The YncE protein of E. coli has been overproduced and purified and preliminary crystallographic analysis has been performed to 2.1 Å resolution. The structure reveals a seven-bladed β-propeller fold.
Keywords: YncE, β-propellers, periplasmic proteins, TonB-dependent outer-membrane receptor, iron regulation, Fur, Escherichia coli
Abstract
The yncE gene of Escherichia coli encodes a predicted periplasmic protein of unknown function. The gene is de-repressed under iron restriction through the action of the global iron regulator Fur. This suggests a role in iron acquisition, which is supported by the presence of the adjacent yncD gene encoding a potential TonB-dependent outer-membrane transporter. Here, the preliminary crystallographic structure of YncE is reported, revealing that it consists of a seven-bladed β-propeller which resembles the corresponding domain of the ‘surface-layer protein’ of Methanosarcina mazei. A full structure determination is under way in order to provide insight into the function of this protein.
1. Introduction
The yncE gene encoding the 327-residue YncE protein was identified in Escherichia coli during studies on global iron-dependent gene expression, where it was found to be up to 18-fold repressed by the Fe2+–Fur complex (McHugh et al., 2003 ▶). A Fur-box-like sequence is suitably located upstream of the yncE promoter, which is consistent with direct transcriptional repression by Fe2+–Fur (McHugh et al., 2003 ▶). YncE possesses a putative N-terminal signal sequence suggestive of export; consistent with this, it has been shown to be secreted into the periplasm via the Sec-dependent pathway (McHugh et al., 2003 ▶; Baars et al., 2006 ▶). yncE is adjacent to the divergently transcribed yncD gene that encodes a potential TonB-dependent outer-membrane (OM) receptor that is possibly involved in the translocation of iron complexes across the outer membrane (McHugh et al., 2003 ▶). In addition, the homologous TieB protein (with a putative role in enterotoxin production) of enteroinvasive E. coli strain O164 is apparently encoded as part of the Fe2+–Fur repressed cjrABC-senB operon (Šmajs & Weinstock, 2001 ▶; Nataro et al., 1995 ▶). This operon also specifies a TonB-dependent OM receptor as well as a TonB homologue, although it is functionally undefined except for a role in colicin Js sensitivity. These observations, together with the iron regulation and periplasmic location, are suggestive of an iron-transport function for YncE.
The primary structure of YncE suggests that it is composed of seven tandemly repeated motifs (∼40 amino acids, four β-strands) forming a so-called ‘YVTN β-propeller’ (as defined in the InterPro database). Similarly repeated motifs are found in ∼200 functionally undefined bacterial and archaeal proteins in the sequence databases. For bacteria, the YVTN β-propeller proteins all consist of a single YVTN β-propeller domain. However, archaeal YVTN β-propeller proteins are often multidomain proteins containing, for instance, TolB-like, multiple PKD-like, peptidase-like or pectinase-like domains together with one or two YVTN β-propeller domains. The archaeal species Methanosarcina mazei and M. acetivorans each have more than ten genes each that encode YVTN β-propeller domains and all are of unknown function. The structure of the YVTN β-propeller domain of the multidomain ‘surface-layer protein’ of M. mazei has been determined, revealing a seven-bladed β-propeller structure (Jing et al., 2002 ▶). This protein currently represents the sole structurally defined member of the YVTN β-propeller family of proteins.
This work reports the overproduction, purification, crystallization and preliminary X-ray crystallographic analysis of YncE from E. coli.
2. Material and methods
2.1. Protein preparation
The yncE gene (GeneID 946006; UniProtKB/Swiss-Prot entry P76116) was PCR-amplified using the high-fidelity DNA polymerase Accuzyme (Bioline) and cloned into the Champion pET Directional TOPO overexpression vector (Invitrogen) to generate plasmid pETyncE-His6. The forward PCR primer was designed such that the initiating ATG codon of yncE directly followed the CACC motif required for TOPO cloning. The reverse PCR primer was designed to exclude the natural stop codon of yncE, thus enabling the production of an in-frame translational fusion between yncE and the downstream vector-encoded V5 epitope and the His6 tag.
