Crystals of an E. coli AcrB contaminant were grown from 95% pure CorA preparations. This very frequently occurring problem in membrane-protein crystallography laboratories is reported, as well as suggestions to avoid it.
Keywords: membrane protein crystallization, contaminants, AcrB
Abstract
In the course of a crystallographic study of the Methanosarcina mazei CorA transporter, the membrane protein was obtained with at least 95% purity and was submitted to crystallization trials. Small crystals (<100 µm) were grown that diffracted to 3.42 Å resolution and belonged to space group R32, with unit-cell parameters a = b = 145.74, c = 514.0 Å. After molecular-replacement attempts using available CorA structures as search models failed to yield a solution, it was discovered that the crystals consisted of an Escherichia coli contaminating protein, acriflavine resistance protein B (AcrB), that was present at less than 5% in the protein preparations. AcrB contamination is a major problem when expressing membrane proteins in E. coli since it binds naturally to immobilized metal-ion affinity chromatography (IMAC) resins. Here, the structure is compared with previously deposited AcrB structures and strategies are proposed to avoid this contamination.
1. Introduction
In the last few years, the number of membrane-protein structures deposited in the Protein Data Bank (PDB) has increased exponentially. Many of these proteins were heterologously overexpressed in Escherichia coli with a polyhistidine fusion tag for subsequent purification. Two well known critical factors in protein crystallization are protein purity and monodispersity. In the case of soluble proteins, reports can be found concerning the unfortunate crystallization of contaminating proteins either from a very low amount of contamination of the purified protein sample (Cámara-Artigas et al., 2006 ▶) or even from a mixture of 50 different proteins with no predominant species (Lohkamp & Dobritzsch, 2008 ▶). In the case of membrane proteins, no report dedicated to the exposition of such situations has been previously published.
The CorA transport protein is the primary Mg2+-uptake system in both bacteria and archaea. Thus, it is a key component of Mg2+ homeostasis in these kingdoms (Kehres & Maguire, 2002 ▶; Knoop et al., 2005 ▶). Recently, three different groups have independently solved the structure of the pentameric Thermotoga maritima CorA, which is believed to be in a closed state (Lunin et al., 2006 ▶; Eshaghi et al., 2006 ▶; Payandeh & Pai, 2006 ▶). These structures, as well as a more recent report (Payandeh et al., 2008 ▶), provide new insights into CorA transporters; however, the mechanism of pore opening and magnesium transport remains to be elucidated. Thus, the structure of CorA in the open state would be of great benefit in order to further understand this fascinating protein family.
During our attempts to crystallize a ligand-free archaeal homologue of CorA, we obtained crystals of an unexpected protein: AcrB from E. coli. This is a proton motive force-dependent multidrug efflux pump that functions via a drug/proton antiport mechanism and belongs to the resistance–nodulation–division (RND) transporter superfamily. It accepts substrates either from the periplasm or from the cytoplasm (Helling et al., 2002 ▶) and works within a tripartite system involving an outer membrane channel (TolC) and a membrane-fusion protein (AcrA). AcrB spontaneously binds to IMAC resins because of the presence of a histidine-rich cluster at its C-terminus. Here, we describe a phenomenon that is very frequent in membrane-protein crystallography laboratories but that has been little documented in the literature. In addition, we present the structure of E. coli AcrB at 3.42 Å resolution from rhombohedral crystals.
2. Materials and methods
2.1. Cloning and expression
The genomic DNA sequence of Methanosarcina mazei CorA was amplified by PCR using specific Gateway primers containing attB sequences, which allow insertion into the pDONR201 cloning vector by the BP recombination reaction, and then introduced into a Gateway home-adapted pET22b vector (Novagen). The resulting construct (pET22b-CorA) bears a C-terminal His6 tag. For protein expression, E. coli BL21 (DE3) strain (Invitrogen) was transformed with pET22b-CorA and cells were grown in a 3 l fermenter at 310 K with batch medium (Xu et al., 1999 ▶) supplemented with 100 µg ml−1 ampicillin. When the OD600 reached 2.5, the temperature was reduced to 293 K and expression was induced overnight by the addition of 0.5 mM IPTG. Cells were harvested by centrifugation and stored at 193 K.
