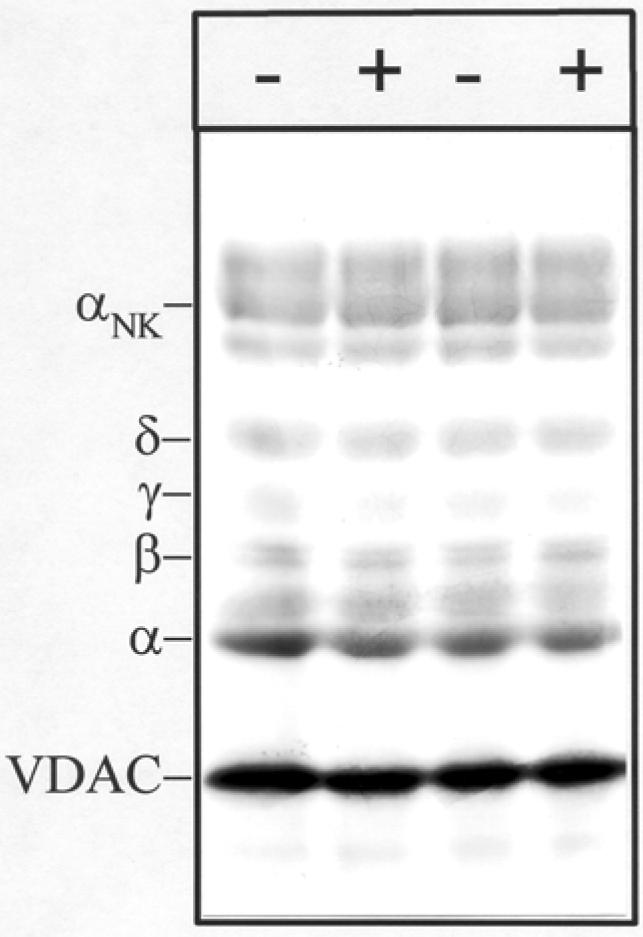
nAChR-enriched membranes were equilibrated (2 h incubation) with 1.25 μM [3H]Azicholesterol in the absence (− lanes) and in the presence (+ lanes) of 400μM carbamylcholine and irradiated at 365 nm for 10 min. After irradiation, polypeptides were resolved by SDS-PAGE and processed for fluorography (3-week exposure). Labeled lipid and free photolysis products were electrophoresed from the gel with the tracking dye. Indicated on the left are the mobility of nAChR subunits (α,β,γ,δ), the α-subunit of Na+/K+-ATPase (αNK, ∼95kDa), and the mitochondrial voltage-dependant anion channel, VDAC (∼34 kDa).
Proteolytic Mapping of [3H]Azicholesterol Photoincorporation into nAChR subunits
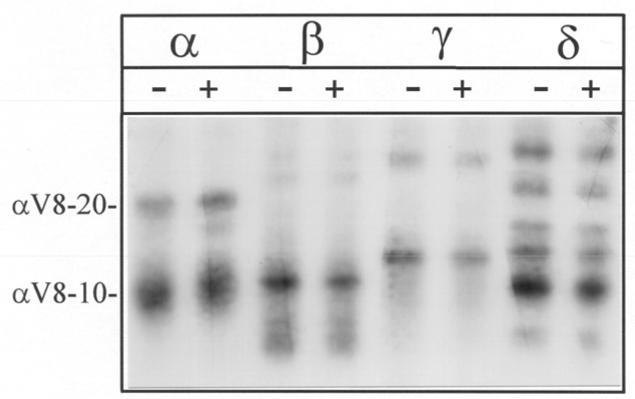
[3H]Azicholesterol incorporation within each of the nAChR subunits was examined by limited digestion with S. aureus V8 protease in a 15% acrylamide mapping gel (12,27). Limited V8 protease digestion of nAChR subunits reproducibly generates a set of non-overlapping fragments (23,12). Inspection of a fluorograph (Fig. 3) of the V8 protease digests reveals likely [3H]Azicholesterol incorporation into the following fragments, using the nomenclature of (12): αV8-20 (Ser-173- Glu-338); αV8-10 (Asn-339-Gly-437); βV8-22 (Ile-173- Glu-383);βV8-12 (Met-384-Ala-469); γV8-24 (Ala-167- Glu-372); γV8-14 (Leu-373- Pro-489); δV8-20 (Ile-192-Glu-372); δV8-12 (Ile-192-Glu-280); δV8-11 (Lys-436-Ala-501). Fragments αV8-20, βV8-22, γV8-24, and δV8-20 contain the transmembrane segments M1, M2, and M3 of each subunit, and fragments αV8-10, βV8-12, γV8-14, and δV8-11 contain the transmembrane segment M4 of each subunit. Based on liquid scintillation counting of excised gel pieces, ∼75% of the total subunit labeling resides within V8 proteolytic fragments that contain the M4 transmembrane segment (e.g. αV8-10); while ∼25% of the labeling resides within V8 proteolytic fragments that contain M1-M2-M3 transmembrane segments (e.g. αV8-20). Addition of agonist (Fig. 3, +lanes) had no significant effect on either the pattern or extent of [3H]Azicholesterol labeling of V8 proteolytic fragments. [Based upon scintillation counting of aliquots of isolated V8 protease fragments from 5 preparative scale labeling experiments the relative incorporation into each fragment was: (absence of agonist) αV8-20 (29%), αV8-10 (71%); βV8-22 (14%), βV8-12 (86%); γV8-24 (26%), γV8-14 (74%); δV8-20 (15%), δV8-11 (85%) and (presence of agonist) αV8-20 (30%), αV8-10 (70%); βV8-22 (11%), βV8-12 (89%); γV8-24 (28%), γV8-14 (72%); δV8-20 (20%), δV8-11 (80%)].
