Abstract
Noncovalent protein-ligand complexes are readily detected by electrospray ionization mass spectrometry (ESI-MS). Ligand binding stoichiometry can be determined easily by the ESI-MS method. The ability to detect noncovalent protein-ligand complexes depends, however, on the stability of the complexes in the gas phase environment. Solution binding affinities may or may not be accurate predictors of their stability in vacuo. Complexes composed of cytidine nucleotides bound to ribonuclease A (RNase A) and ribonuclease S (RNase S) were detected by ESI-MS and were further analyzed by MS/MS. RNase A and RNase S share similar structures and biological activity. Subtilisin-cleavage of RNase A yields an S-peptide and an S-protein; the S-peptide and S-protein interact through hydrophobic interactions with a solution binding constant in the nanomolar range to generate an active RNase S. Cytidine nucleotides bind to the ribonucleases through electrostatic interactions with a solution binding constant in the micromolar range. Collisionally activated dissociation (CAD) of the 1:1 RNase A-CDP and CTP complexes yields cleavage of the covalent phosphate bonds of the nucleotide ligands, releasing CMP from the complex. CAD of the RNase S-CDP and CTP complexes dissociates the S-peptide from the remaining S-protein/nucleotide complex; further dissociation of the S-protein/nucleotide complex fragments a covalent phosphate bond of the nucleotide with subsequent release of CMP. Despite a solution binding constant favoring the S-protein/S-peptide complex, CDP/CTP remains electrostatically bound to the S-protein in the gas phase dissociation experiment. This study highlights the intrinsic stability of electrostatic interactions in the gas phase and the significant differences in solution and gas phase stabilities of noncovalent complexes that can result.
Keywords: electrospray ionization, noncovalent complexes, electrostatic interactions, protein-ligand binding, ribonuclease
Introduction
The biological function of many macromolecules often requires the binding of small molecules and ions. For proteins in general, such binding involves the formation of noncovalent bonds between the small molecule or ion (i.e., ligand) and protein at a specific region or surface of the protein. Most biological macromolecules possess binding sites of varying degrees of strength and specificity for a variety of ligands. The binding of ligands could be a complex process, as the binding event may induce a conformational change in the structure of the protein. Mass spectrometry (MS), especially with electrospray ionization (ESI), has been applied for the analysis of protein complexes involving noncovalent interactions with a variety of partners (e.g., other peptides and proteins, small molecule ligands, metal ions, oligonucleotides, etc) since the early 1990s [1–4], and especially during the past few years [5–10]. Very large protein assemblies, such as the 700 kDa 20S proteasome [11–13], the 2 MDa ribosome, and other large megaDalton complexes [8, 10, 14, 15] are sufficiently stable for ESI mass spectrometry detection. The molecular weight range for the detection of protein complexes can be extended by coupling ESI with ion mobility analysis, for example, for the detection of MDa-sized viral particles and other large assemblies greater than 10 MDa [16–18].
As ESI-MS is being applied for the characterization of noncovalent protein complexes, a better understanding of the relative stability of protein-ligand complexes both in solution and in the gas phase becomes very important for the interpretation of such measurements. In general, the sites of protein ligand binding can be identified or inferred by a number of biophysical methods, such as using hydrogen-deuterium exchange coupled with MS detection, high resolution x-ray crystallography and NMR, chemical and photo cross-linking, and other techniques. The direct detection of protein-ligand noncovalent complexes using ESI mass spectrometry can be challenging, depending on the types of compounds tested and the types of physical interactions necessary to maintain a stable complex in both solution and in the gas phase.
In principle, one can envision that an ESI-MS-based assay could be developed to assess the binding capabilities of small molecule compounds (of less than 1000 Da) to a targeted protein. In fact, with moderate-to-high resolving mass analyzers, such as a time-of-flight (TOF), Fourier-transform ion cyclotron resonance (FT-ICR) system, or an OrbiTrap, throughput and capacity could be greatly increased by multiplexing either the compounds in the form of compound mixtures or libraries and/or the protein targets for known protein and compound entities. Pharmaceutical and biotechnology organizations could apply such a moderate throughput assay for more targeted compound screens to support structure-activity relationship (SAR) studies. The technology for such an instrument platform is readily available with automated liquid handling robots and nanoESI-chip devices for analyte introduction and ionization. One should be able to acquire a mass spectrum for the protein-ligand complex every 30 sec or less; compound concentrations can be varied to construct a titration curve for solution phase equilibrium binding constant determinations. Previous literature suggests that the concept is feasible [19]. However, it has not been put into large-scale practice for a variety of reasons. In our opinion, one of the primary reasons is that the avidity of the ligand in the gas phase may not mirror its behavior in solution.
Removal of water from a protein (i.e., dehydration) has unknown consequences on its structural integrity and its interactions with ligands and other macromolecules [20]. The fact that many noncovalently-bound protein complexes can be observed intact suggests that the removal of water itself may not cause dissociation of the protein complex in vacuo. Potentially, there are competing factors that can stabilize and destabilize protein interactions in the gaseous state. The electrostatic potential between two point charges in vacuum is given by Coulomb’s law. In homogeneous environments, the electrostatic interaction is inversely proportional to the dielectric constant of the medium; the dielectric constant of water is 78.5 (25 °C), and values for the dielectric constant for the interior of a protein range from 1–20. Removing water may serve to increase the electrostatic interaction between two oppositely charged molecules, but the degree of this increase is unclear [21]. The stability of some macromolecules and protein complexes may derive in large part from hydrophobic bonding. It is unlikely that hydrophobic interactions persist in the vacuum state. Indeed, it has been suggested by several studies that protein-protein and protein-ligand complexes driven by hydrophobic interactions are sufficiently unstable in the gas phase to preclude their detection by ESI-MS [7, 22]. However, there are limited reports of successful detection of such complexes [3, 23], including lipid-peptide complexes by Cole’s group [24]. Thus, the stability of the gas phase protein-ligand complex may not always mirror precisely its stability in the solvated state [7, 25].
In this report, we highlight the differences between electrostatic and hydrophobic interactions by our ESI-MS study of ribonuclease A (RNase A) and ribonuclease S (RNase S). RNase A has been a primary object of landmark work on the folding, stability, and chemistry of proteins, and in enzymology [26]. RNase A catalyzes the cleavage of the P–O 5′-bond of single-stranded ribonucleic acids on the 3′-side of cytidine and uridine residues. This cleavage reaction requires the 2′-hydroxyl group of RNA; RNase A does not hydrolyze DNA. Protein-nucleotide complexes have been detected using ESI-MS [14, 27–30]. More specifically, the complexation of mono- and oligonucleotide ligands with ribonuclease A has been measured using ESI-MS by many laboratories [14, 19, 31–33].
RNase S is formed by the cleavage of a single bond in native RNase A by the protease, subtilisin [34]. The product of subtilisin cleavage generally generates two tightly associated fragments, S-peptide (residues 1–20 of RNase A) and S-protein (residues 21–124), that are held together largely through hydrophobic interactions. Although neither fragment alone has any ribonucleolytic activity, RNase S has enzymatic activity similar to that of RNase A. In general, nucleotide-binding proteins have important biological function in a number of cellular processes, as nucleotides are co-factors or substrates for enzymes, regulators of protein function, or act in concert with structural binding motifs.
From our studies, we show that the stability of gas phase noncovalent protein-ligand complexes may not reflect their solution affinities for interactions with a significant electrostatic component. Specifically, the gas phase intermolecular interactions of the S-peptide/S-protein complex are significantly weaker than the cytidine nucleotide-RNase S complex, even though the interactions in solution are stronger for the S-peptide/S-protein complex. We find that protein-nucleotide binding can be nearly covalent, or “possesses a covalent-like stability” in the gas phase [35].