2.2. Purification
All steps were conducted at 277 K. Cells were resuspended in lysis buffer [50 mM Tris pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, 1 mM EDTA, 0.5 mg ml−1 lysozyme, 20 µg ml−1 DNAse I, 20 mM MgSO4, phenylmethylsulfonyl fluoride (PMSF) and antiproteases (Complete EDTA-free antiproteases, Roche)] and lysed by three passages through a French press (Thermo) at 124 MPa. Two low-speed centrifugation steps removed cellular debris, inclusion bodies and precipitated proteins. The resulting supernatant was ultracentrifuged in a Ti-45 rotor (Beckman-Coulter) at 40 000 rev min−1 for 1.5 h. The pellet (membranes) was resuspended in buffer A [50 mM Tris pH 8.0, 300 mM NaCl, 5%(v/v) glycerol, PMSF and antiproteases (Roche)], flash-frozen in liquid nitrogen and stored at 193 K. Solubilization of CorA was accomplished by stirring for 30 min in buffer A + 1% n-dodecyl-β-d-maltopyranoside (DDM; Solgrade, Anatrace) before ultracentrifugation at 40 000 rev min−1 for 1.5 h. The supernatant was then incubated with agitation for 30 min with 15 ml Ni–NTA resin (Qiagen) equilibrated in buffer A + 0.05% DDM (Solgrade, Anatrace). Washing and elution steps took place on a gravity column with buffer A + 0.05% DDM (Anagrade, Anatrace) containing 50 and 250 mM imidazole, respectively. CorA-containing fractions were pooled based on SDS–PAGE analysis. The protein was then injected onto a Superdex 200 16/60 column (Amersham) and run in a buffer containing 10 mM Tris pH 8.0, 300 mM NaCl, 5%(v/v) glycerol and 0.05% DDM (Anagrade, Anatrace). Two peaks were observed: the first was in the void volume (aggregated CorA) and the second corresponded to the pentameric form of CorA. Fractions from the second peak were pooled and concentrated to 6 mg ml−1 using a centrifugal filter device (Millipore, 100 kDa molecular-weight cutoff). Before crystallogenesis, the protein was ultracentrifuged in a Ti-42.2 rotor (Beckman-Coulter) at 42 000 rev min−1 for 30 min. Protein purity and integrity were checked by Coomassie-stained SDS–PAGE. The single observed band was excised for protease treatment. Protein digestion with trypsin took place as described previously (Shevchenko et al., 1996 ▶) before peptide mass-fingerprint MALDI–TOF (PMF–MALDI–TOF) mass-spectrometric analysis. In all mass-spectrometric studies, spectra were acquired with an Autoflex spectrometer (Bruker Daltonics).
2.3. Crystallization
Crystallization screening was performed using a sitting-drop vapour-diffusion setup in 96-well plates at 293 K. Initial hits were obtained with the commercial screens Membrane Class I and II (Qiagen). A lead condition (0.1 M MES pH 6.5, 30% PEG 400) was optimized in 24-well plates with 0.5 ml of precipitant solution in the reservoir to a range of final crystallization conditions: 0.1 M MES pH 6.0–7.0, 20–40% PEG 400. By varying the pH, the temperature, the protein and precipitant concentrations, the drop size and the protein:precipitant solution ratio, the best condition was found to be 3 µl protein solution + 2 µl mother liquor (0.1 M MES pH 6.6, 31% PEG 400). Crystals appeared after 10 d and reached maximum dimensions of 95 × 80 × 40 µm within about two months (Fig. 1 ▶). To improve crystal quality, various detergents were added to the reservoir solution. Most of those with a glucoside or a maltoside headgroup produced similar but significantly smaller crystals. However, the addition of 0.1% n-octyl-β-d-thioglucopyranoside (Anagrade, Anatrace) allowed us to obtain crystals as large as those obtained in DDM, but with a dramatic loss of diffraction power. For cryocooling, crystals were directly flash-frozen in a stream of boiling nitrogen.