Figure 3. Proteolytic mapping of the sites of [3H]Azicholesterol incorporation into nAChR subunits using “in-the-gel” digestion with S. aureus V8 protease.
nAChR-enriched membranes were labeled with 1.25 μM [3H]Azicholesterol in the absence and in the presence of 400 μM carbamylcholine. After photolysis (365 nm for 10 min), membranes were resolved by SDS-PAGE (1.0 mm thick, 8% acrylamide). nAChR subunit bands were excised following identification by staining (Coomassie Blue) and transferred to the wells of a 15% acrylamide mapping for digestion with S. aureus V8 protease. Following electrophoresis, the mapping gel was stained with Coomassie Blue and processed for fluorography (16-week exposure). The principal [3H]Azicholesterol labeled proteolytic fragments, following the nomenclature of (12), are: αV8-20 (Ser-173- Glu-338); αV8-10 (Asn-339-Gly-437); βV8-22 (Ile-173/ Asn-183- Glu-383); βV8-12 (Met-384-Ala-469); γV8-24 (Ala-167/ Trp-170- Glu-372); γV8-14 (Leu-373/ Ile-413-Pro-489); δV8-20 (Ile-192- Glu-372); δV8-12 (Ile-192-Glu-280); δV8-11 (Lys-436-Ala-501).
Mapping the Sites of [3H]Azicholesterol incorporation in M4 Segments of each nAChR subunit
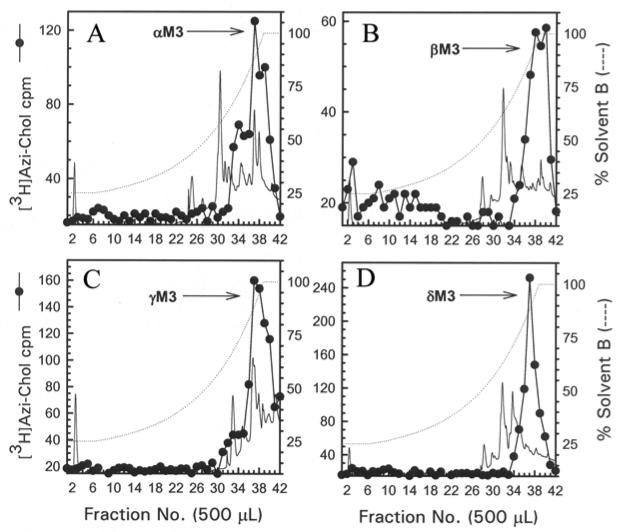
The M4 segments were isolated from exhaustive tryptic digests of relatively large V8 protease fragments of each nAChR subunit (αV8-10, βV8-12, γV8-14, and δV8-11). When an aliquot of each tryptic digestion was resolved by Tricine SDS-PAGE, the corresponding fluorograph revealed in each case a principal band migrating with an apparent molecular weight of ∼5 kDa (Fig. 4). When the bulk of each digest was resolved by reversed-phase HPLC (Fig. 5), for the α, β, and γ subunit digests >90% of 3H eluted as a hydrophobic peak at the end of the gradient, while for the δ subunit the 3H was distributed in two hydrophobic peaks.
Figure 5. Reversed-phase HPLC purification of [3H]Azicholesterol labeled fragments from tryptic digests of αV8-10, βV8-12, γV8-14, and δV8-11.
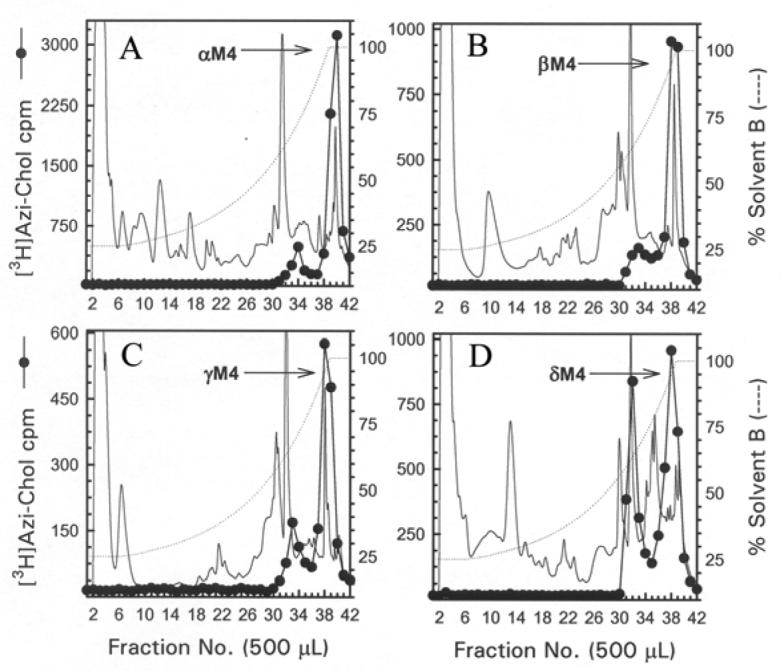
For αV8-10 (A), βV8-12 (B), γV8-14 (C), and δV8-11 (D) isolated from nAChRs labeled with 1.25 μM [3H]Azicholesterol in the absence of agonist, tryptic digests were fractionated by reversed-phase HPLC on a Brownlee Aquapore C4 column (100 × 2.1 mm) as described in the Experimental Procedures. The elution of peptides was monitored by absorbance at 210 nm (solid line) and elution of 3H by scintillation counting of aliquots (25 μL) of each 500 μL fraction (●). The point of elution of the M4 peptide based on previous work (and results from Fig. 6) is indicated with an arrow. Based upon recovery of radioactivity, >90% of the material was recovered from the HPLC column. HPLC fractions 38-41 (A) or 37-40 (B-D) were pooled for sequence analyses (Figure 6).