Experimental
Positive ion electrospray ionization mass spectra were acquired with a QqTOF mass spectrometer (Applied Biosystems/Sciex QSTAR Pulsar XL, Concord, ON, Canada). A nanoESI source (Proxeon Biosystems, Odense, Denmark) using borosilicate glass capillaries with Au/Pd coatings (Proxeon) was operated at low analyte flow conditions (50 nL/min). A metal sleeve used to restrict pumping in the initial ca. 62-mm region of the 20-cm long Q0-focusing quadrupole was used to allow for improved trapping and transmission of the noncovalent complexes [12, 13, 17].
For single MS measurements, the Q1 mass filter is operated in the radiofrequency (r.f.)-only mode, while the TOF analyzer is used to record mass spectra. For MS/MS, Q1 is operated as a mass filter to transmit only the selected precursor ion, and the precursor ion is then accelerated before it enters the collision cell (Q2), where it undergoes CAD with neutral argon molecules (4 mTorr). The products are focused and transmitted to the TOF analyzer. (An excellent account of the operating principles of the QqTOF is found in Chernushevish et al [36].) The declustering potential is increased to induce ion fragmentation in the orifice-skimmer region of the source area. The declustering potential is the difference between the orifice voltage and the skimmer voltage.
Bovine pancreatic RNase A, subtilisin-modified ribonuclease A (RNase S), cytidine 5′-triphosphate (CTP), cytidine 5′-diphosphate (CDP), cytidine 2′-monophosphate (2′-CMP), and cytidine 3′-monophosphate (3′-CMP) were purchased from Sigma-Aldrich (St. Louis, MO, USA). All protein samples were desalted with 10 mM ammonium acetate prior to analysis using centrifugal filter devices (10,000 molecular weight cutoff, Microcon and Amicon Ultra, Millipore Corporation, Billerica, MA, USA). The final protein concentration for ESI-MS measurements was 5 μM in 10 mM ammonium acetate, pH 6.6 unless noted otherwise.
Matrix-assisted laser desorption/ionization (MALDI)-MS experiments were performed with a Voyager DE-STR TOF analyzer (Applied Biosystems, Foster City, CA USA). Sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid; Sigma-Aldrich) was used as the matrix.
Results and Discussion
ESI-MS and MS/MS of Ribonuclease A Binding to Cytidine Nucleotide Ligands
Consistent with previous ESI-MS studies for the complexation of proteins to small nucleic acids [14, 19, 27–33], the noncovalent binding of RNase A to cytidine nucleotides was readily observed. Zhang et al previously measured using automated ESI-MS a solution equilibrium dissociation constant (Kd) for the RNase A association with 2′-CMP and CTP to be approximately 1–2 μM [19].
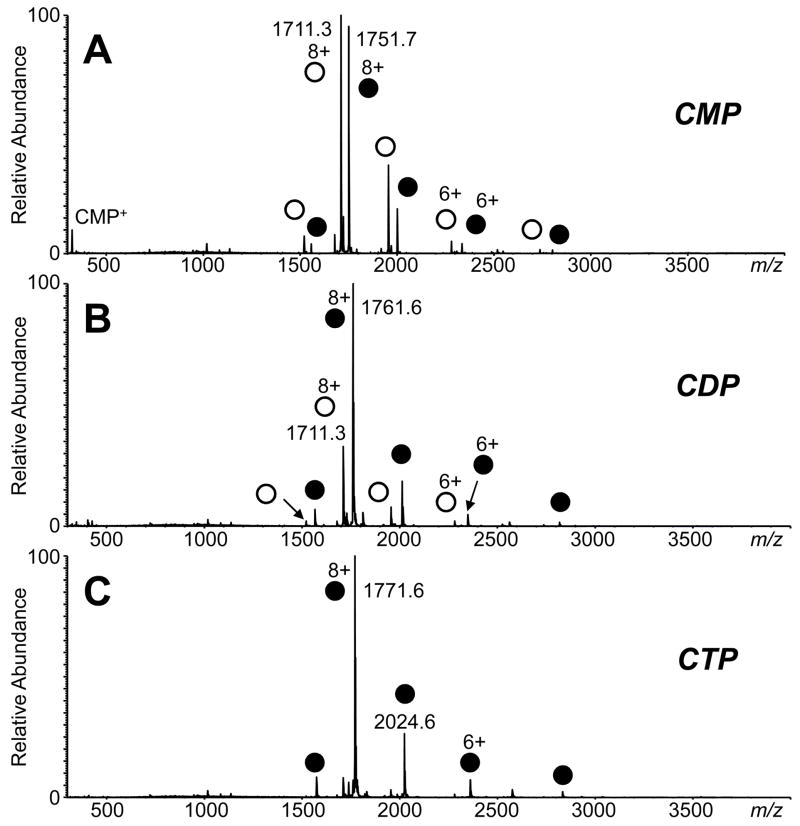
Figure 1 shows the ESI mass spectra of RNase A (13682 Da) complexed to 2′-CMP (323 Da), CDP (403 Da), and CTP (483 Da) in pH 6.6 solution. A protein-ligand stoichiometry of 1:1 was observed for each of the nucleotide ligands. The charge state of the most abundant protein-ligand complex molecule was 8+ for each of the ligands studied. (The 8+ charge state is the most abundant species for RNase A alone at pH 6.6 (data not shown).) However, the proportion of the formation of the holo-complex increased with increasing number of phosphate groups (for the same 1:10 protein-ligand concentration ratio). The measured holo-apo ratio was 0.8, 2.9, and 11.6 for 2′-CMP, CDP, and CTP, respectively. A small amount of the RNase A-ligand dimer complex (less than 3% of the total protein signal) was observed for CDP and CTP as well. Using a low declustering potential of +60 V in the atmospheric pressure-vacuum interface, little dissociation of the complexes were observed. A small amount of dissociation may have occurred for 2′-CMP binding to RNase A, as an ion at m/z 324 for liberated 2′-CMP was observed (Figure 1A).
Figure 1.
Positive ion ESI mass spectra of RNase A with (A) 2′-CMP, (B) CDP, (C) CTP at a 1:10 protein-ligand concentration ratio in 10 mM ammonium acetate, pH 6.6. A declustering potential of +60 V in the atmospheric pressure-vacuum interface was used. Peaks representing the nucleotide-bound holo-form of the protein are labeled with the filled circles, and the peaks for the apo-protein are labeled with open circles. (Free, unbound 2′-CMP was detected at m/z 324 in Figure A.)
Consistent with previous studies of the stability of protein-DNA ionic complexes [37, 38], increasing salt concentration reduces the relative abundance of the RNase A-nucleotide complexes detected by ESI-MS. Increasing the ammonium acetate concentration from 10 mM to 100 mM reduced the holo/apo ratio to 0.3, 0.9, and 2.3 for 2′-CMP, CDP, and CTP binding, respectively. Similarly, increasing the ammonium acetate concentration further to 200 mM further reduced the holo/apo ratio to 0.1, 0.1, and 0.3.