Figure 1.
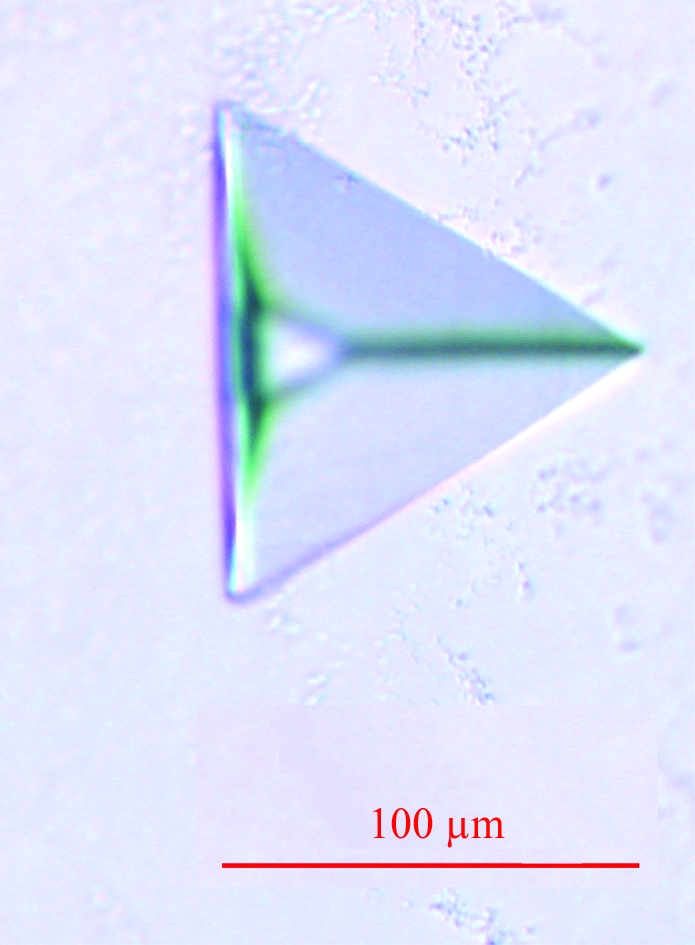
Rhombohedral crystal of E. coli AcrB.
2.4. Data collection and processing
Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beamline ID-23 EH1 with a beam size of 100 × 100 µm using an ADSC Quantum Q315r detector. The sample temperature was 100 K and the wavelength was 0.98 Å. Each frame was taken with an oscillation of 1° and 1 s exposure at 10% transmission. Data from 100 images were indexed and integrated with XDS and scaled with XSCALE (Kabsch, 1993 ▶). The AcrB crystals diffracted to 3.42 Å resolution and belonged to the rhombohedral space group R32, with unit-cell parameters a = b = 145.74, c = 514.0 Å. The calculated solvent content with one AcrB molecule in the asymmetric unit was 73.73% (V M = 4.72 Å3 Da−1). A random selection of 4% of the data (test set) were assigned for calculation of the free R factor (Brünger, 1992 ▶) and were not included in the refinement. Data-collection and processing statistics are reported in Table 1 ▶.
Table 1. Crystallographic data processing.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 20–3.42 (3.6–3.42) |
| Total observations | 176182 (25406) |
| Unique observations | 28788 (4072) |
| Completeness (%) | 99.3 (100) |
| Rmerge† (%) | 12.4 (61.9) |
| Multiplicity | 6.12 (6.24) |
| I/σ(I) | 11.97 (3.25) |
R
merge = 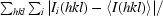
 .
.
