A putative nicotianamine synthase from M. thermoautotrophicus was cloned, expressed, purified and crystallized. The native crystals diffracted to a resolution of 1.7 Å.
Keywords: nicotianamine synthase, Methanothermobacter thermoautotrophicus
Abstract
In plants, nicotianamine synthase (NAS) plays a key role in metal homeostasis as it catalyzes the formation of nicotianamine, an important iron and nickel chelator and a precursor of plant phytosiderophores. Here, the crystallization of a protein from Methanothermobacter thermoautotrophicus (MTH675; referred to here as MtNAS) that appears to be homologous to plant NAS is reported. Purification of this protein showed a monomer–dimer equilibrium that could be displaced by using a reducing agent such as DTT. Crystals belonging to space group P212121 and containing dimers of MtNAS were grown by the vapour-diffusion method using polyethylene glycol 3350 as precipitant. A complete native X-ray data set was collected to 1.7 Å resolution at a synchrotron source.
1. Introduction
Nicotianamine (NA) is a ubiquitous nonproteinogenic amino acid found in all higher plants that is involved in the homeostasis of essential metals ions such as Fe, Cu and Zn (Takahashi et al., 2003 ▶). In vitro, NA can bind several metal ions (Fe, Mn, Zn, Ni, Cu) with high affinity (Mari et al., 2006 ▶). In graminaceous plants, NA is also a structural analogue and a precursor of phytosiderophores, which are essential in iron-acquisition strategy (Mizuno et al., 2003 ▶). NA is characterized in planta by high inter-organ mobility and is thus found at a micromolar concentration in the circulating saps of the xylem and the phloem (Le Jean et al., 2005 ▶). The role of NA has been established in the loading of copper to the xylem for root-to-shoot translocation (Mari et al., 2006 ▶; Le Jean et al., 2005 ▶) and also in the unloading of Fe in the leaves from the vascular system, with subsequent distribution in the surrounding mesophyll cells (Herbik et al., 1996 ▶). NA is also involved in seed loading of Fe (Le Jean et al., 2005 ▶). In addition, NA has been shown to inhibit angiotensin I-converting enzyme in humans and consequently reduces high blood pressure (Wada et al., 2006 ▶).
The biosynthesis of NA is ensured by nicotianamine synthase (EC 2.5.1.43), which catalyses the condensation of three S-adenosylmethionine (SAM) molecules (Higuchi et al., 1999 ▶). NAS is unique among the family of enzymes that use SAM as a substrate as it does not transfer the methyl group from SAM but instead its aminopropyl moiety. This is also the case in polyamine synthases such as spermidine or spermine synthase, which use putrescine or spermidine, respectively, as the aminopropyl acceptor (Cheng et al., 2007 ▶). One peculiarity of NAS is that it uses three molecules of SAM without any other acceptor. Three covalent bonds are formed in the course of NA synthesis: one is generated by the self-cyclization of an aminopropyl moiety, leading to an azetidine ring, and two others result from the polymerization of the remaining aminopropyl moiety to the azetidine ring (Higuchi et al., 1999 ▶). This process does not require additional energy (e.g. from ATP). Although nicotianamine synthase has to date been widely considered to be a plant-specific gene family, recent studies have reported the detection of both NA and functional NAS in a filamentous fungus, namely Neurospora crassa (Trampczynska et al., 2006 ▶). While the key role of NAS in metal homeostasis has been demonstrated in vivo by genetic approaches, few data are available on the molecular characterization of this class of enzymes and enzymatic parameters or structural data are still lacking.
Recent sequencing has revealed putative nas genes in the genomes of various organisms, including plants, fungi and archaea (Trampczynska et al., 2006 ▶; Herbik et al., 1999 ▶). The encoded protein from Methanothermobacter thermoautotrophicus, which shows sequence homology to eukaryotic NAS (about 20%), could therefore possess NAS activity (Trampczynska et al., 2006 ▶). This protein contains 266 residues with a calculated molecular weight of 30 263.7 Da.