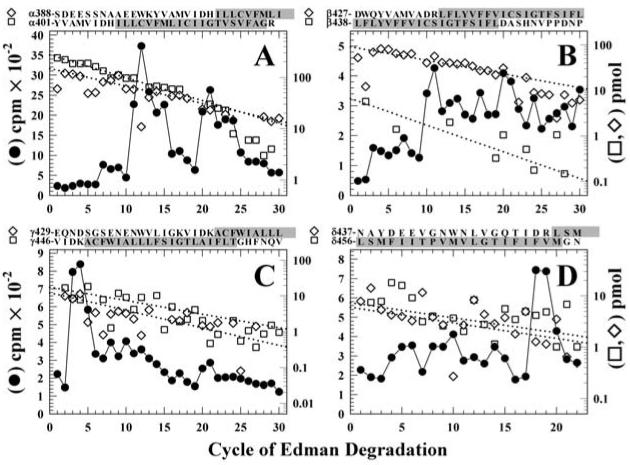
For the αV8-10 tryptic digest, HPLC fractions 38-41 (Fig. 5A) were pooled and subjected to N-terminal sequence analysis. As shown in Figure 6A, sequence analysis revealed the presence of fragments beginning at αTyr-401 (274 pmol) and αSer-388 (148 pmol), overlapping sequences, each of which contains the αM4 segment beginning at αIle-409. The 3H release pattern was rather complex, with the largest release occurring in cycles 11 (1830 cpm),12 (1460 cpm), and 20 (1460 cpm)2. The presence of two abundant peptides precluded a definitive assignment of 3H release in a particular cycle with [3H]Azicholesterol incorporation into a single amino acid. However, the release in cycles 11, 12 and 20 would correspond to labeling of αGlu-398, αTrp-399, and αAsp-407 in the fragment beginning at αSer-388, and/or of αLeu-411, αCys-412 and αIle-420 in the fragment beginning at αTyr-401. Since other aliphatic diazirines have been shown to efficiently photolabel acidic side chains in the nAChR agonist binding site and ion channel (21,22), and we have not seen efficient photolabeling of aliphatic side chains, we consider it most likely that the 3H release in cycles 11 and 20 result from [3H]Azicholesterol photolabeling of αGlu-398 and αAsp-4073 at the N-terminus of αM4. Since αCys-412 is the primary amino acid in αM4 photolabeled by [3H]Azioctanol (21), and the 3H release in cycle12 is as large as that in cycle 11, it is likely that the cycle 12 release results at least in part from labeling of αCys-412, as well as from the lag from the release in cycle 11.
Figure 6. Sequence analysis of [3H]Azicholesterol labeled fragments containing the M4 segment.
Peaks of 3H from the HPLC fractionations of tryptic digests of nAChR subunit fragments (Figure 5) were pooled for sequencing. Panel A, the digest of αV8-10. Fragments were detected beginning at αTyr-401 (□, Io, 274 pmol; R, 90%) and αSer-388 (◇, Io, 148 pmol; R, 92.5%) (254,000 cpm loaded on the filter and 25,600 cpm remaining after 30 cycles), with other unidentified peptides present at <10 pmol. Panel B, the digest of βV8-12. The primary sequence began at βAsp-427 (◇, Io, 86 pmol; R, 94%), with secondary sequences beginning at βLeu-438 (□, Io, 9 pmol; R, 91%), βLys-216 (βM1, Io, 9 pmol; R, 94%, not shown), and βMet-249 (βM2, Io, 4 pmol; R, 92%, not shown) (90,000 cpm loaded, 5,500 cpm remaining after 40 cycles). Panel C, the trypsin digest of γV8-14. Four fragments were present beginning at γVal-446 (□, Io, 18 pmol; R, 92%), γGlu-429 (◇, Io, 12 pmol; R, 89%), γThr-276 (γM3, Io, 28 pmol; R, 90%, not shown), and γLys-218 (γM1, ∼5 pmol) with (53,000 cpm loaded on the filter, 3,300 cpm remaining after 30 cycles). Panel D, the trypsin digest of δV8-11. Peptides were identified beginning at δAsn-437 (□, Io, 6 pmol; R, 93%), δLeu-456 (◇, Io, 7 pmol; R, 93%), δLys-224 (δM1, Io, 12 pmol; R, 94%, not shown), δMet-257 (δM2, Io, 17 pmol; R, 89%, not shown), and Thr-28 from the β-subunit of the Na+/K+-ATPase (Io, 10 pmol; R, 96%, not shown) (90,000 cpm loaded on the filter, 5,500 cpm remaining after 22 cycles). For each sample, ∼83% of each cycle of Edman degradation was analyzed for released 3H (●) and ∼17% for PTH-derivatives (□,◇), with the dotted line corresponding to the fit of the amount of detected PTH-derivatives. The amino acid sequences of the fragments containing the M4 segments are shown above each panel, with the limits of the M4 regions shaded.
For the tryptic digestion of βV8-12, HPLC fractions 37-40 (Fig 5B) were pooled and sequenced (Fig 6B). The primary sequence began at βAsp-427 (89 pmol), 12 amino acids before the N-terminus of βM4 (βLeu-438), with secondary sequences beginning at βLeu-438 (9 pmol) as well as at βLys-216 (9 pmol) and βMet-249 (4 pmol) at the N-termini of βM1 and βM2, respectively. The 3H release profile was more complex than that seen for αM4, with the largest 3H release occurring in cycles 10 (215 cpm) and 20 (140 cpm). Release in these cycles could result from [3H]Azicholesterol incorporation into βAsp-436 and βIle-446 in the primary sequence, or from βCys-447 and βAsp-557 in the secondary sequence beginning at βLeu-438. Thus it is most likely that βAsp-436 and βAsp-457 at the N- and C-termini of the βM4 segment are both labeled.
For the tryptic digest of γV8-14, HPLC fractions 37-40 were pooled and sequenced (Fig. 6C). Fragments containing γM4 began at γVal-446 (18 pmol), 4 amino acids before the N-terminus of γM4, and at γGlu-429 (12 pmol), and there were also fragments containing γM3 (γThr-276 (28 pmol) and γM1 (γLys-218 (5 pmol), the N-terminus of γM1). The dominant 3H release in cycle 3 (650 cpm) would correspond to [3H]Azicholesterol incorporation into γAsp-448 at the N-terminus of γM4 of the γVal-446 peptide, and there was no clear evidence of labeling of any of the uncharged amino acids in γM4 such as γCys-451 which was labeled by [125I]TID (12). Consistent with this assignment is the much smaller amount of 3H release in cycle 20 (100 cpm), corresponding to γAsp-448 for the γGlu-429 peptide.