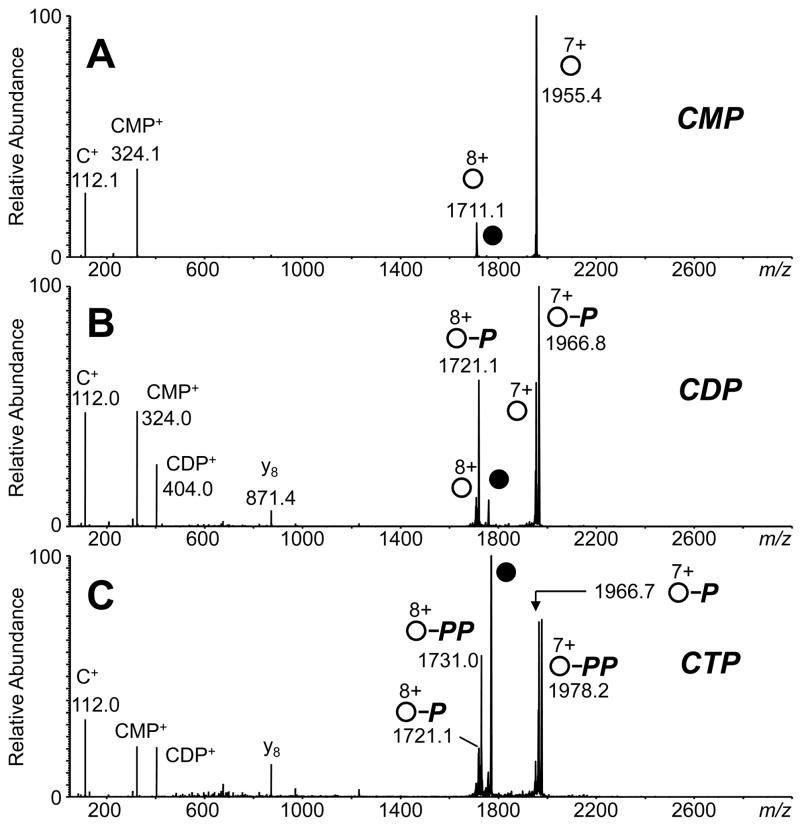
To investigate the stability of the protein-ligand complexes, collisionally activated dissociation (CAD) mass spectra were recorded for each of the multiply charged complexes. Figure 2 shows the MS/MS spectra of the 8+-charged RNase A complex with 2′-CMP, CDP, and CTP. Dissociation of the (M + 8H + CMP)8+ complex (where “M” is the RNase A protein) yields the complementary (M + 7H)7+ and (CMP + H)+ product ion pair. In addition, the (M + 8H)8+ apo-protein with concurrent loss of neutral 2′-CMP was observed as well (Figure 2A). MS/MS of the complex with 3′-CMP shows similar behavior (data not shown). For most weakly-bound noncovalent protein complex, regardless of their solution binding constants, CAD results in ready dissociation of the protein-binding partner from the complex.
Figure 2.
CAD mass spectra of the 8+-charged complex between RNase A and (A) 2′-CMP (40 V collision energy, CE), (B) CDP (CE = 50 V), and (C) CTP (CE = 50 V) in 10 mM ammonium acetate, pH 6.6. The 8+-charged precursor ions are labeled with filled circles. Product ions for the apo-protein are labeled with open circles. Retention of one phosphate group by the product ion is represented by “P”, and retention of two phosphates is labeled with “PP.”
The CAD behavior of RNase A binding to CDP and CTP, however, is substantially different than for 2′-CMP binding. Dissociation of the (M + 8H + CDP)8+ complex yields the complementary (M + 7H)7+ (m/z 1955) and (CDP + H)+ (m/z 404) product ion pair as one dissociation pathway, and a minor pathway yielding the (M + 8H)8+ protein with loss of neutral CDP (Figure 2B). (The y8 product ion from dissociation of the N-terminal bond to Pro-117 is readily observed in CAD mass spectra of RNase A [39].) However, the predominant route is interpreted as the dissociation of a covalent phosphate bond within the diphosphate group of CDP and the subsequent release of charged CMP. Assuming that the phosphate group is the primary contact site for attachment to RNase A, as it is in x-ray crystallographic data for oligonucleotide binding to the enzyme [26, 40], the terminal phosphate group remains bound to the protein and the leaving group is a molecule of CMP. An ion at m/z 324 is the (CMP + H)+, and its complement is the (M + 7H + phosphate)7+ (m/z 1967). Loss of neutral CMP and formation of the (M + 8H + phosphate)8+ (m/z 1721) is observed as well.
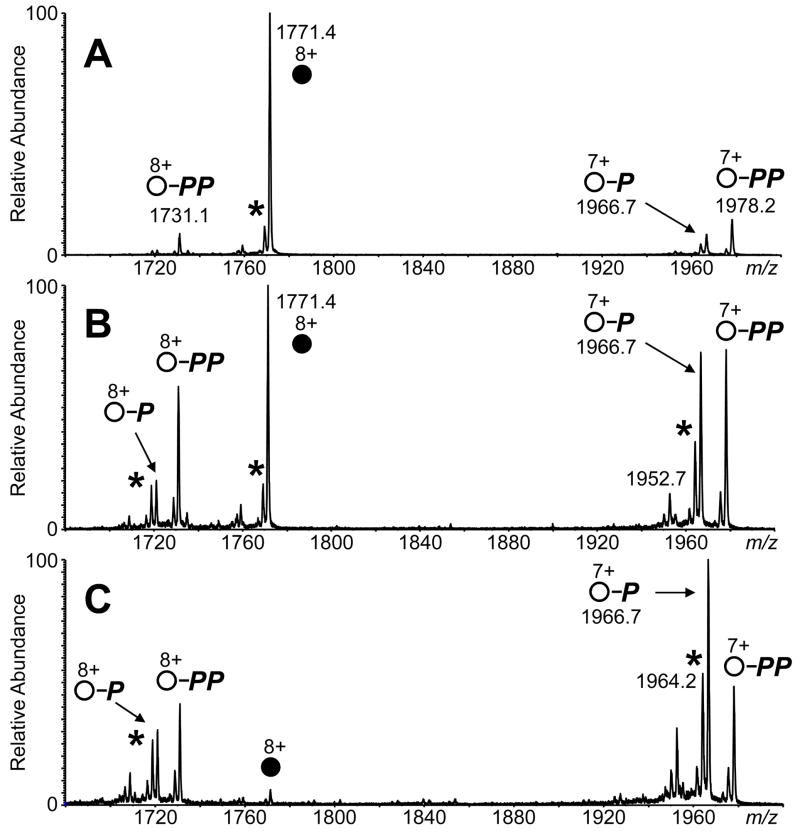
Dissociation of the (M + 8H + CTP)8+ complex yields predominantly products from the cleavage of the terminal phosphate group(s) of CTP, i.e., loss of intact CTP is not observed. Both (CMP + H)+ and (CDP + H)+ leaving groups were observed at m/z 324 and 404, respectively. One and two phosphate binding to RNase A (for both 7+ and 8+-charged products) was measured (Figure 2C and Scheme 1). Loss of H2O from the 7+ and 8+-charged products is observed also. Increasing the collision energy results in the increase in relative abundance of the protein bound to one phosphate group and the protonated CDP molecule (Figure 3); conceivably this increase of the abundance of the one phosphate-bound protein could result from secondary dissociation of the two phosphate-bound protein with increasing collision energies.
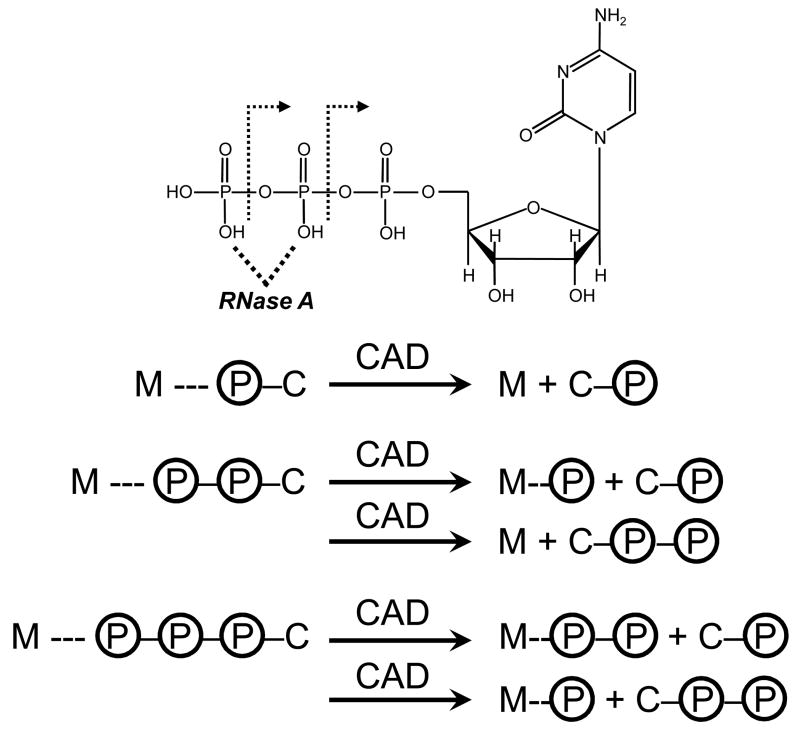
Scheme 1.
