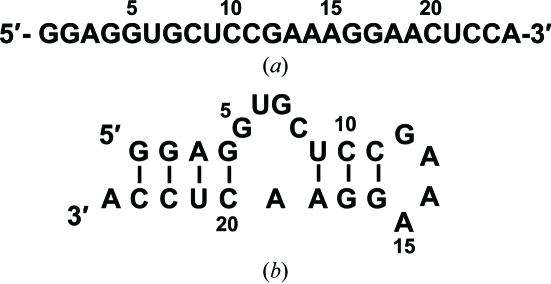An RNA aptamer in complex with the human IgG Fc fragment have been crystallized. The stirring technique with a rotary shaker was used to improve the crystals and to ensure that they were of high quality and single, resulting in crystals that diffracted to 2.2 Å resolution.
Keywords: RNA aptamers, Fc fragments, immunoglobulin G
Abstract
Aptamers, which are folded DNA or RNA molecules, bind to target molecules with high affinity and specificity. An RNA aptamer specific for the Fc fragment of human immunoglobulin G (IgG) has recently been identified and it has been demonstrated that an optimized 24-nucleotide RNA aptamer binds to the Fc fragment of human IgG and not to other species. In order to clarify the structural basis of the high specificity of the RNA aptamer, it was crystallized in complex with the Fc fragment of human IgG1. Preliminary X-ray diffraction studies revealed that the crystals belonged to the orthorhombic space group P21212, with unit-cell parameters a = 83.7, b = 107.2, c = 79.0 Å. A data set has been collected to 2.2 Å resolution.
1. Introduction
Aptamers are folded DNA or RNA molecules that exhibit high affinity for a target molecule. The concept of using nucleic acids (aptamers) as affinity molecules for protein binding was first described in 1990 (Tuerk & Gold, 1990 ▶; Ellington & Szostak, 1990 ▶, 1992 ▶). The concept is based on the ability of sequences to fold in the presence of a target into unique three-dimensional structures that bind the target with high affinity and specificity. The field of functional RNA is rapidly expanding and includes applications that are not only based on the molecule’s primary nucleotide sequence (e.g. RNAi) but are also based on RNA aptamers, which can suppress the activity of any target molecule. The first aptamer-based therapeutic, Pegaptanib (Macugen), which targets vascular endothelial growth factor, was approved by the FDA in 2004 for the treatment of age-related macular degeneration (AMD; Ng et al., 2006 ▶; Zhou & Wang, 2006 ▶). Aptamers are generated by a process that combines combinatorial chemistry with in vitro evolution, known as SELEX (systematic evolution of ligands by exponential enrichment), from a complex library of randomized sequences (Oguro et al., 2003 ▶; Miyakawa et al., 2006 ▶; Ohuchi et al., 2006 ▶; Klussmann, 2006 ▶; Keefe & Schaub, 2008 ▶). Using SELEX, we have recently identified an RNA aptamer that binds the Fc fragment of human IgG with the purpose of creating an alternative to protein A for the mass purification of therapeutic antibodies. This RNA aptamer (Fig. 1 ▶) is highly specific for human IgG, whereas protein A can bind to IgGs from a broader range of species such as mouse, rat, rabbit and so on. Furthermore, one of the key disadvantages of protein A resin in the manufacture of therapeutic antibodies is the requirement of acidic conditions for the elution of bound IgG. Some antibodies are known to adopt an inactive conformation under these conditions (Tsumoto et al., 2004 ▶; Ghose et al., 2005 ▶; Cromwell et al., 2006 ▶). In our previous study, we have shown that the aptamer resin is able to purify IgG from human sera with equivalent efficacy and purity to protein A resin. Importantly, the aptamer resin is able to release bound IgG under neutral pH conditions using an EDTA-based or 0.2 M KCl-based elution buffer (Miyakawa et al., 2008 ▶). Biochemical assays have indicated that the sixth U from the 5′-end must be 2′-fluoro-modified uridine, raising the question of the role of this nucleotide.
Figure 1.
The sequence (a) and secondary structure (b) of the optimized RNA aptamer used in the cocrystallization experiment. All uridines and cytidines are 2′-fluoro-modified. The secondary structure was predicted with the MFold program (Zuker, 2003 ▶).