For the tryptic digest of δV8-11, sequence analysis (Fig 6D) of HPLC fractions 37-40 revealed the presence of overlapping fragments beginning at δAsn-437 (6 pmol) and δLeu-456 (7 pmol), the N-terminus of δM4, as well as fragments beginning at the N-termini of δM2 (δMet-257 (17 pmol) and δM1 (δLys-224 (12 pmol), and beginning at Thr-28 of the Na+/K+-ATPase β-subunit (10 pmol). The 3H release of 550 cpm in cycle 18 would correspond to [3H]Azicholesterol incorporation into δAsp-454 in the fragment beginning at the δAsn-437 (or in uncharged side chains in any of the other fragments), and there was no clear evidence of labeling of any of the amino acids in δM4 sequenced in the first 17 cycles.
Mapping [3H]Azicholesterol incorporation to the M3 segment of each nAChR subunit
Previous studies have established that in addition to the M4 segment, amino acids within the M1 and M3 segments also have contact with the lipid bilayer (12,14). To determine if there is [3H]Azicholesterol incorporation into the M3 segment of each nAChR subunit, the large V8 protease fragments αV8-20, βV8-22, γV8-24, and δV8-20, each of which contains M1, M2, and M3, were further digested with V8 protease. When aliquots of each digest (∼10%) were fractionated on a Tricine SDS-PAGE gel and analyzed by fluorography (Fig. 7), the following fragments were evident, based on the nomenclature of (12): αV8-8.5, βV8-7 (just barely visible in this particular fluorograph), γV8-7.5, and δV8-7.5. These bands have previously been shown to contain the M3 segment of each nAChR subunit (12). When the bulk of material from each V8 digest was resolved on a preparative scale Tricine SDS-PAGE gel, the bands corresponding to αV8-8.5, βV8-7, γV8-7.5, and δV8-7.5 were excised (with the aid of prestained low molecular weight range standards). When the material from each band was isolated and further purified by reversed-phase HPLC (Fig. 8), for each subunit the 3H eluted as a hydrophobic peak at the end of the gradient (fractions 36-40). For each V8 fragment, sequence analysis of the pool of HPLC fractions (36-40) revealed the presence of a primary sequence (present at a five- to ten-fold greater abundance than any other sequences) with an amino terminus near the beginning of the M3 segment: αLeu-263 (363 pmol), βThr-273 (149 pmol), γThr-276 (63 pmol), δThr-281 (58 pmol)). While the extent of [3H]Azicholesterol incorporation into the M3 segments was too low to effectively allow for the determination of individually labeled amino acids by radio-sequencing, the results suggest that [3H]Azicholesterol labels amino acids within the M3 segment of each nAChR subunit.
Figure 7. V8 protease digestion of [3H]Azicholesterol labeled V8 protease fragments αV8-20, βV8-22, γV8-24, and δV8-20.
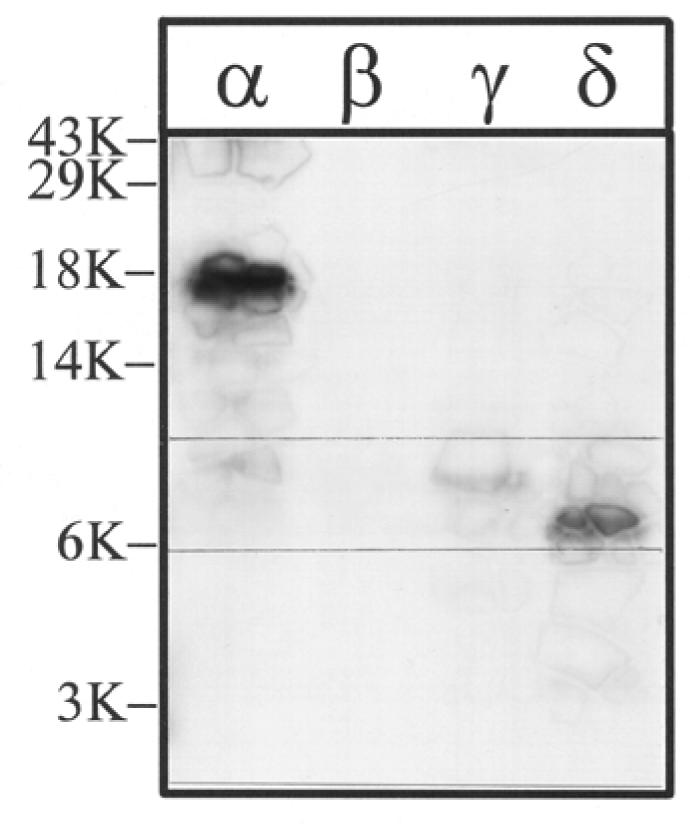
The V8 protease fragments αV8-20, βV8-22, γV8-24, and δV8-20, isolated from nAChRs labeled with 1.25 μM [3H]Azicholesterol in the presence of agonist, were exhaustively digested with S.aureus V8 protease (400% w/w) for six days. Aliquots of the total digests (∼10%) were fractionated by Tricine SDS-PAGE and then subjected to fluorography for 12 weeks. The migration of prestained molecular weight standards are indicated on the left. For each digest, there is a principal band of 3H which migrates with apparent molecular masses of: ∼ 8.5 kDa (α), 7 kDa (β), 7.5 kDa (γ), and 7.5 kDa (δ) (bracketed gel region). For the digest of αV8-20 there remains a substantial amount of undigested material and this band has approximately the same electrophoretic mobility as the 18 kDa standard.
Figure 8. Reversed-phase HPLC purification of [3H]Azicholesterol labeled fragments from V8 protease digests of αV8-20, βV8-22, γV8-24, and δV8-20.