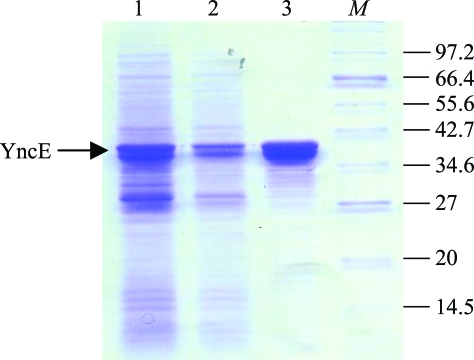
Overproduction of YncE-His6 was achieved using BL21 (DE3) (pETyncE-His6) grown in L-broth plus ampicillin (100 µg ml−1) at 310 K, with the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the culture achieved an optical density of 0.5 at 650 nm. IPTG-induced cells were grown for a further 4 h, harvested, resuspended in 3 ml of binding buffer [25 mM HEPES pH 7.4, 10 mM imidazole, 150 mM NaCl, 20 mM mannitol, 10%(v/v) glycerol] per gram of cell weight and lysed at 76 MPa with a French pressure cell. YncE-His6 was then purified from the soluble supernatant by chromatography with an Ni2+-charged HiTrap Affinity resin column and a linear gradient of 0.01–0.5 M imidazole in binding buffer. The resulting protein was >95% pure as judged by SDS–PAGE analysis (Fig. 1 ▶) and was dialysed against storage buffer [50 mM HEPES pH 7.4, 100 mM NaCl, 10%(v/v) glycerol] and concentrated using a Vivaspin system (10 kDa cutoff; Vivascience) to 3.4 mg ml−1 prior to storage at 193 K.
Figure 1.
Purification of YncE-His6. SDS–PAGE (12% acrylamide) analysis of samples at different stages of YncE-His6 purification. Lane 1, crude whole-cell extract following cell lysis; lane 2, soluble cell extract derived from the crude extract by centrifugation; lane 3, YncE-His6 after purification by Ni2+-affinity chromatography; lane M, molecular-weight markers (sizes indicated in kDa). Approximately 50, 25 and 5 µg of protein were loaded in lanes 1, 2 and 3, respectively.
2.2. Crystallization
Purified YncE-His6 was buffer-exchanged and concentrated to 15 mg ml−1 in 50 mM HEPES pH 7.4 in preparation for crystallization trials. Initial crystallization screening was performed manually using the sitting-drop vapour-diffusion method in 24-well Linbro plates against the following commercial screens at 291 K: Crystal Structure Screens I and II, Structure Screens I and II, Stura Footprint Screen, Macrosol I and II and PEG/Ion Screen (all from Molecular Dimensions Ltd). The drop size was 2 µl plus 2 µl in all cases.
2.3. Diffraction analysis
The YncE-His6 crystals could be sufficiently cryoprotected in the mother-liquor solution [which contained 22%(v/v) PEG 550 MME] and therefore could be flash-cooled directly from the hanging drop. Intensity data were collected on an ADSC Quantum 4 CCD detector at 100 K on the macromolecular beamline PX9.6 at SRS Daresbury, UK. Data were integrated and scaled using the programs MOSFLM v.6.2.4 (Leslie, 1992 ▶) and SCALA (Evans, 1997 ▶), respectively, from the CCP4 program package (Collaborative Computational Project, Number 4, 1994 ▶).
2.4. Phasing
According to WU-Blast2 analysis, the protein that shared the highest amino-acid sequence identity (25% over 183 residues) for which a structure has been reported was found to be the tandem YVTN β-propeller domain from the archaeal surface-layer protein (PDB code 1l0q). The CCP4 program CHAINSAW was used to create an initial model based on 1l0q, pruning the nonconserved residues to the last common atom. The resulting model, consisting of residues 22–323 of YncE-His6, was used as the search model for molecular replacement against all data between 50 and 3.0 Å resolution using Phaser (McCoy et al., 2007 ▶).