Products from the CAD of RNase A (M) bound to CMP, CDP, and CTP. The structure of CTP and the P-O bonds cleaved upon CAD are shown (top).
Figure 3.
MS/MS spectra of the (M + 8H + CTP)8+ complex (M = RNase A) at (A) 45 V, (B) 50 V, and (C) 55 V collision energies. The 8+-charged precursor ions are labeled with filled circles. Product ions for the apo-protein are labeled with open circles. Retention of one phosphate group by the product ion is represented by “P”, and retention of two phosphates is labeled with “PP.” Peaks for the loss of water are labeled with an asterisk symbol (*).
The interaction between the cytidine nucleotides with RNase A is largely through ionic, noncovalent forces [33, 41]. ESI mass spectra of RNase A with CDP and CTP in acidic pH (less than 4) and with 50% (v/v) methanol or acetonitrile shows the apo-protein and the free nucleotide ligand as the major components. Likewise, MALDI-TOF mass spectra of RNase A incubated with CTP in 10 mM ammonium acetate, pH 6.6 for over 1-hr and spotted onto a MALDI plate with sinapinic acid matrix and 0.1% trifluoroacetic acid shows only the apo-protein, i.e., transfer of a phosphate group to RNase A does not result in a covalent product from a solution reaction. In vacuo, the electrostatic interaction of the positively charged, desolvated protein and the negatively charged ligand is greatly strengthened compared to its properties in solution. The strength of charge-charge interactions is approximately inversely proportional to the dielectric constant of the medium. Removal of solvent (i.e., water) replaces the dielectric term to that of either vacuum (dielectric constant of one) or of the protein itself (estimated to be approximately 3–4), resulting in an increase in the electrostatic forces.
The increased avidity of noncovalent complexes driven by charge-charge interactions has been observed by others [28, 32, 35, 42–47]. Removal of water results in a near-covalent strength interaction between the phosphate groups of CDP and CTP to RNase A. It is not clear why CAD of the protein bound to the monophosphate nucleotide CMP does not result in transfer of a phosphate group to RNase A. Zhang et al measured the solution binding constants for RNase A and 2′-CMP and CTP to be 2.0 μM and 0.7 μM, respectively [19]. Whether the 2–3 fold higher binding affinity for CTP-binding than for 2′-CMP-binding can be rationalized by the additional diphosphate and/or the different attachment position of the phosphate group to ribose (5′ versus 2′) is unclear.
We performed separate CAD experiments for CMP, CDP, and CTP to investigate the relative stabilities of their P-O bonds. Cleavage of a P-O bond resulting in the loss of one and two phosphate groups is readily observed for CTP, and the loss of one phosphate group is observed for CDP. MS/MS of CMP results primarily in loss of the cytidine base. Thus, the general intrinsic stabilities of the P-O bonds for the cytidylic acid ligands appear to be carried forward into the RNase A-cytidylic acid complexes.
ESI-MS and MS/MS of Ribonuclease S-Nucleotide Complexes
RNase A treated with subtilisin yields a modified RNase A, RNase S. RNase S has nearly the same enzymatic activity as RNase A. Subtilisin cleaves RNase A between Ala-20 and Ser-21 to generate the S-peptide (residues 1–20, 2166 Da) and the S-protein (residues 21–124, 11534 Da). The S-peptide is necessary for maintaining enzymatic activity, as His-12 has a fundamental role in its catalytic activity [3, 23, 26]. S-peptide and S-protein interact mostly through hydrophobic interactions with a solution equilibrium dissociation constant of approximately 1 nM to form active RNase S [26].
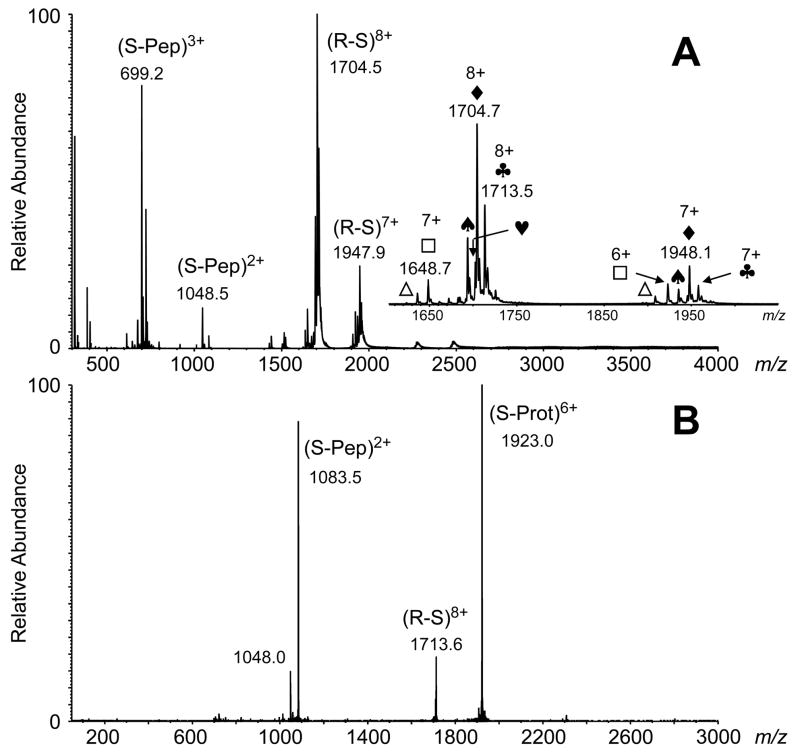
The direct detection of the noncovalent RNase S complex by ESI-MS has been reported by us some 15-years prior [3, 23], but to our knowledge it has not been reported by any laboratory since. Although the solution binding constant may suggest a stable complex, the gas phase S-peptide/S-protein complex is extremely fragile, primarily because of the hydrophobic forces that maintain the complex. Removal of water should diminish greatly the avidity of the complex in the gas phase. However, similar to the prototype, in-house built single quadrupole mass spectrometer used to measure the RNase S complex originally [3, 23], the commercial QqTOF instrument used for the present study was modified to increase the local pressure in the initial ion transfer stage. Improvements in sensitivity for large noncovalently-bound protein complexes appear to derive from improved cooling and focusing in Q0 produced by raising the pressure in this region [12, 13]. A metal sleeve (now called IonCooler™ for the Applied Biosystems QSTAR instruments) that is fitted around the initial ion entrance region of Q0 increases the local pressure to greater than 30 mTorr and greatly improves detection of weakly-bound complexes. This is demonstrated in Figure 4A, showing primarily the detection of intact RNase S complex with 7+ and 8+ charges. Without the metal sleeve to increase the pressure in the Q0 region, ions for the intact RNase S measured to less than 30% relative abundance of the dissociated S-protein and S-peptide (data not shown). CAD of the 8+-charged complex results in its dissociation into the expected S-protein (6+-charged) and S-peptide (2+-charged) complementary products, exclusively (Figure 4B).
Figure 4.