In order to clarify the structural basis of the high specificity of the RNA aptamer, we have performed crystallization and preliminary crystallographic studies of the aptamer in complex with the Fc fragment of human IgG1.
2. Materials and methods
2.1. Sample preparation
Human IgG1 was purchased from Calbiochem (USA) and was digested with papain (Wako Pure Chemical Industries Ltd, Japan) to produce Fc fragments. The procedure was as follows: 10 mg human IgG1 was incubated at 310 K with 10 mM l-cysteine, 2 mM EDTA and papain in 100 mM sodium phosphate buffer pH 7.2. The reaction was stopped after 1 h by the addition of protease inhibitor (Complete EDTA-free Protease Inhibitor Cocktail Tablets, Roche Diagnostics, Japan). The Fc fragment was purified on a Protein A column (GE Healthcare Bioscience Corp., USA). The chemically synthesized RNA aptamer containing 2′-fluoropyrimidines was purchased from GeneDesign Inc. (Japan). The aptamer was purified by PAGE using 30 × 40 cm glass plates under denaturing conditions with 7 M urea and extensive desalting by ultrafiltration (Centricon YM-3, Millipore Inc., USA) was carried out. The aptamer was annealed by heating for 5 min at 368 K followed by snap-cooling on ice. To confirm the formation of the stem-loop structure, we subjected the aptamer to native PAGE. The Fc fragment and the aptamer were dissolved in 20 mM Tris–HCl pH 7.5 containing 150 mM NaCl, 2 mM CaCl2 and 1 mM MgCl2. We confirmed in a previous NMR study that the interaction between the Fc fragment and the aptamer has a 1:2 (Fc fragment:aptamer) stoichiometry (Miyakawa et al., 2008 ▶). Thus, the aptamer was combined with the Fc fragment in a molar ratio of 1:2.2 (Fc fragment:aptamer) for crystallization. The final concentration of the Fc fragment was 7 mg ml−1 and it was stored at 193 K.
2.2. Crystallization
The initial search for crystallization conditions was performed by the sitting-drop vapour-diffusion method using 96-well microplates (Corning Inc., USA) and optimization was performed in CombiClover crystallization plates (Emerald BioSystems Inc., USA). Preliminary crystallization screening conditions were conducted using Crystal Screens 1 and 2 (Hampton Research, USA) and Wizard Screens I and II (Emerald Biosystems Inc., USA). Typically, a 0.5 µl droplet containing approximately 7 mg ml−1 aptamer–Fc fragment complex dissolved in 20 mM Tris–HCl pH 7.4, 50 mM NaCl, 1 mM MgCl2 and 2 mM CaCl2 was mixed with an equal volume of reservoir solution and the droplet was allowed to equilibrate against 100 µl reservoir solution. All crystallizations were carried out at 293 K. To improve the quality of the crystals, the stirring technique was applied. In this technique, we stirred the protein solution using a speed of 50 rev min−1. Details of this technique have been described elsewhere (Adachi, Matsumura et al., 2004 ▶; Adachi, Takano et al., 2004 ▶; Adachi et al., 2005 ▶).
3. Results and discussion
Crystals of the aptamer–Fc fragment complex were obtained with Crystal Screen 1 condition Nos. 6, 18 and 46, Wizard Screen I condition Nos. 12 and 36, and Wizard Screen II condition Nos. 3 and 28. We confirmed that these crystals contained the Fc fragment and the aptamer by SDS–PAGE and denaturing PAGE, respectively. Initially, the crystals grew as clusters of thin plates from reagents that contained both polyethylene glycol and salt as precipitants. However, the crystals from the cluster showed poor diffraction quality and were too thin for X-ray experiments. Based on these results, the crystallization conditions were optimized by changing the precipitant (from PEG 1000 to PEG 10 000), the pH value and the additives. After optimization of the crystallization conditions, the best crystals were obtained at 293 K using 0.1 M Tris–HCl buffer pH 8.0 containing 20%(w/v) PEG 1000 and 0.2 M calcium acetate as the precipitant solution (Fig. 2 ▶). Several single crystals diffracted to 2.5 Å resolution using synchrotron radiation. The stirring technique with a rotary shaker (Adachi, Matsumura et al., 2004 ▶; Adachi, Takano et al., 2004 ▶; Adachi et al., 2005 ▶) was used to improve the crystals and to ensure that they were of high quality and single, resulting in crystals that diffracted to 2.2 Å resolution.