Here, we report the crystallization and preliminary X-ray analysis of an NAS-like protein from M. thermoautotrophicus (MtNAS). This is a first step towards the description of the structural determinants involved in NAS activity.
2. Materials and methods
2.1. Cloning, expression and purification
The gene encoding M. thermoautotrophicus hypothetical protein MtNAS (MTH675) was PCR-amplified from M. thermoautotrophicus genomic DNA (a kind gift from Edward L. Bolt of the Medical School, University of Nottingham) using the following PCR primers: forward, 5′-CACCATGAGCTGCTACATCTACTGGGA-3′, and reverse, 5′-ATCAGGACATTTAAAAACAA-3′. The PCR product was cloned in pET101/D-TOPO, leading to a fusion containing the gene of interest followed by a nucleotide sequence encoding a C-terminal six-histidine tag (5′-CATCATCATCATCATCAT-3′). After ensuring verification of the amplified sequence and plasmid transformation, the recombinant protein was overexpressed in Escherichia coli BL21 (DE3) strain and cells were grown semi-aerobically at 310 K on LB medium. Protein expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the optical density at 600 nm reached a value of about 0.6. After 4 h growth, cells were pelleted, resuspended in buffer A (20 mM HEPES buffer pH 7.0, 500 mM NaCl, 20 mM imidazole) and disrupted with a French press at 6.9 MPa. The crude extract was first centrifuged at 10 000g for 10 min to remove unbroken cells and inclusion bodies and the supernatant was then centrifuged at 100 000g for 1 h at 277 K to remove cell-wall debris and membrane proteins.
The resulting soluble fraction was loaded onto a nickel-charged column (1 ml HisTrap column, GE Healthcare) and the protein was eluted by imidazole steps (75 mM wash and 250 mM elution). The collected fractions were loaded onto a gel-filtration column (Hiload 26/60 Superdex 200, GE Healthcare) equilibrated with buffer B (20 mM HEPES, 50 mM NaCl pH 7.5). At this stage, no attempt was made to remove the His tag and peak fractions corresponding to the monomer and the dimer (see §3) were concentrated separately using an Amicon Ultra 30K centrifugal filter (Millipore, Billerica, USA) prior to crystallization trials.
2.2. Crystallization
The protein from the monomeric and dimeric fractions was initially concentrated to 7 mg ml−1 as estimated by the Bradford method (Bradford, 1976 ▶) in buffer B without DTT. All crystallization experiments were carried out at 293 K using the hanging-drop vapour-diffusion technique. Hanging drops consisted of 2 µl concentrated protein solution and 2 µl reservoir solution. Preliminary crystallization trials were conducted using commercially available crystallization screens for proteins: PEG/Ion, Index Screen, SaltRX (Hampton Research, Aliso Viejo, USA) and PACT premier (Molecular Dimensions, Suffolk, England).
2.3. X-ray data collection
MtNAS crystals were directly flash-cooled in liquid nitrogen after a quick soak in paraffin oil. Diffraction data were collected using synchrotron radiation (ID23-2 beamline at the ESRF, France). An ADSC Quantum IV detector and X-ray radiation of 0.98 Å wavelength were used. For a complete data set, collection was performed using a 180° oscillation range and a Δϕ value of 1°. The data sets were processed with MOSFLM (Leslie, 1993 ▶) and subsequent data reduction was carried out using SCALA from the CCP4 suite (Evans, 1997 ▶; Collaborative Computational Project, Number 4, 1994 ▶). The statistics of the X-ray data are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 64.41, b = 68.28, c = 147.54 |
| Resolution (Å) | 44.68–1.70 (1.79–1.70) |
| Total No. of observations | 339168 (50200) |
| No. of unique observations | 71075 (10418) |
| Multiplicity | 4.8 (4.8) |
| Data completeness (%) | 98.5 (99.9) |
| 〈I/σ(I)〉 | 12.4 (3.7) |
| Rmerge† | 0.091 (0.337) |
| Rp.i.m.‡ | 0.050 (0.201) |
R
merge = 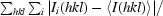
 , where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
, where Ii(hkl) is the ith observation of reflection hkl and 〈I(hkl)〉 is the weighted average intensity for all observations i of reflection hkl.