(A) ESI mass spectra of RNase S (R-S, 5 M in 10 mM ammonium acetate, pH 6.6). A declustering potential of +0 V in the atmospheric pressure-vacuum interface was used. The commercial sample of RNase S is a mixture of two S-peptide (S-Pep) components, residues 1–19 and 1–20 (2095 Da and 2166 Da, respectively), and two S-protein (S-Prot) polypeptides encompassing residues 21–124 and 22–124 (11534 Da and 11447 Da), consistent with our previous reports [3, 23]. The inset shows an expanded view of the RNase S complex peaks [◆, (1–19)/(21–124) complex; ♣ (1–20)/(21–124) complex; ♠ (1–19)/(22–124) complex; ♥ (1–20)/(22–124) complex; □ S-protein 21–124; △ S-protein 22–124]. (B) ESI-MS/MS of the 8+-charged RNase S complex (m/z 1713.5; CE = 10 V), generating the S-peptide (residues 1–20) and S-protein (residues 21–124) product pair.
As RNase S has similar enzymatic activity to RNase A in solution, it would be expected that RNase S should interact similarly to cytidine nucleotides. Using a concentration ratio of 1:10 protein:nucleotide in solution, the cytidine nucleotides was observed to form a 1:1 complex with intact RNase S. However, as observed for RNase A (Figure 1), CDP- and CTP-binding promote the formation of the 1:1 complex more readily than 2′-CMP-binding (data not shown). The measured holo-apo ratio was 1.3, 3.8, and 4.7 for 2′-CMP, CDP, and CTP, respectively. The full S-peptide/S-protein complex is required for enzymatic activity, and also for cytidine nucleotide binding. HPLC-separated and purified S-peptide and S-protein were reconstituted separately with 10-fold excess of CTP (pH 6.6) and measured by ESI-MS. No detectable CTP-binding was observed for the S-peptide. A holo-apo ratio of only 0.1 was measured for S-protein binding to CTP, significantly less than for the full RNase S complex.
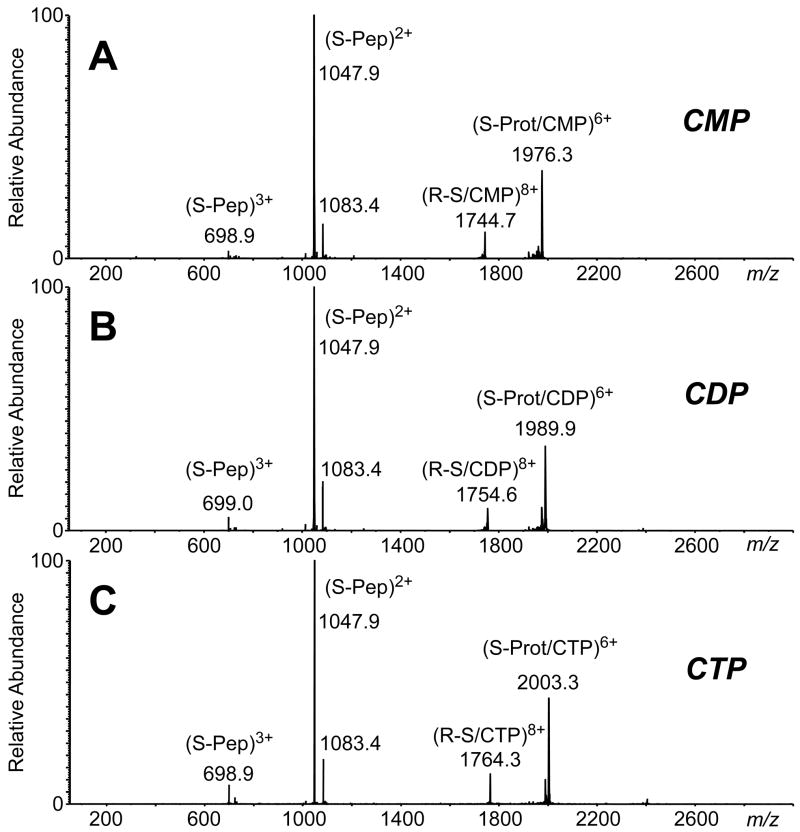
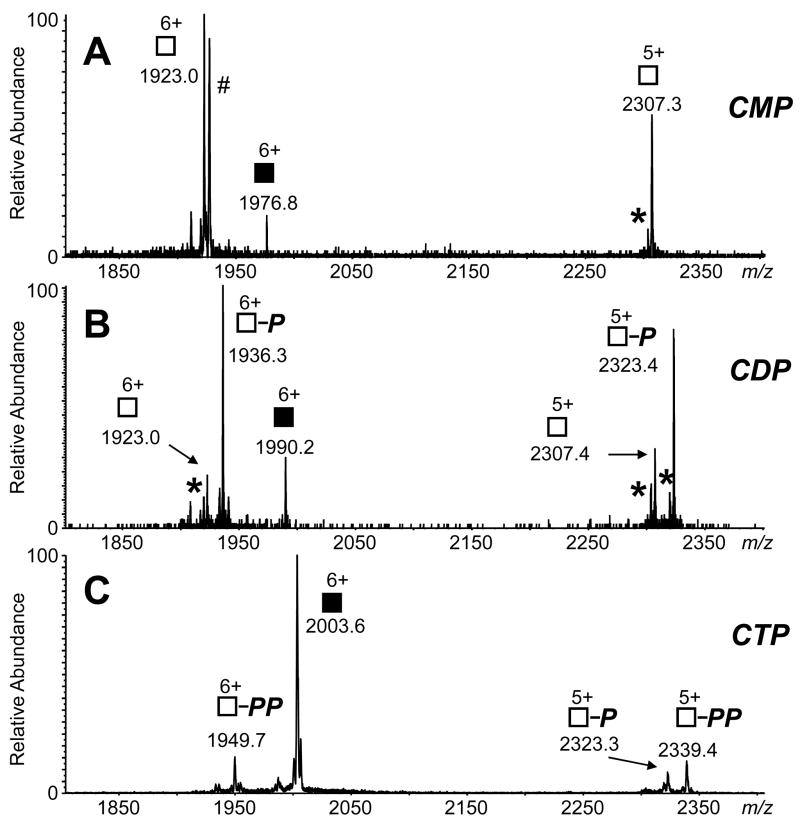
CAD of the RNase S-nucleotide complexes highlights the significant differences between gas phase hydrophobic and electrostatic interactions and the differences observed in solution phase and gas phase binding. For the RNase S-nucleotide gas phase complexes, the reduction of hydrophobic forces and the increasing strength of the electrostatic forces contribute to their behavior upon CAD. In solution, the equilibrium binding constant for the S-protein/S-peptide complexation fall in the nanomolar range, whereas cytidine-nucleotide binding to RNase S is expected to be in the micromolar concentration range. CAD of the gas phase 8+-charged RNase S-nucleotide complex yields products that are contrary to that expected from solution; S-protein and S-peptide dissociate as separated products, and the nucleotide remains bound to the S-protein (Figure 5). The active site of RNase A binding to nucleotides involves the side chains of His-12, His-119, Lys-41, and Gln-11 [26, 40, 48]. The electrostatic interactions between the phosphate groups of CMP, CDP, and CTP and the S-protein appear to be retained in the CAD experiment. Fragmentation of the 6+-charged S-protein/nucleotide complex in a “pseudo-MS3” experiment, in which the declustering potential in the atmospheric pressure-vacuum interface is elevated to +60-70 V to induce dissociation of the S-peptide from the RNase S-nucleotide complex [39, 49], and selection of the targeted S-protein/nucleotide precursor ion for further CAD results in the separation of CMP as the leaving group and one and two phosphate groups being retained on the S-protein for the S-protein/CDP and S-protein/CTP complexes, respectively. Pseudo-MS3 of the S-protein/2′-CMP generates only the separated S-protein and 2′-CMP product ion pair (Figure 6). Thus, nucleotide binding to S-protein behaves similarly to RNase A in the gas phase, and electrostatic interactions predominate.
Figure 5.