2.5. Molecular-replacement attempts with CorA
The reported full-length structures of T. maritima CorA and the two soluble-domain structures of T. maritima and Archaeoglobus fulgidus CorA were used as molecular-replacement search models (PDB codes 2hn2, 2bbj, 2iub, 2hn1 and 2bbh). Unmodified and polyalanine models as well as various truncated structures based on these PDB entries were produced to generate potentially invariable search models. Attempts to solve the M. mazei CorA structure were made using MOLREP (Vagin & Teplyakov, 1997 ▶), Phaser (McCoy et al., 2007 ▶), FSEARCH (Hao, 2001 ▶) and RSEARCH (Collaborative Computational Project, Number 4, 1994 ▶) using several resolution cutoffs. Moreover, we generated models predicting putative motions of CorA by normal-mode analysis of the full length, the transmembrane pore and the pentameric soluble domain of the T. maritima CorA structure (PDB code 2iub) using the elNémo server (Suhre & Sanejouand, 2004 ▶).
2.6. AcrB structure determination and refinement
We finally used the structure of E. coli AcrB solved at 3.1 Å resolution (PDB code 2i6w) without modification as the molecular-replacement search model for phasing with MOLREP. Refinement was carried out with REFMAC5 (Murshudov et al., 1997 ▶) and model building was performed using Coot (Emsley & Cowtan, 2004 ▶). TLS groups were generated with the TLS Motion Determination server (Painter & Merritt, 2006 ▶). The structure was refined to R cryst and R free factors of 28.8% and 35.4%, respectively. The final AcrB model contained 1045 amino-acid residues and did not include the last four residues (1046–1049). Refinement and model validation statistics are reported in Table 2 ▶.
Table 2. Crystallographic refinement and model-validation statistics.
Values in parentheses are for the highest resolution shell.
| Resolution range | 20–3.42 (3.51–3.42) |
| No. of reflections used in refinement | 27636 (1961) |
| Final overall R factor (%) | 28.8 (34.1) |
| Atomic displacement model | Isotropic |
| Overall average B factor excluding solvent (Å2) | 67.7 |
| No. of atoms | 7951 |
| No. of reflections in test set for Rfree | 1152 (82) |
| Final Rfree (%) | 35.4 (39.4) |
| R.m.s.d. deviation from ideal values | |
| R.m.s.d. bond lengths (Å) | 0.01 |
| R.m.s.d. bond angles (°) | 1.40 |
| R.m.s.d. dihedral angles (°) | 1.19 |
| Ramachandran plot analysis (excluding Gly and Pro) | |
| Most favoured (%) | 82.8 |
| Additional allowed (%) | 15.4 |
| Generously allowed (%) | 1.4 |
| Disallowed (%) | 0.4 |
3. Results and discussion
3.1. CorA purity analysis
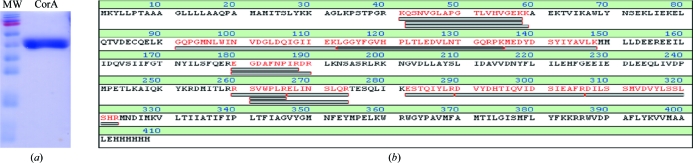
Classical Coomassie-stained SDS–PAGE analysis of the two-step purified CorA showed only one band at the expected molecular weight for the CorA monomer (46.9 kDa, Fig. 2 ▶ a). PMF–MALDI–TOF mass spectrometry was used to unambiguously identify the protein in the unique observed band as M. mazei CorA (Fig. 2 ▶ b).
Figure 2.
CorA purification. (a) Coomassie-stained SDS–PAGE of purified M. mazei CorA. Lane MW, molecular-weight markers (170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDa); lane CorA, two-step purified M. mazei CorA. (b) Peptide mass-fingerprint MALDI–TOF identification using the sequence of M. mazei CorA and the band shown in Fig. 2 ▶(a). The sequence coverage was 35.5%. Grey bars indicate matching peptides.
3.2. AcrB identification
Before contamination was detected, we performed extensive molecular-replacement trials using CorA-based search models without success, despite the 46% sequence identity between T. maritima and M. mazei CorA. We were able to identify that the crystallized protein was E. coli AcrB instead of M. mazei CorA thanks to the helpful suggestions of Dr Liz Carpenter (Membrane Protein Laboratory, Diamond, Oxford, United Kingdom) and Dr Susanna Törnroth-Horsefield (University of Gothenburg, Sweden). The crystal morphology, space group and unit-cell parameters were typical of E. coli AcrB rhombohedral crystals.