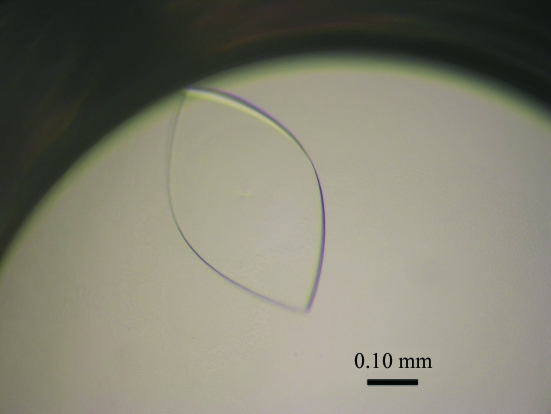
Figure 2.
Crystal of the RNA aptamer in complex with the Fc fragment of human IgG1 grown by sitting-drop vapour diffusion. Plate-shaped crystals with maximum dimensions of 0.5 × 0.3 × 0.03 mm appeared within two weeks.
X-ray diffraction experiments were performed under liquid-nitrogen-cooled conditions at 100 K. A crystal was mounted in a nylon loop, soaked briefly in Paratone-N oil and then frozen by rapidly submerging it in liquid nitrogen. X-ray diffraction data were collected on both beamline BL26B utilizing the mail-in data-collection system (Okazaki et al., 2008 ▶) and beamline BL41XU of the SPring-8 synchrotron-radiation source (Harima, Japan). A total of 120 frames were recorded with an oscillation angle of 1.0°, an exposure time of 10 s per frame and a crystal-to-detector distance of 250 mm. Diffraction data were processed and scaled using MOSFLM (Leslie, 1992 ▶). Analysis of symmetry and systematic absences in the recorded diffraction pattern indicated that the crystals belonged to the orthorhombic space group P21212, with unit-cell parameters a = 83.7, b = 107.2, c = 79.0 Å. Assuming the presence of two aptamers (2 × 7 kDa) and one Fc fragment (60 kDa) in the complex in the asymmetric unit, the V M value was calculated to be 2.4 Å3 Da−1, with an estimated solvent content of 48.6%; these values are within the range commonly observed for protein crystals (Matthews, 1968 ▶). A total of 158 276 observed reflections were merged to 36 180 unique reflections in the resolution range 50.0–2.2 Å. The data-collection and processing statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics for the RNA aptamer in complex with the Fc fragment of human IgG1.
Values in parentheses are for the highest resolution shell.
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 83.7, b = 107.2, c = 79.0 |
| VM† (Å3 Da−1) | 2.4 |
| VS‡ (%) | 48.6 |
| Z | 4 |
| Resolution (Å) | 2.20 (2.32–2.20) |
| No. of observations | 158276 |
| No. of unique reflections | 36180 |
| Completeness (%) | 98.4 (89.4) |
| Rmerge§ (%) | 6.1 (28.0) |
| Redundancy | 4.4 (3.2) |
| I/σ(I) | 16.0 (2.6) |
V M = V c/ZM, where V c is the unit-cell volume and M is the molecular weight.
Solvent content V S = 1 − 1.23/V M.
R
merge = 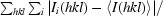
 .
.
An attempt to solve the structure using the molecular-replacement method with CNS (Brünger et al., 1998 ▶) and MOLREP (Vagin & Teplyakov, 1997 ▶) as implemented within the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) was carried out using the structure of the human Fc fragment (PDB code 1fc1; Deisenhofer, 1981 ▶) as a model. The results of molecular replacement suggested that the asymmetric unit of the crystal contains two aptamers complexed with one Fc fragment. The preliminary structure model gives a crystallographic R factor of 0.38 for data between 50.0 and 3.0 Å resolution. Structure analysis and refinement are now in progress.