CAD mass spectra of the 8+-charged 1:1 complex between RNase S (residues 1–19 S-peptide and residues 21–124 S-protein) and (A) 2′-CMP, (B) CDP, and (C) CTP (11 V collision energy, CE). (The low abundance peak at m/z 1083 is the S-peptide 1–20 resulting from low resolution precursor ion selection.)
Figure 6.
Pseudo-MS3 of the 6+-charged S-protein (residues 21–124) complex with (A) 2′-CMP, (B) CTP, and (C) CTP. A declustering potential of +60–70 V was used to induce dissociation of the RNase S/nucleotide complex. The 6+-charged precursor ions for the S-protein/2′-CMP (m/z 1976), S-protein/CDP (m/z 1990) and S-protein/CTP (m/z 2003, 6+) were selected for further MS/MS (55 V collision energy, CE). The 6+-charged precursors are labeled with filled squares. Product ions are labeled with open squares. Retention of one phosphate group by the product ion is represented by “P”, and retention of two phosphates is labeled with “PP.” Peaks for the loss of water are labeled with an asterisk symbol (*). [The #-labeled peak at m/z 1927 is assigned as the singly-sodiated version of (S-protein)6+ appearing at m/z 1923.]
Unusual Stability of Gas Phase Charge-Charge Interactions
The dissociation of covalent bonds over noncovalent bonds in the CAD of noncovalent complexes, as demonstrated by the RNase A/S-nucleotides examples, has been reported previously for other systems. An early example was presented by Feng on the unusual gas phase stability of a peptide-polyamine complex [50]. The polyamine spermine and a 14-residue spermine-binding peptide (SBP) with 4 glutamic residues and no basic residues form a complex, as measured by ESI-MS. Its binding constant in solution is approximately 0.1 mM, but the 1:1 complex was readily detected. However, dissociation of the complex by CAD was not found; at high collision energy cleavage of a covalent peptide bond was observed. It was hypothesized that its “stability in the gas phase may be due to increased Coulombic stabilization energy of the opposite charges in the solvent-free environment.”
The near covalent stability of gas phase noncovalent electrostatic interactions has been featured in reports by Woods’ laboratory [44]. In one publication, they report on the ESI-MS/MS of the 1:1 complex formed between a highly basic peptide, VLRRRRKRVN, and phosphorylated SVSTDpTpSAE. Two dissociation pathways were observed for the complex: disruption of the electrostatic interactions between the Arg residues and the phosphate groups, and separation of the peptides with concurrent dissociation of the C-O sidechain covalent bond and transfer of HPO3 from either pT or pSi to the basic peptide. Terrier et al recently reported a study of noncovalent complexes involving single-stranded DNA and polybasic compounds, including poly-lysine and penta-arginine [51]. For some of the complexes, MS/MS of the DNA-polybasic complex yielded fragmentation of DNA covalent bonds and an oligonucleotide fragment remain bound to the polybasic compound. This phenomena is similar to an earlier report by Griffey and coworkers, in which CAD of oligoribonucleotide-ligand complexes generated product ions representing RNA fragments bound to the ligand [52].
Numerous studies have suggested an apparent link between the energy required for dissociation of gas phase noncovalent complexes and the solution phase binding constants or activation energies [21, 23, 46]. Despite such studies, a number of others have reported on the enhanced gas phase stability of mono- and oligonucleotides complexed with proteins compared to their solution phase characteristics [28, 31, 32]. Benkestock et al noted that upon CAD of a RNase A-dC6 complex, dissociation of covalent bonds within dC8 occurred, with loss of dC6 from the gas phase complex [31]. They observed also the ready dissociation of the noncovalent RNase A-CMP complex (without breaking covalent bonds), as we observed in the present study. Huang et al studied the effect of the declustering energy in the atmospheric pressure-vacuum interface of their triple quadrupole instrument on the stability of the noncovalent protein-ligand ras-GDP complex [28]. It was found that a substantial amount of energy was required to dissociate the gas-phase ras-GDP complex. In fact, the resulting mass spectrum shows peaks that could be assigned as the transfer of phosphate to the ras protein at CAD energy; however, the authors could not assign these peaks in the original report.
Our laboratory recently reported the complexation of polyamine spermine and spermidine with an acidic protein implicated in Parkinson’s disease, α-synuclein [53]. The solution Kd for the 1:1 protein:spermine complex of 1 mM was confirmed by ligand titration and ESI-MS. The charge-charge interaction between polybasic spermine and spermidine and the acidic C-terminal tail of the protein likely contributed to the enhanced gas phase stability of the complex that allowed ESI-MS detection for a weak solution phase complex. Unlike the behavior observed for RNase A/S binding to CDP and CTP, CAD of the protein-spermine complex yielded only dissociation of the noncovalent bonds, separating the spermine ligand from the 140-residue, 14.5 kDa α-synuclein protein. However, we showed also that electron capture dissociation (ECD) favors cleavage of covalent backbone bonds of protein molecules primarily [53]. This characteristic of ECD translates directly to noncovalent protein-ligand complexes, as covalent backbone bonds of protein complexes are dissociated, but the noncovalent ligand interaction is retained. For the α-synuclein-spermine complex, ECD generates c-/z·-product ions that retain the protein-spermine noncovalent interaction. Spermine binding was localized to residues 106–138. Thus, the enhancement of the gas phase stabilities of noncovalent complexes by electrostatic interactions can be useful to detect weak solution phase complexes by ESI-MS.
The results of this study highlight several important points for measuring noncovalent protein-ligand complexes, and perhaps noncovalent complexes in general by ESI-MS. The ability of ESI-MS to detect a specific noncovalent complex depends on factors related to both its solution phase and gas phase properties. Schug and Linder define a relationship, termed relative transmission factor, as “the relationship between measured ion intensities and the initial concentration of each component” [54, 55]. The relative transmission factor will depend on the binding affinities the molecular partners have for each other in solution, and this in-turn is influenced by the structures of the components and the types and strengths of the forces holding them together in solution. Once these specific interactions are formed in solution, the process of transferring them to the gas phase will then dictate whether they will be observed by ESI-MS.
Unlike in solution, processes in the gas phase are not at true equilibrium, as noncovalent bonds disrupted in the gas phase generally remain disrupted throughout the ESI-MS process. This has implications for applying ESI-MS to measure absolute ligand binding affinities and to screen compounds for binding to drug targets in the pharmaceutical and biotechnology sectors. If the gas phase binding affinities are not sufficiently low to preclude their detection or significantly reduces their apparent abundances as measured by MS, then ESI-MS could be used to assess relative or absolute binding affinities for a range of compounds. However, it would be difficult to envision that ESI-MS could be considered as a general tool to measure protein-ligand complexes because of the apparent differences in solution phase and gas phase binding affinities, as found for the RNase S-nucleotide example presented in the current study. An ideal situation for comparing binding affinities for a variety of compounds would be cases in which the dominant type of interaction in the gas phase is similar.
Conclusions
The magnitudes of noncovalent protein binding in aqueous solution can be determined largely by hydrophobic effects. However, the stability of gas phase noncovalent complexes depends on the binding mode employed in holding complexes together. In the gas phase, electrostatic interactions are strengthened and may be difficult to disrupt. For RNase A/S binding to nucleotides, although CMP and CTP have similar solution binding affinities, RNase A/S-CTP is significantly more stable in the gas phase, as probed by ESI-MS and MS/MS experiments. ESI-MS/MS of RNase A-CMP results in facile loss of CMP, whereas CAD of RNase A-CTP leads to dissociation of covalent amide bonds of the RNase A backbone and fragmentation of the cytidine-nucleotide structure. The di-phosphate group from CTP remains bound to the protein because of strengthened electrostatic interactions in the gas phase. CAD-MS/MS of RNase S-CDP/CTP dissociates S-peptide from the remaining S-protein/nucleotide complex, despite the 3 orders of magnitude difference in solution binding favoring S-peptide/S-protein association. Thus, CAD-MS/MS may not be a generally applicable method for gauging or predicting solution binding constants, especially for complexes held together by different types of noncovalent forces.