The [3H]Azicholesterol labeled proteolytic fragments αV8-8.5 (A), βV8-7 (B), γV8-7.5 (C), and δV8-7.5 (D) isolated from exhaustive V8 protease digests of αV8-20, βV8-22, γV8-24, and δV8-20 (Fig. 7) were fractionated by reversed-phase HPLC as described in the Experimental Procedures and in the legend for Fig.5. The elution of peptides was monitored by absorbance at 210 nm (solid line) and elution of 3H by scintillation counting of aliquots (50 μL) of each 500 μL fraction (●). The point of elution of the M3 peptide based on previous work (and from sequencing results from this report) is indicated with an arrow. Based upon recovery of radioactivity, >90% of the material was recovered from the HPLC column.
Mapping [3H]Azicholesterol incorporation to the M1 segment of each nAChR subunit
Previous work has established that tryptic digestion of αV8-20 produces an ∼4-kDa tryptic peptide which contains the αM1 segment (12). Therefore the M1 segment of each nAChR subunit was isolated from tryptic digests of αV8-20, βV8-22, γV8-24, and δV8-20. The tryptic digests were resolved by Tricine SDS-PAGE and in each case the region of the gel between the 4- and 6-kDa molecular weight standards was excised. Also excised was the gel region containing an 8 kDa band that appeared prominently labeled in a fluorograph of an analytical Tricine SDS-PAGE gel containing a small aliquot of each digest (not shown). The material from each band was isolated and further purified by reversed-phase HPLC (Fig. 9). For the αV8-20 tryptic digest, HPLC fractions 39-41 were pooled, and sequence analysis revealed the presence of a peptide beginning at αIle-210 (30 pmol), the amino terminus of the αM1 segment as well as two secondary sequences (αAsp-180, 20 pmol; αMet-243,10 pmol). Based on a molecular mass of 4-6 kDa the αIle-210 peptide extends through the αM1 segment and likely terminates at αLys-242. For the other tryptic digests, HPLC fractions 37-40 were pooled and in each case sequence analysis revealed the presence of a primary peptide, although secondary sequences were present, which contains the M1 segment (the amino terminus of each peptide was: βLys-216 (211 pmol), γLys-218 (109 pmol), and δLys-224 (70 pmol)). The material from the 8 kDa band was also isolated and further purified by reversed-phase HPLC (data not shown). In each case a single peak of 3H-containing HPLC fractions was evident and sequence analysis of the pooled fractions (39-42) revealed the presence of a primary sequence present in at least a ten-fold greater abundance than any secondary sequences: αIle-210 (132 pmol), βLys-216 (32 pmol), γLys-218 (19 pmol), and δLys-224 (74 pmol). Based on a molecular mass of 8 kDa, each peptide extends through both the M1 and M2 segments. The extent of [3H]Azicholesterol incorporation into the M1 segments was too low to effectively allow for determination of individually labeled amino acids by radio-sequencing. It should be noted that in the absence of radio-sequencing, [3H]Azicholesterol incorporation into the M1 segments (as well as M3 segments) can only be tentatively assigned based on peptide mapping results. However, the results do suggest the presence of [3H]Azicholestrol incorporation into residues within the M1 segment of each nAChR subunit, a conclusion that is supported by previous photolabeling studies in which [125I]Azido-cholesterol labeling was mapped to the αM1 segment (18).
Figure 9. Reversed-phase HPLC purification of [3H]Azicholesterol labeled fragments from tryptic digests of αV8-20, βV8-22, γV8-24, and δV8-20.
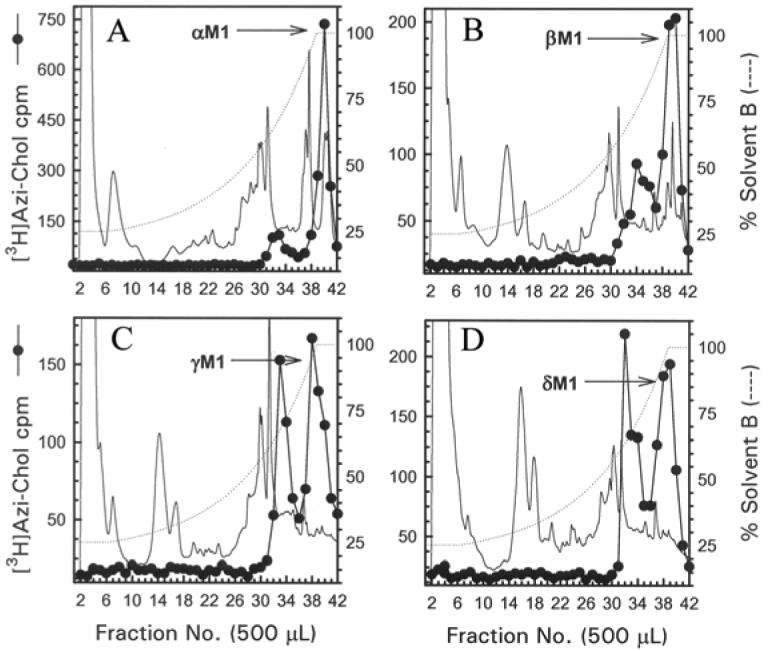
The [3H]Azicholesterol labeled tryptic peptides (∼ 4 kDa) isolated from exhaustive digests of αV8-20 (A), βV8-22 (B), γV8-24 (C), and δV8-20 (D) that were resolved by Tricine SDS-PAGE were further purified by reversed-phase HPLC as described in the Experimental Procedures and in the legend for Fig.5. The elution of peptides was monitored by absorbance at 210 nm (solid line) and elution of 3H by scintillation counting of aliquots (50 μL) of each 500 μL fraction (●). The point of elution of the M1 peptide based on previous work (and from sequencing results from this report) is indicated with an arrow. Based upon recovery of radioactivity, >90% of the material was recovered from the HPLC column.