See Weiss &, Hilgenfeld (1997 ▶).
3. Results and discussion
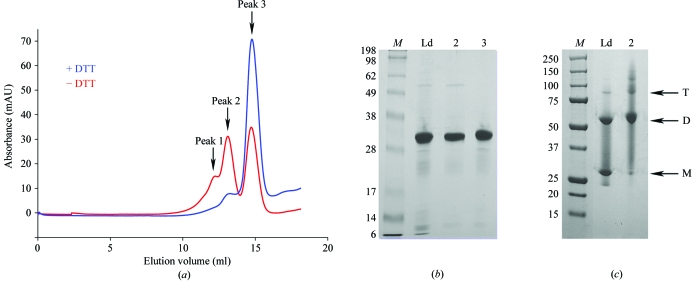
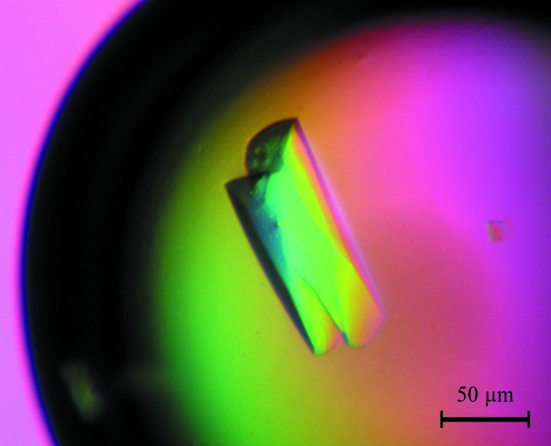
The genes encoding MtNAS was cloned in pET101 vector. A size-exclusion chromatographic step followed by SDS–PAGE analysis revealed that MtNAS was present as either a monomer or a dimer (Fig. 1 ▶, red curve). Therefore, a novel size-exclusion experiment was carried out using the same buffer but containing 10 mM DTT. Under these conditions, MtNAS eluted almost exclusively as a single peak with an elution volume corresponding to an apparent molecular weight of 30 kDa (the theoretical MW including the tag is 33 803.7 Da; see Fig. 1 ▶). The MtNAS dimers could therefore be reduced to monomers (Fig. 1 ▶, blue curve), suggesting that the dimers were linked by at least one disulfide bridge (there are two cysteine residues per monomer in the MtNAS sequence). The yield of MtNAS was low, typically 1.7–2 mg per litre of culture, with a dimer:monomer ratio of approximately 1:3. Initial screenings with the monomeric and dimeric fractions of MtNAS revealed that only the dimeric fraction crystallized, although after a few weeks small crystals also appeared in the setups containing the monomeric fraction. We believe that the crystallization of the monomeric form is a consequence of oxidation and dimerization of the protein. This hypothesis is sustained by the similarity of the crystals forms obtained from these two fractions. Using the dimeric fraction of MtNAS, thin small crystals unsuitable for crystallographic study were obtained within two weeks from many conditions of the screens, particularly in conditions containing PEG 3350. Finally, the best crystals of the native protein were obtained by mixing 2 µl dimeric MtNAS solution (10 mg ml−1 in 20 mM HEPES, 50 mM NaCl pH 7.5) with 2 µl of a reservoir solution containing 100 mM HEPES pH 7.5, 22%(w/v) PEG3350 and 400 mM NaBr and equilibrating against 1 ml reservoir solution. Elongated crystals grew in 1 d and reached their final dimensions (50 × 50 × 150 µm) in about two weeks (Fig. 2 ▶). Some of the crystals contained a defect at both ends, but we found that these crystals diffracted X-rays better than the regular crystals and one of them was successfully used to collect a complete data set to 1.7 Å resolution. Analysis of symmetry-related reflections and systematic absences indicated that the crystal belonged to space group P212121, with unit-cell parameters a = 64.41, b = 68.28, c = 147.54 Å. The asymmetric unit most probably contains two molecules, giving a crystal volume per protein weight (V M) of 2.39 Å3 Da−1 (Matthews, 1968 ▶) and a solvent content of 48.7% by volume. There is no peak in the self-rotation function (in particular, there is no peak corresponding to κ = 180° as expected for twofold NCS) and also no significant peak in the self Patterson (as expected for translational NCS). This suggests the presence of a twofold NCS axis parallel to a crystallographic axis.