E. coli AcrB spontaneously binds to IMAC resins because of the presence of a histidine-rich cluster at its C-terminus. Because of the low amount of contamination, the presence of AcrB was not detected by classical Coomassie-stained SDS–PAGE. Furthermore, MALDI–TOF mass spectrometry carried out on dissolved crystals did not allow us to observe AcrB. We estimated the AcrB concentration in our sample to be approximately 0.1 mg ml−1.
3.3. AcrB crystallization conditions summary
We crystallized E. coli AcrB in a previously unreported condition using only PEG 400 as the precipitating agent. A summary of the crystallization conditions, crystal space groups and unit-cell parameters as well as resolutions for all AcrB structures reported to date is shown in Table 3 ▶. The precipitating agent involved was PEG in all cases. Various buffers covering a wide pH range allowed E. coli AcrB crystal growth. The detergent used for crystallization was DDM in 23 of the 25 deposited structures (including this work). The two remaining structures were obtained with cyclohexyl-n-hexyl-β-d-maltoside (Cymal-6; Seeger et al., 2006 ▶) as the crystallization detergent. Among these structures, 76% (19 of 25) were solved from rhombohedral crystals (R32) and always with similar unit-cell parameters. This provides a major criterion for the identification of AcrB rhombohedral crystals. To conclude, E. coli AcrB crystallizes under a variety of conditions and is probably not limited to those reported to date. Thus, this possible issue should always be kept in mind when crystallization screening is carried out for membrane proteins that have been heterologously overexpressed in E. coli and purified by IMAC.
Table 3. Summary of crystallization conditions for the 25 structures of E. coli AcrB deposited to date in the PDB.
For each structure, the crystal space group, unit-cell parameters and resolution are given.
| Crystallization conditions | Space group | Unit-cell angles (°) | Unit-cell dimensions (Å) | PDB code | Resolution (Å) |
|---|---|---|---|---|---|
| 7% PEG 4000, 20–40 mM sodium (or potassium) citrate pH 5.6 or 6.5, 20–50 mM KNO3 (in some cases) and 10% glycerol | R32 | α = β = 90, γ = 120 | a = b = 143.56, c = 519.64 | 1oy6 | 3.68 |
| a = b = 144.80, c = 518.61 | 1oy8 | 3.63 | |||
| a = b = 144.68, c = 517.51 | 1oy9 | 3.80 | |||
| a = b = 144.77, c = 517.89 | 1oyd | 3.80 | |||
| a = b = 145.11, c = 517.16 | 1oye | 3.48 | |||
| 14% PEG 4000, 100 mM sodium phosphate pH 6.2 and 50 mM NaCl | C2 | α = γ = 90, β = 97.71 | a = 225.87, b = 134.42, c = 163.19 | 2dhh | 2.80 |
| α = γ = 90, β = 98.08 | a = 227.05, b = 134.56, c = 161.70 | 2dr6 | 3.30 | ||