Thus, we find that protein-nucleotide binding can be nearly covalent or irreversible in the gas phase. This suggests a possible new strategy to functionally probe for and identify new protein kinases, enzymes that modify other proteins by phosphorylation. Protein kinases represent perhaps the single largest mammalian enzyme family with more than 500 members in the human proteome. The ATP-binding sites of protein kinases are highly conserved. Protein binding to ATP (adenosine 5′-triphosphate) has been measured directly using ESI-MS by several laboratories [14, 27, 29, 30]. Preliminary ESI-MS experiments by our lab suggest that the kinase-ATP noncovalent interaction is exceedingly stable in the gas phase, and it may be possible to design strategies to identify the ATP-binding sites of protein kinases using mass spectrometry exclusively.
Acknowledgments
The UCLA Functional Proteomics Center was established and equipped by a grant to UCLA from the W. M. Keck Foundation. JAL acknowledges support from the UCLA Jonsson Comprehensive Cancer Center, the U. S. Department of Energy for funding of the UCLA-DOE Institute for Genomics and Proteomics, and the National Institutes of Health (RR 20004).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ganem B, Li YT, Henion JD. Detection of Noncovalent Receptor-Ligand Complexes by Mass Spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 2.Ganem B, Li YT, Henion JD. Observation of Noncovalent Enzyme-Substrate and Enzyme-Product Complexes by Ion-Spray Mass Spectrometry. J Am Chem Soc. 1991;113:7818–7819. [Google Scholar]
- 3.Ogorzalek Loo RR, Goodlett DR, Smith RD, Loo JA. Observation of a Noncovalent Ribonuclease S-Protein/S-Peptide Complex by Electrospray Ionization Mass Spectrometry. J Am Chem Soc. 1993;115:4391–4392. [Google Scholar]
- 4.Rockwood AL, Busman M, Smith RD. Coulombic Effects in the Dissociatlon of Large Highly Charged Ions. Int J Mass Spectrom Ion Proc. 1991;111:103–129. [Google Scholar]
- 5.Breuker K. The Study of Protein-Ligand Interactions by Mass Spectrometry - a Personal View. Int J Mass Spectrom. 2004;239:33–41. [Google Scholar]
- 6.Loo JA. Studying Noncovalent Protein Complexes by Electrospray Ionization Mass Spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Loo JA. Electrospray Ionization Mass Spectrometry: A Technology for Studying Noncovalent Macromolecular Complexes. Int J Mass Spectrom. 2000;200:175–186. [Google Scholar]
- 8.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Kitova EN, Wang W, Klassen JS. Method for Distinguishing Specific from Nonspecific Protein-Ligand Complexes in Nanoelectrospray Ionization Mass Spectrometry. Anal Chem. 2006;78:3010–3018. doi: 10.1021/ac0522005. [DOI] [PubMed] [Google Scholar]
- 10.van den Heuvel RHH, Heck AJR. Native Protein Mass Spectrometry: From Intact Oligomers to Functional Machineries. Curr Opin Chem Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Sharon M, Witt S, Felderer K, Rockel B, Baumeister W, Robinson CV. 20S Proteasomes Have the Potential to Keep Substrates in Store for Continual Degradation. J Biol Chem. 2006;281:9569–9575. doi: 10.1074/jbc.M511951200. [DOI] [PubMed] [Google Scholar]
- 12.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie Y, Kaufman SL, Chernushevich IV. Electrospray Ionization Mass Spectrometry and Ion Mobility Analysis of the 20S Proteasome Complex. J Am Soc Mass Spectrom. 2005;16:998–1008. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Chernushevich IV, Thomson BA. Collisional Cooling of Large Ions in Electrospray Mass Spectrometry. Anal Chem. 2004;76:1754–1760. doi: 10.1021/ac035406j. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin SH, Sobott F, Yao Z-P, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The Co-Chaperone p23 Arrests the Hsp90 ATPase Cycle to Trap Client Proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 15.Sharon M, Robinson CV. The Role of Mass Spectrometry in Structure Elucidation of Dynamic Protein Complexes. Annu Rev Biochem. 2007;76:167–193. doi: 10.1146/annurev.biochem.76.061005.090816. [DOI] [PubMed] [Google Scholar]
- 16.Hogan CJ, Kettleson EM, Ramaswami B, Chen DR, Biswas P. Charge Reduced Electrospray Size Spectrometry of Mega- and Gigadalton Complexes: Whole Viruses and Virus Fragments. Anal Chem. 2006;78:844–852. doi: 10.1021/ac051571i. [DOI] [PubMed] [Google Scholar]
- 17.Kaddis CS, Lomeli SH, Yin S, Berhane B, Apostol MI, Kickhoefer VA, Rome LH, Loo JA. Sizing Large Proteins and Protein Complexes by Electrospray Ionization Mass Spectrometry and Ion Mobility. J Am Soc Mass Spectrom. 2007;18:1206–1216. doi: 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaddis CS, Loo JA. Native Protein Mass Spectrometry: From Intact Oligomers to Functional Machineries. Anal Chem. 2007;79:1779–1784. [Google Scholar]
- 19.Zhang S, Van Pelt CK, Wilson DB. Quantitative Determination of Noncovalent Binding Interactions Using Automated Nanoelectrospray Mass Spectrometry. Anal Chem. 2003;75:3010–3018. doi: 10.1021/ac034089d. [DOI] [PubMed] [Google Scholar]
- 20.Patriksson A, Marklund E, van der Spoel D. Protein Structures under Electrospray Conditions. Biochemistry. 2007;46:933–945. doi: 10.1021/bi061182y. [DOI] [PubMed] [Google Scholar]
- 21.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative Determination of Noncovalent Binding Interactions Using Soft Ionization Mass Spectrometry. Int J Mass Spectrom. 2002;216:1–27. [Google Scholar]
- 22.Robinson CV, Chung EW, Kragelund BB, Knudsen J, Aplin RT, Poulsen FM, Dobson CM. Probing the Nature of Noncovalent Interactions by Mass Spectrometry. A Study of Protein-CoA Ligand Binding and Assembly. J Am Chem Soc. 1996;118:8646–8653. [Google Scholar]
- 23.Goodlett DR, Ogorzalek Loo RR, Loo JA, Wahl JH, Udseth HR, Smith RD. A Study of the Thermal Denaturation of Ribonuclease S by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 1994;5:614–622. doi: 10.1016/1044-0305(94)85002-X. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Heitz F, Le Grimellec C, Cole R. Hydrophobic Component in Noncovalent Binding of Fusion Peptides to Lipids as Observed by Electrospray Mass Spectrometry. Rapid Commun Mass Spectrom. 2004;18:135–137. doi: 10.1002/rcm.1288. [DOI] [PubMed] [Google Scholar]
- 25.Krishnaswamy SR, Williams ER, Kirsch JF. Free Energies of Protein–Protein Association Determined by Electrospray Ionization Mass Spectrometry Correlate Accurately with Values Obtained by Solution Methods. Protein Sci. 2006;15:1465–1475. doi: 10.1110/ps.062083406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 27.Daubenfeld T, Bouin A-P, Van der Rest GA. Deconvolution Method for the Separation of Specific Versus Nonspecific Interactions in Noncovalent Protein-Ligand Complexes Analyzed by ESI-FT-ICR Mass Spectrometry. J Am Soc Mass Spectrom. 2006;17:1239–1248. doi: 10.1016/j.jasms.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Huang EC, Pramanik BN, Tsarbopoulos A, Reichert P, Ganguly AK, Trotta PP, Nagabhushan LT, Covey TR. Application of Electrospray Mass Spectrometry in Probing Protein-Protein and Protein-Ligand Noncovalent Interactions. J Am Soc Mass Spectrom. 1993;4:624–630. doi: 10.1016/1044-0305(93)85026-T. [DOI] [PubMed] [Google Scholar]