Acknowledgments
We are grateful to Dr M. Kawamoto and Dr N. Shimizu for their kind help during data collection on BL41XU at SPring-8. The synchrotron-radiation experiments were performed at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI; 2008A1418-CL-np). We also thank Mrs S. Okada for helpful discussion. This research was supported by a Core Research for Evolution Science and Technology (CREST) grant (YM), the CREST grant (YN) and Science and Technology Incubation Program from the Japan Science and Technology Agency, and the Industrial Technology Research Grant Program from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
References
- Adachi, H., Matsumura, H., Niino, A., Takano, K., Kinoshita, T., Warizaya, M., Inoue, T., Mori, Y. & Sasaki, T. (2004). Jpn. J. Appl. Phys.43, L522–L525.
- Adachi, H., Takano, K., Matsumura, H., Inoue, T., Mori, Y. & Sasaki, T. (2004). J. Synchrotron Rad.11, 121–124. [DOI] [PubMed]
- Adachi, H., Takano, K., Niino, A., Matsumura, H., Kinoshita, T., Warizaya, M., Inoue, T., Mori, Y. & Sasaki, T. (2005). Acta Cryst. D61, 759–762. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cromwell, M. E. M., Hilario, E. & Jacobson, F. (2006). AAPS J.8, E572–E579. [DOI] [PMC free article] [PubMed]
- Deisenhofer, J. (1981). Biochemistry, 20, 2361–2370. [PubMed]
- Ellington, A. D. & Szostak, J. W. (1990). Nature (London), 346, 818–822. [DOI] [PubMed]
- Ellington, A. D. & Szostak, J. W. (1992). Nature (London), 355, 850–852. [DOI] [PubMed]
- Ghose, S., Allen, M., Hubbard, B., Brooks, C. & Cramer, S. M. (2005). Biotechnol. Bioeng.92, 665–673. [DOI] [PubMed]
- Keefe, A. D. & Schaub, R. G. (2008). Curr. Opin. Pharmacol.8, 1–6. [DOI] [PubMed]
- Klussmann, S. (2006). The Aptamer Handbook. Weinheim: Wiley–VCH.
- Leslie, A. G. W. (1992). Jnt CCP4/ESF-EACBM Newslett. Protein Crystallogr. No. 26.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Miyakawa, S., Nomura, Y., Sakamoto, T., Yamaguchi, Y., Kato, K., Yamazaki, S. & Nakamura, Y. (2008). RNA, 14, 1154–1163. [DOI] [PMC free article] [PubMed]
- Miyakawa, S., Oguro, A., Ohtsu, T., Imataka, H., Sonenberg, N. & Nakamura, Y. (2006). RNA, 12, 1825–1834. [DOI] [PMC free article] [PubMed]
- Ng, E. W., Shima, D. T., Calias, P., Cunningham, E. T. Jr, Guyer, D. R. & Adamis, A. P. (2006). Nature Rev. Drug Discov.5, 123–132. [DOI] [PubMed]
- Oguro, A., Ohtsu, T., Svitkin, Y. V., Sonenberg, N. & Nakamura, Y. (2003). RNA, 9, 394–407. [DOI] [PMC free article] [PubMed]
- Ohuchi, S. P., Ohtsu, T. & Nakamura, Y. (2006). Biochimie, 88, 897–904. [DOI] [PubMed]
- Okazaki, N., Hasegawa, K., Ueno, G., Murakami, H., Kumasaka, T. & Yamamoto, M. (2008). J. Synchrotron Rad.15, 288–291. [DOI] [PMC free article] [PubMed]
- Tsumoto, K., Umetsu, M., Kumagai, I., Ejima, D., Philo, J. S. & Arakawa, T. (2004). Biotechnol. Prog.20, 1301–1308. [DOI] [PubMed]
- Tuerk, C. & Gold, L. (1990). Science, 249, 505–510. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Zhou, B. & Wang, B. (2006). Exp. Eye Res. 83, 615–619. [DOI] [PubMed]
- Zuker, M. (2003). Nucleic Acids Res.31, 3406–3415. [DOI] [PMC free article] [PubMed]