DISCUSSION
The purpose of the work presented here was to probe the binding domain for membrane cholesterol in the nAChR using the photoactivatable sterol [3α-3H]6-Azi-5α-cholestan-3β-ol ([3H]Azicholesterol). [3H]Azicholesterol inserts efficiently into nAChR-enriched membranes (>98%), photoincorporates into nAChR subunits on an equal molar basis, and neither the pattern nor the extent of labeling is affected by the inclusion of the agonist carbamylcholine (Fig. 2). In addition, concentrations of nonradioactive Azicholesterol up to 100 μM had no effect on the high affinity binding of either [3H]TCP (in the presence of agonist) or [3H]tetracaine (in the absence of agonist) to the nAChR, indicating little or no direct interaction with the receptor channel (data not shown). Therefore, there is a single component of [3H]Azicholesterol labeling that is consistent with incorporation at the nAChR lipid-protein interface.
Sites of [3H]Azicholesterol labeling in M4 segments
Approximately three-quarters of the total [3H]Azicholesterol incorporation into each nAChR subunit resides within V8 fragments that contain the M4 segment (e.g. γV8-14, 74%), suggesting that the M4 segment has the greatest interaction with membrane cholesterol. Residues incorporating [3H]Azicholesterol in the M4 segments (Fig. 6) could in most cases only be tentatively assigned due the presence of more than one peptide during sequencing. In βM4, it is most likely that [3H]Azicholesterol reacted with the charged amino acids (βAsp-436 and βAsp-457) which are situated on either end of the βM4 transmembrane helix (Fig. 6 and 10). In γM4, sequencing revealed the presence of two primary peptides, both of which contain the γM4 segment (Fig. 6), and the observed 3H release pattern is consistent with [3H]Azicholesterol incorporation into γAsp-448. Like for βM4, γAsp-448 is a charged amino acids located at the amino-terminus of the γM4 helix. The pattern of [3H]Azicholesterol incorporation into charged residues is continued in δM4, the 3H release pattern (Fig. 6) is consistent with labeling of δAsp-454 which is located at the amino-terminus of the δM4 helix. For αM4, the 3H release pattern is consistent with [3H]Azicholesterol incorporation into αGlu-398, αAsp-407, and αCys-412. (Fig 6). These labeled residues are at the amino-terminus of αM4 (αGlu-398, αAsp-407) or contained within the αM4 segment (αCys-412; Fig. 10).
Figure 10. AziCholesterol Sites of Photoincorporation in the Torpedo nAChR Structure.
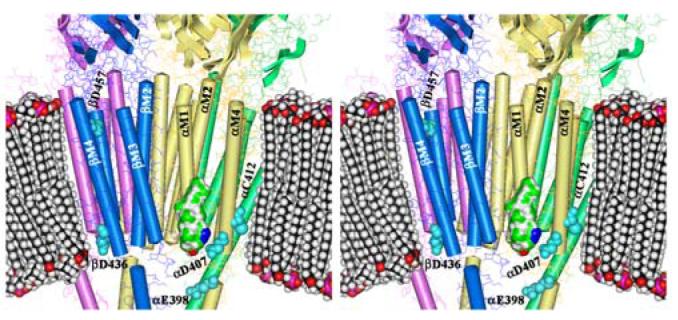
Shown is a stereo figure of the Torpedo nAChR cryo-electron microscopy structure (15; pdb #2BG9) focusing on the membrane spanning region. The α subunits are yellow, β is blue, γ is green, and δ is magenta. The pdb-designated α-helices and β-sheets are shown. A space-filling model of a phospholipid bilayer is included for scale as is a Connelly surface model of AziCholesterol with the Azide colored blue. Residues identified as labeled by [3H]AziCholesterol in the α, and β subunits are shown as cyan spaced-filled amino acids.
It is striking that the conserved Asp at the N-terminus of each M4 segment (αAsp-407, βAsp-436, γAsp-448, δAsp-454) is labeled, as well as the only acidic side chain at the C-terminus of the M4 segments (βAsp-457). One of the potential pitfalls of using [3H]Azicholesterol as a photoaffinity probe is that the photoactivatable alkyldiazirine group (Fig. 1) may undergo photoisomerization to yield diazo compounds which react preferentially with carboxylic acids (30), such as those found in the side chains of Asp and Glu residues, and could explain the observed labeling pattern in the M4 segments. However [3H]Azicholesterol incorporation into αCys-412 (and perhaps additional residues) is more consistent with the expected photoinduced reactive alkylcarbene. The latter is supported by recent studies demonstrating that alkyldiazirine containing photoaffinity probes exhibit a broad range of side chain reactivities (21,22) and that [3H]Azioctanol incorporates into αHis-408 and αCys-412 within the αM4 segment, but there is no reaction with αAsp-407(21). Therefore it remains a question as to whether the residues labeled by [3H]Azicholesterol in the M4 segments are driven more by proximity to the photoreactive ligand (Azicholesterol) or by diffusional encounters with generally reactive side chains. If the labeling pattern is driven more by proximity to the photoreactive sterol molecule this implies that [3H]Azicholesterol, and more specifically the B-ring of the sterol molecule, is generally positioned in the lipid bilayer near the charged amino acids (e.g. βAsp-436, βAsp-457) that flank each end of the M4 transmembrane helices. The charged amino acids at each end of the M4 helix are positioned proximal to the phospholipid headgroups within the lipid bilayer in the 4Å structure of the nAChR (14; Fig. 10). Positioning of the cholesterol B-ring (in particular position 6 of the sterol molecule) proximal to the phospholipid headgroups, places the sterol molecule at a more shallow depth in the lipid bilayer than expected (31). Further studies are needed to determine if the proposed positioning of cholesterol in the bilayer is a true reflection of its interaction with the M4 helix or rather that [3H]Azicholesterol labeling of the acidic residues at either end of the M4 helices is a reflection of the photoreactive properties of the alkyldiazirine and vertical fluctuations of cholesterol in the bilayer.