Figure 1.
A redox-dependent dimer. (a) Elution diagram of MtNAS in a gel-filtration experiment (Hiload 10/30 Superdex 200, GE Healthcare). The red curve represent the elution diagram without reducing agent whereas the blue curve is the elution diagram obtained after the addition of 10 mM DTT to the sample and elution buffer. The first peak on the red and blue curves corresponds to small amount of trimer and dimer, respectively. (b) SDS–PAGE gel (10%) of purified fractions of MtNAS with reducing agent. (c) SDS–PAGE gel (10%) of purified fractions of MtNAS without reducing agent. Lane M, molecular-weight markers (kDa). Lane Ld, sample loaded onto size-exclusion chromatography after Ni–NTA purification. Lane 2, sample collected from the second peak. Lane 3, sample collected from the third peak. T, trimer. D, dimer. M, monomer.
Figure 2.
Crystal of MtNAS, space group P212121, with approximate dimensions of 50 × 50 × 150 µm.
Determination of the phases is in progress using a classical search for heavy-atom derivatives and the production of selenomethionine-labelled recombinant protein.
Acknowledgments
We would like to thank the ESRF (Grenoble, France) for the provision of data-collection and processing facilities and the ID23-2 team for technical assistance in synchrotron data collection. We also thank Dr S. Mari for his collaboration and continuous support of the project.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Cheng, L., Wang, F., Shou, H., Huang, F., Zheng, L., He, F., Li, J., Zhao, F. J., Ueno, D., Ma, J. F. & Wu, P. (2007). Plant Physiol.145, 1647–1657. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- Herbik, A., Giritch, A., Horstmann, C., Becker, R., Balzer, H. J., Baumlein, H. & Stephan, U. W. (1996). Plant Physiol.111, 533–540. [DOI] [PMC free article] [PubMed]
- Herbik, A., Koch, G., Mock, H. P., Dushkov, D., Czihal, A., Thielmann, J., Stephan, U. W. & Baumlein, H. (1999). Eur. J. Biochem.265, 231–239. [DOI] [PubMed]
- Higuchi, K., Suzuki, K., Nakanishi, H., Yamaguchi, H., Nishizawa, N. K. & Mori, S. (1999). Plant Physiol.119, 471–480. [DOI] [PMC free article] [PubMed]
- Le Jean, M., Schikora, A., Mari, S., Briat, J. F. & Curie, C. (2005). Plant J.44, 769–782. [DOI] [PubMed]
- Leslie, A. G. W. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 44–51. Warrington: Daresbury Laboratory.
- Mari, S., Gendre, D., Pianelli, K., Ouerdane, L., Lobinski, R., Briat, J. F., Lebrun, M. & Czernic, P. (2006). J. Exp. Bot.57, 4111–4122. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mizuno, D., Higuchi, K., Sakamoto, T., Nakanishi, H., Mori, S. & Nishizawa, N. K. (2003). Plant Physiol.132, 1989–1997. [DOI] [PMC free article] [PubMed]
- Takahashi, M., Terada, Y., Nakai, I., Nakanishi, H., Yoshimura, E., Mori, S. & Nishizawa, N. K. (2003). Plant Cell, 15, 1263–1280. [DOI] [PMC free article] [PubMed]
- Trampczynska, A., Bottcher, C. & Clemens, S. (2006). FEBS Lett.580, 3173–3178. [DOI] [PubMed]
- Wada, Y., Kobayashi, T., Takahashi, M., Nakanishi, H., Mori, S. & Nishizawa, N. K. (2006). Biosci. Biotechnol. Biochem.70, 1408–1415. [DOI] [PubMed]
- Weiss, M. S. & Hilgenfeld, R. (1997). J. Appl. Cryst.30, 203–205.