3. Results and discussion
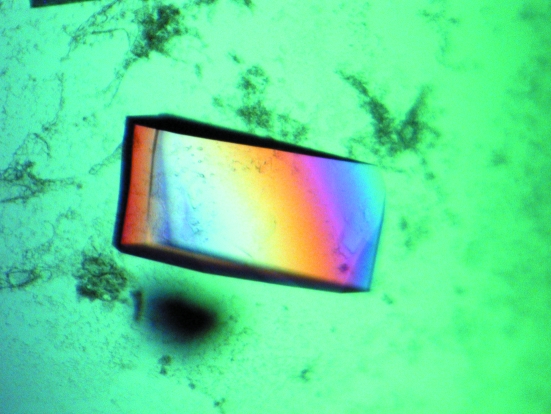
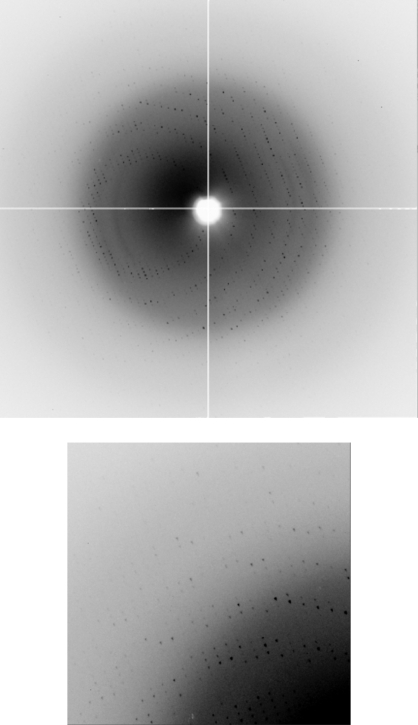
The YncE protein of E. coli was purified to homogeneity and crystallized for structure determination. From the 480 conditions screened, optimization of Stura Footprint Screen condition C1 [0.1 M Na HEPES pH 8.2, 30%(v/v) PEG 550 MME] gave rise to large single crystals of typical dimensions 0.30 × 0.15 × 0.05 mm (Fig. 2 ▶). The final optimized conditions for reliable production of diffraction-quality crystals of YncE-His6 were 0.1 M Tris pH 7.5, 22%(v/v) PEG 550 MME. The crystals appeared within two weeks. The crystals diffracted to 1.9 Å resolution and belonged to space group P21, with unit-cell parameters a = 69.7, b = 108.8, c = 85.3 Å, β = 105.03°, giving rise to four monomers in the asymmetric unit with a solvent content of 48%. A typical image showing diffraction intensities to 2.1 Å resolution for native YncE-His6 (PX9.6, SRS Daresbury) is shown in Fig. 3 ▶ and the data-processing statistics are presented in Table 1 ▶.
Figure 2.
Crystallization of YncE-His6 produced large (typical dimensions 0.3 × 0.15 × 0.05 mm) diffraction-quality crystals from optimization of Stura Footprint Screen condition C1 [0.1 M HEPES pH 8.2, 30%(v/v) PEG 550 MME]. Optimal conditions for YncE-His6 were found to be 0.1 M Tris pH 7.5, 22%(v/v) PEG 550 MME. The drop size was 2 µl plus 2 µl and the protein concentration was 8 mg ml−1.
Figure 3.
Typical diffraction image obtained for YncE-His6 recorded at station PX9.6, SRS Daresbury. The enlarged region is taken from the upper-left corner of the image, showing diffraction close to the edge of the detector. The resolution at the edge is 2.0 Å.
Table 1. Data-collection and processing statistics for YncE-His6 .
Values in parentheses are for the highest resolution shell.
| Synchrotron beamline | SRS PX9.6 |
| Wavelength (Å) | 0.87 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 69.7, b = 108.8, c = 85.3, β = 105.03 |
| Resolution range (Å) | 50.0–2.1 (2.21–2.10) |
| Rmerge† | 0.107 (0.396) |
| No. of observations | 254997 (36245) |
| No. of unique reflections | 71543 (10361) |
| Mean I/σ(I)‡ | 13.5 (4.0) |
| Completeness (%) | 99.8 (99.4) |
| Multiplicity | 3.6 (3.5) |
| Solvent content (%) | 44.9 |
| Molecules per ASU | 4 |
R
merge = 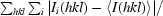
 , where the outer summation is over all unique reflections with multiple observations and the inner summation is over all observations of each reflection.
, where the outer summation is over all unique reflections with multiple observations and the inner summation is over all observations of each reflection.
σ(I) is the standard deviation of I.