Sites of [3H]Azicholesterol labeling in M1 and M3 segments
Approximately one-quarter of the total [3H]Azicholesterol incorporation into each nAChR subunit resides within V8 fragments that contain the M1-M2-M3 segments (e.g. γV8-24, 26%). Further mapping suggests that there is [3H]Azicholesterol incorporation into peptides that contain either the M1 or M3 segments of each nAChR subunit. The extent of [3H]Azicholesterol incorporation into either M1 or M3 containing peptides was however too low to effectively allow the determination of individually labeled amino acids by radio-sequencing. Collectively, these results argue that while the M1, M3, and M4 segments are all in contact with [3H]Azicholesterol, the M1 and M3 segments interact with membrane cholesterol to a significantly lesser degree than M4.
Relevance to the nAChR lipid-protein interface and receptor function
The salient new finding in this study is that while the M4 segments of each receptor subunit may have the greatest interaction with membrane cholesterol, each of the transmembrane segments that contribute to the nAChR lipid-protein interface (M1,M3,M4) likely interact with cholesterol. That is the cholesterol binding domain fully overlaps the nAChR lipid-protein interface. An earlier study using a photoreactive analog of phosphatidylcholine ([125I]TIDPC/16) mapped the labeling to M1 and M4 segments of the nAChR (it was not determined if there was labeling in M3 segments, Ref. 26). Therefore further mapping (i.e. identify labeled amino acids) with both [3H]Azicholesterol and [125I]TIDPC/16 will be necessary to verify the existence of non-annular lipid binding sites at the nAChR lipid-protein interface, sites that bind cholesterol but not phospholipids (32). On the other hand the existence of non-annular vs. annular lipid binding sites may be a temporal rather than a static phenomenon. At the lipid-protein interface, lipids have a lateral diffusion coefficient 50-100 times slower than that of the fluid lipid bilayer and lipids that selectively interact with nAChR (e.g. cholesterol) spend on average a greater amount of time in proximity to the receptor (4). While the present work suggests that cholesterol interacts with the entire lipid-protein interface, on a dynamic time-scale cholesterol may interact preferentially with a sub-region of the lipid-protein interface. Along these same lines it is worth noting that [3H]Azicholesterol was added to Torpedo nAChR-enriched membranes that already contain ∼35% cholesterol on a molar basis (6), which may limit contact with some cholesterol sites that are slowly exchanging.
The molecular interactions that underlie the effect of cholesterol on nAChR function are not understood. Cholesterol has no detectable effect on the overall secondary structure composition of the receptor protein (9) and yet nAChR state transitions are preserved in the presence of a single shell of lipids (∼45 lipids) surrounding the receptor protein (10) and amino acid substitutions at residues that contact lipid have significant effects on channel gating (reviewed in 4). These results suggest that direct interactions between cholesterol and the nAChR lipid-protein interface exert subtle yet critical structural alterations that are necessary for receptor functionality. In a recent analysis of the effects of amino acid substitutions in the αM4 segment on channel gating, the authors concluded that the αM4 helix undergoes a rigid body motion during channel opening (11). The details of this rigid body motion are unclear, particularly given that the surface of the αM4 helix that is exposed to lipid/sterol does not appear to change during channel activation (12,33). Any proposed model of how cholesterol interacts with the nAChR lipid-protein interface to facilitate gating-induced movements of the transmembrane segments must account for these seemingly conflicting results.
1 Abbreviations
- nAChR
nicotinic acetylcholine receptor
- [3H]Azicholesterol
[3α-3H]6-Azi-5α-cholestan-3β-ol
- Carb
carbamylcholine
- MOPS
4-Morpholinopropanesulfonic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- HPLC
high-performance liquid chromatography
- TFA
trifluoroacetic acid
- PTH
phenylthiohydantoin
- Tricine
N-tris(hydroxymethyl) methylglycine
- VDB
vesicle dialysis buffer
- V8 protease
S. aureus glutamyl endopeptidase
- VDAC
voltage-dependent anion channel
Footnotes
This research was supported in part by National Institutes of Health Grants R29-NS35786 (M.P.B.), NS19522 and GM58448 (JBC), and by an Intramural Grant from Texas Tech University Health Sciences Center School of Medicine (M.P.B.).
As discussed in Methods, we feel that the additional 3H release seen in cycles 12 and 21, following the major releases in cycles 11 and 20, probably contains contributions from the inefficient transfer to the fraction collector of the hydrophobic, [3H]Azicholesterol-labeled PTH amino acids cleaved at cycles 11 and 20.
Association of the 3H release in cycle 20 with αAsp-407 in the fragment beginning at αSer-388 is not unambiguous, because the 3H release in cycle 20 is substantially greater than that in cycle 7 that would correspond to αAsp-407 in the primary sequence beginning at αTyr-401 peptide. Such a discrepancy is not unexpected, however, if the incorporation of the hydrophobic [3H]Azicholesterol shifts subtly the HPLC elution profile of the labeled fragments relative to the unlabeled.