- 29.Koenig S, Hasche A, Pallast S, Krieglstein J, Klumpp S. Detection of ATP-Binding to Growth Factors. J Am Soc Mass Spectrom. 2008;19:91–95. doi: 10.1016/j.jasms.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 30.White HD, Ashcroft AE. Real-Time Measurement of Myosin-Nucleotide Noncovalent Complexes by Electrospray Ionization Mass Spectrometry. Biophys J. 2007;93:914–919. doi: 10.1529/biophysj.106.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benkestock K, Sundqvist G, Edlund P-O, Roeraade J. Influence of Droplet Size, Capillary-Cone Distance and Selected Instrumental Parameters for the Analysis of Noncovalent Protein-Ligand Complexes by Nano-Electrospray Ionization Mass Spectrometry. J Mass Spectrom. 2004;39:1059–1067. doi: 10.1002/jms.685. [DOI] [PubMed] [Google Scholar]
- 32.Sundqvist G, Benkestock K, Roeraade J. Investigation of Multiple Binding Sites on Ribonuclease A Using Nano-Electrospray Ionization Mass Spectrometry. Rapid Commun Mass Spectrom. 2005;19:1011–1016. doi: 10.1002/rcm.1880. [DOI] [PubMed] [Google Scholar]
- 33.Camilleri P, Haskins NJ. Investigating the Non-Covalent Interaction Cytidylic Acids, with Ribonuclease A by Electrospray Mass Spectrometry. Rapid Commun Mass Spectrom. 1993;7:603–604. [Google Scholar]
- 34.Richards FM, Vithayathil PJ. Preparation of Subtilisin-Modified Ribonuclease and the Separation of the Peptide and Protein Components. J Biol Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 35.Woods AS, Ferre S. Amazing Stability of the Arginine-Phosphate Electrostatic Interaction. J Proteome Res. 2005;4:1397–1402. doi: 10.1021/pr050077s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chernushevich IV, Loboda AV, Thomson BA. An Introduction to Quadrupole-Time-of-Flight Mass Spectrometry. J Mass Spectrom. 2001;35:849–865. doi: 10.1002/jms.207. [DOI] [PubMed] [Google Scholar]
- 37.Akashi S, Osawa R, Nishimura Y. Evaluation of Protein-DNA Binding Affinity by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2005;16:116–125. doi: 10.1016/j.jasms.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Kapur A, Beck JL, Brown SE, Dixon NE, Sheil MM. Use of Electrospray Ionization Mass Spectrometry to Study Binding Interactions between a Replication Terminator Protein and DNA. Protein Sci. 2002;11:147–157. doi: 10.1110/ps.27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo JA, Edmonds CG, Smith RD. Primary Sequence Information from Intact Proteins by Electrospray Ionization Tandem Mass Spectrometry. Science. 1990;248:201–204. doi: 10.1126/science.2326633. [DOI] [PubMed] [Google Scholar]
- 40.Leonidas DD, Chavali GB, Oikonomakos NG, Chrysina ED, Kosmopoulou MN, Vlassi M, Frankling C, Acharya KR. High-Resolution Crystal Structures of Ribonuclease A Complexed with Adenylic and Uridylic Nucleotide Inhibitors. Implications for Structure-Based Design of Ribonucleolytic Inhibitors. Protein Sci. 2003;12:2559–2574. doi: 10.1110/ps.03196603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das M, Rao BV, Ghosh S, Varadarajan R. Attempts to Delineate the Relative Contributions of Changes in Hydrophobicity and Packing to Changes in Stability of Ribonuclease S Mutants. Biochemistry. 2005;44:5923–5930. doi: 10.1021/bi050001+. [DOI] [PubMed] [Google Scholar]
- 42.Alves S, Woods A, Tabet JC. Charge State Effect on the Zwitterion Influence on Stability of Non-Covalent Interaction of Single-Stranded DNA with Peptides. J Mass Spectrom. 2007;42:1613–1622. doi: 10.1002/jms.1359. [DOI] [PubMed] [Google Scholar]
- 43.Jackson SN, Wang H-YJ, Yergey A, Woods AS. Phosphate Stabilization of Intermolecular Interactions. J Proteome Res. 2006;5:122–126. doi: 10.1021/pr0503578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson SN, Wang H-YJ, Woods AS. Study of the Fragmentation Patterns of the Phosphate-Arginine Noncovalent Bond. J Proteome Res. 2005;4:2360–2363. doi: 10.1021/pr050261d. [DOI] [PubMed] [Google Scholar]
- 45.Woods AS. The Mighty Arginine, the Stable Quaternary Amines, the Powerful Aromatics, and the Aggressive Phosphate: Their Role in the Noncovalent Minuet. J Proteome Res. 2004;3:478–484. doi: 10.1021/pr034091l. [DOI] [PubMed] [Google Scholar]
- 46.Hunter CL, Mauk AG, Douglas DJ. Dissociation of Heme from Myoglobin and Cytochrome B5: Comparison of Behavior in Solution and the Gas Phase. Biochemistry. 1997;36:1018–1025. doi: 10.1021/bi961993+. [DOI] [PubMed] [Google Scholar]
- 47.Loo JA, Hu P, McConnell P, Mueller WT. A Study of Src SH2 Domain Protein-Phosphopeptide Binding Interactions by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 1997;8:234–243. [Google Scholar]
- 48.Vitagliano L, Merlino A, Zagari A, Mazzarella L. Reversible Substrate-Induced Domain Motions in Ribonuclease A. Proteins: Struc Func Genet. 2002;46:97–104. doi: 10.1002/prot.10033. [DOI] [PubMed] [Google Scholar]
- 49.Loo JA, Edmonds CG, Smith RD. Tandem Mass Spectrometry of Very Large Molecules: Serum Albumin Sequence Information from Multiply Charged Ions Formed by Electrospray Ionization. Anal Chem. 1991;63:2488–2499. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 50.Feng R. Unusually Strong Binding of a Noncovalent Gas-Phase Spermine-Peptide Complex and Its Dramatic Temperature Dependence. Proceedings of the 43rd ASMS Conference on Mass Spectrometry and Allied Topics; Atlanta, GA. May 21–26, 1995; p. 1264. [Google Scholar]
- 51.Terrier P, Tortajada J, Buchmann W. A Study of Noncovalent Complexes Involving Single-Stranded DNA and Polybasic Compounds Using Nanospray Mass Spectrometry. J Am Soc Mass Spectrom. 2007;18:346–358. doi: 10.1016/j.jasms.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 52.Griffey RH, Greig MJ, An H, Sasmor H, Manalili S. Targeted Site-Specific Gas-Phase Cleavage of Oligoribonucleotides. Application in Mass Spectrometry-Based Identification of Ligand Binding Sites. J Am Chem Soc. 1999;121:474–475. [Google Scholar]
- 53.Xie Y, Zhang J, Yin S, Loo JA. Top-Down ESI-ECD-FT-ICR Mass Spectrometry Localizes Noncovalent Protein-Ligand Binding Sites. J Am Chem Soc. 2006;128:14432–14433. doi: 10.1021/ja063197p. [DOI] [PubMed] [Google Scholar]
- 54.Schug K, Lindner W. Development of a Screening Technique for Noncovalent Complex Formation between Guanidinium- and Phosphonate-Functionalized Amino Acids by Electrospray Ionization Ion Trap Mass Spectrometry: Assessing Ionization and Functional Group Interaction. Int J Mass Spectrom. 2004;235:213–222. [Google Scholar]
- 55.Schug K, Lindner W. Using Electrospray Ionization-Mass Spectrometry/Tandem Mass Spectrometry and Small Molecules to Study Guanidinium-Anion Interactions. Int J Mass Spectrom. 2005;241:11–23. [Google Scholar]