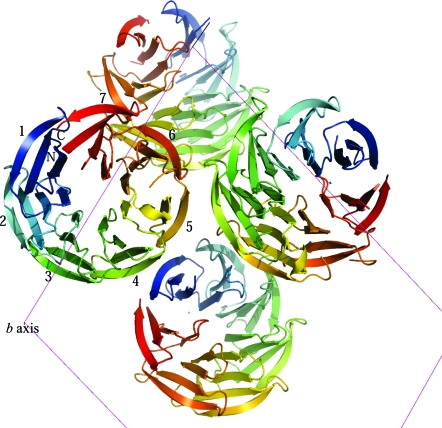
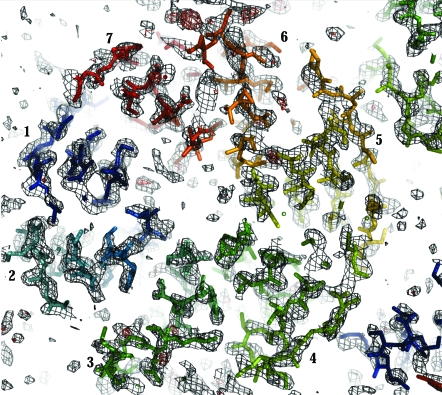
The structure solution was obtained using Phaser (Fig. 4 ▶), based on a model derived from the β-propeller domain of the archeal surface-layer protein (PDB code 1l0q). The program DM (Cowtan & Main, 1996 ▶) was used for noncrystallographic symmetry (NCS) averaging and solvent flattening of the electron-density map (as implemented in the CCP4 program NCSREF). The resulting electron-density map clearly shows distinguishable secondary-structural features for at least 70% of the protein (Fig. 5 ▶). The R factor and R free for the current model (residues 22–323) are 41.1% and 51.5%, respectively. The two C-terminal blades of the β-propeller structure show the most poorly defined density; consequently, iterative manual rebuilding and maximum-likelihood refinement are in progress using the programs Coot (Emsley & Cowtan, 2004 ▶) and REFMAC (Murshudov et al., 1997 ▶), respectively.
Figure 4.
The molecular-replacement model from Phaser of YncE-His6 (residues 22–323) exhibits seven four-stranded β-sheets forming a seven-bladed β-propeller fold. The seven blades of the monomer (numbered 1–7) are shown colour coded from blue to red from the N-terminus to the C-terminus, respectively. The view is down the b axis (unit cell shown in magenta), showing four molecules in the asymmetric unit.
Figure 5.
The 2F o − F c (grey) and F o − F c (red, positive) electron-density maps for the current model of YncE-His6 (residues 22–323) contoured at 1σ and 3σ, respectively, calculated using NCSREF. The R and R free for the current model are 41.1% and 51.5%, respectively, following ten cycles of restrained refinement using REFMAC. The view and labelling are similar to those in Fig. 4 ▶, highlighting the β-propeller fold and showing clear secondary-structural features for the majority of the protein. Manual rebuilding and refinement are in progress.
The crystal structure of YncE is only the second structurally characterized protein belonging to the YVTN β-propeller family and confirms the presence of a single β-propeller domain that contains seven four-stranded β-sheets. The completed YncE structure will allow comparisons with other members of the β-propeller superfamily, which will provide insights into the possible molecular function of this protein as well as those of other YVTN β-propeller proteins.
Acknowledgments
The authors wish to express their thanks to the staff at SRS Daresbury for providing excellent beamline facilities and support. This work was supported by the Lister Institute of Preventive Medicine (personal fellowship to KAW), the BBSRC (project grant to SCA) and the Borno State Scholarship Board (studentship to AB-D).
References
- Baars, L., Ytterberg, A. J., Drew, D., Wagner, S., Thilo, C., van Wijk, K. J. & de Gier, J. W. (2006). J. Biol. Chem.281, 10024–10034. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. D. & Main, P. (1996). Acta Cryst. D52, 43–48. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- Jing, H., Takagi, J., Liu, J. H., Lindgren, S., Zhang, R. G., Joachimiak, A., Wang, J. H. & Springer, T. A. (2002). Structure, 10, 1453–1464. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- McHugh, J. P., Rodriguez-Quinones, F., Abdul-Tehrani, H., Svistunenko, D. A., Poole, R. K., Cooper, C. E. & Andrews, S. C. (2003). J. Biol. Chem.278, 29478–29486. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nataro, J. P., Seriwatana, J., Fasano, A., Maneval, D. R., Guers, L. D., Noriega, F., Dubovsky, F., Levine, M. M. & Morris, J. G. Jr (1995). Infect. Immun.63, 4721–4728. [DOI] [PMC free article] [PubMed]
- Šmajs, D. & Weinstock, G. M. (2001). J. Bacteriol.183, 3958–3966. [DOI] [PMC free article] [PubMed]