REFERENCES
- 1.Corringer PJ, Le Novere N, Changeux J-P. Nicotinic receptors at the amino acid level. Annu. Rev. Pharmacol. Toxicol. 2000;40:431–458. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 2.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature Rev. Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 3.daCosta CJB, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE. Lipid-protein interactions at the nicotinic acetylcholine receptor. J. Biol. Chem. 2002;277:201–208. doi: 10.1074/jbc.M108341200. [DOI] [PubMed] [Google Scholar]
- 4.Barrantes FJ. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Research Reviews. 2004;47:71–95. doi: 10.1016/j.brainresrev.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Hamouda AK, Sauls D, Vardanyan N, Sanghvi M, Blanton MP. Assessing the lipid requirements of the nicotinic acetylcholine receptor. Biophys. J. 2005;88:624a. doi: 10.1021/bi052281z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantipragada SB, Horvath LI, Arias HR, Schwarzmann G, Sandhoff K, Barrantes FJ, Marsh D. Lipid-protein interactions and effect of local anesthetics in acetylcholine receptor-rich membranes from Torpedo marmorata electric organ. Biochemistry. 2003;42:9167–9175. doi: 10.1021/bi034485q. [DOI] [PubMed] [Google Scholar]
- 7.Marsh D, Barrantes FJ. Immobilized lipid in acetylcholine receptor-rich membranes from Torpedo marmorata. Proc. Natl. Acad. Sci. U.S.A. 1978;75:4329–4333. doi: 10.1073/pnas.75.9.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellena JF, Blazing MA, McNamee MG. Biochemistry. 1983;22:5523–5535. doi: 10.1021/bi00293a012. [DOI] [PubMed] [Google Scholar]
- 9.Methot N, Demers CN, Baenziger JE. Structure of both the ligand- and lipid-dependent channel-inactive states of the nicotinic acetylcholine receptor probed by FTIR spectroscopy and hydrogen exchange. Biochemistry. 1995;34:15142–15149. doi: 10.1021/bi00046a021. [DOI] [PubMed] [Google Scholar]
- 10.Jones OT, Eubanks JH, Earnest JP, McNamee MG. A minimum number of lipids are required to support the functional properties of the nicotinic acetylcholine receptor. Biochemistry. 1988;27:3733–3742. doi: 10.1021/bi00410a032. [DOI] [PubMed] [Google Scholar]
- 11.Mitra A, Bailey TD, Auerbach AL. Structural dynamics of the M4 transmembrane segment during acetylcholine receptor gating. Structure. 2004;12:1909–1918. doi: 10.1016/j.str.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 13.Blanton MP, Dangott LJ, Raja SK, Lala AK, Cohen JB. Probing the structure of the nicotinic acetylcholine receptor ion channel with the uncharged photoactivatable compound [3H]diazofluorene. J. Biol. Chem. 1998;273:8659–8668. doi: 10.1074/jbc.273.15.8659. [DOI] [PubMed] [Google Scholar]
- 14.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 15.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J. Mol. Biol. 2004;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Middlemas DS, Raftery MA. Identification of subunits of acetylcholine receptor that interact with a cholesterol photoaffinity probe. Biochemistry. 1987;26:1219–1223. doi: 10.1021/bi00379a003. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez AM, Fernandez-Ballester G, Ferragut JA, Gonzalez-Ros JM. Labeling of the nicotinic acetylcholine receptor by a photoactivatable steroid probe:effects of cholesterol and cholinergic ligands. Biochim. Biophys. Acta. 1993;1149:135–144. doi: 10.1016/0005-2736(93)90034-w. [DOI] [PubMed] [Google Scholar]
- 18.Corbin J, Wang HH, Blanton MP. Identifying the cholesterol binding domain in the nicotinic acetylcholine receptor with [125I]azido-cholesterol. Biochim. Biophys. Acta. 1998;1414:65–74. doi: 10.1016/s0005-2736(98)00153-9. [DOI] [PubMed] [Google Scholar]
- 19.Blanton MP, Xie Y, Dangott LJ, Cohen JB. The steroid promegestone is a noncompetitive antagonist of the Torpedo nicotinic acetylcholine receptor that interacts with the lipid-protein interface. Mol. Pharmacol. 1999;55:269–278. doi: 10.1124/mol.55.2.269. [DOI] [PubMed] [Google Scholar]
- 20.Mintzer EA, Waarts B-L, Wilschut J, Bittman R. Behavior of a photoactivatable analog of cholesterol, 6-photocholesterol, in model membranes. FEBS Lett. 2002;510:181–184. doi: 10.1016/s0014-5793(01)03271-9. [DOI] [PubMed] [Google Scholar]
- 21.Pratt MB, Husain SS, Miller KW, Cohen JB. Identification of sites of incorporation in the nicotinic acetylcholine receptor of a photoactivatable general anesthetic. J. Biol. Chem. 2000;275:29441–29451. doi: 10.1074/jbc.M004710200. [DOI] [PubMed] [Google Scholar]
- 22.Ziebell MR, Nirthanan S, Husain SS, Miller KW, Cohen JB. Identification of binding sites in the nicotinic acetylcholine receptor for [3H]Azietomidate, a photoactivatable general anesthetic. J. Biol. Chem. 2004;279:17640–17649. doi: 10.1074/jbc.M313886200. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen SE, Dreyer EB, Cohen JB. Location of ligand-binding sites on the nicotinic acetylcholine receptor alpha-subunit. J. Biol. Chem. 1986;261:13735–13743. [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 26.Blanton MP, McCardy EA, Huggins A, Parikh D. Probing the structure of the nicotinic acetylcholine receptor with the hydrophobic photoreactive probes [125I]TID-BE and [125I]TIDPC/16. Biochemistry. 1998;37:14545–14555. doi: 10.1021/bi981435q. [DOI] [PubMed] [Google Scholar]
- 27.Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 28.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Blanton MP, Lala AK, Cohen JB. Identification and characterization of membrane-associated polypeptides in Torpedo nicotinic receptor-rich membranes by hydrophobic photolabeling. Biochim. Biophys. Acta. 2001;78091:1–10. doi: 10.1016/s0005-2736(01)00321-2. [DOI] [PubMed] [Google Scholar]
- 30.Morgan S, Jackson JE, Platz MS. Laser flash photolysis study of adamantanylidene. J. Am. Chem. Soc. 1991;113:2782–2783. [Google Scholar]
- 31.Kessel A, Ben-Tal N, May S. Interactions of cholesterol with lipid bilayers: the preferred configuration and fluctuations. Biophys. J. 2001;81:643–658. doi: 10.1016/S0006-3495(01)75729-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones OT, McNamee MG. Annular and nonannular binding sites for cholesterol associated with the nicotinic acetylcholine receptor. Biochemistry. 1988;27:2364–2374. doi: 10.1021/bi00407a018. [DOI] [PubMed] [Google Scholar]
- 33.Leite JF, Blanton MP, Shahgholi M, Dougherty DA, Lester HA. Conformation-dependent hydrophobic photolabeling of the nicotinic receptor: electrophysiology-coordinated photochemistry and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13054–13059. doi: 10.1073/pnas.2133028100. [DOI] [PMC free article] [PubMed] [Google Scholar]