| α = γ = 90, β = 98.17 | a = 225.80, b = 134.47, c = 162.12 | 2drd | 3.10 | ||
| 8% PEG 4000, 50 mM ADA pH 6.5 and 200 mM (NH4)2SO4 | P212121 | a = 146.18, b = 157.41, c = 246.04 | 2j8s | 2.54 | |
| 12% PEG 4000, 0.1 M HEPES pH 7.5, 0.1 M NaCl and 0.3 M Li2SO4 | R32 | α = β = 90, γ = 120 | a = b = 146.30, c = 514.30 | 2i6w | 3.10 |
| 8–10% PEG 3000, 40 mM potassium citrate pH 6.5 and 10% glycerol | R32 | α = β = 90, γ = 120 | a = b = 144.37, c = 518.93 | 1t9t | 3.23 |
| a = b = 144.09, c = 518.80 | 1t9u | 3.11 | |||
| a = b = 144.37, c = 518.44 | 1t9v | 3.80 | |||
| a = b = 144.92, c = 516.67 | 1t9y | 3.64 | |||
| a = b = 144.97, c = 519.37 | 1t9x | 3.08 | |||
| a = b = 144.51, c = 519.36 | 1t9w | 3.23 | |||
| 6.5–8.5% PEG 3000, 30 mM sodium citrate pH 5.6 or 30 mM potassium citrate pH 6.5 and 8% glycerol | R32 | α = β = 90, γ = 120 | a = b = 145.40, c = 514.51 | 2hqc | 3.56 |
| a = b = 145.04, c = 513.67 | 2hqd | 3.65 | |||
| a = b = 145.65, c = 519.71 | 2hqf | 3.38 | |||
| a = b = 144.90, c = 518.64 | 2hqg | 3.38 | |||
| 15–16% PEG 2000, 80 mM sodium phosphate pH 6.2, 20 mM sodium citrate–HCl pH 5.6 | R32 | α = β = 90, γ = 120 | a = b = 144.54, c = 519.18 | 1iwg | 3.50 |
| 14–28% PEG 1000 or PEG 1500, 0.1 M Tris pH 7.5, 0.1 M Li2SO4, 18 mMn-octyl-β-D-thioglucopyranoside and 20% 1,2,3-heptanetriol | R32 | α = β = 90, γ = 120 | a = b = 145.10, c = 511.64 | 2rdd | 3.50 |
| 5% PEG 400, 16–22% PEG 300, 70 mM sodium citrate pH 4.6 and 8–11% glycerol | C2 | α = γ = 90, β = 98.21 | a = 222.80, b = 134.10, c = 161.01 | 2gif | 2.90 |
| P1 | α = 103.9, β = 94.64, γ = 90.11 | a = 127.33, b = 134.87, c = 140.84 | 2hrt | 3.00 | |
| 31% PEG 400, 100 mM MES pH 6.6 | R32 | α = β = 90, γ = 120 | a = b = 145.74, c = 514.00 | 3d9b | 3.42 |
3.4. Crystal-packing analysis
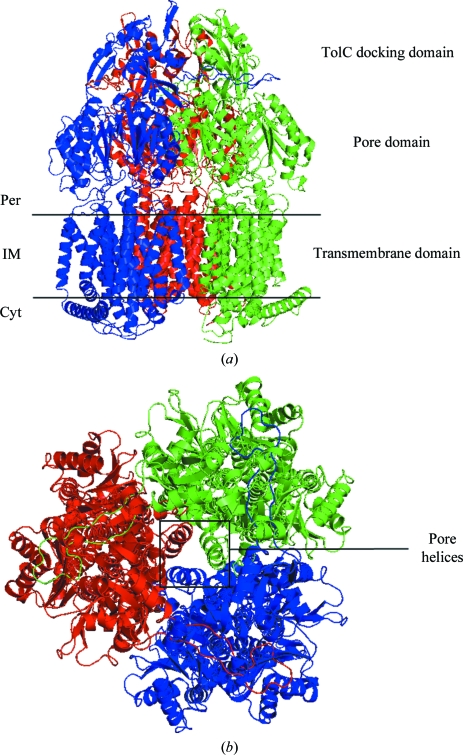
The molecular-replacement solution obtained from MOLREP was of good quality (R = 0.38). Typically, AcrB rhombohedral crystals contain one monomer in the asymmetric unit, with the functional AcrB trimer generated by the crystallographic threefold axis. The resulting AcrB structure is symmetric and comprises three conformationally identical monomers (Figs. 3 ▶ a and 3 ▶ b).
Figure 3.
AcrB structure. (a) Side view of the AcrB trimer. The three protomers constituting the functional AcrB trimer are individually coloured (blue, red and green). The different domains are indicated. Per, periplasm; IM, inner membrane; Cyt, cytoplasm. (b) Periplasmic view of the AcrB trimer. The three central pore helices involved in the peristaltic pump mechanism (Murakami et al., 2006 ▶; Seeger et al., 2006 ▶; Sennhauser et al., 2007 ▶) are indicated. Figs. 3–5 were created using PyMOL (DeLano, 2002 ▶).
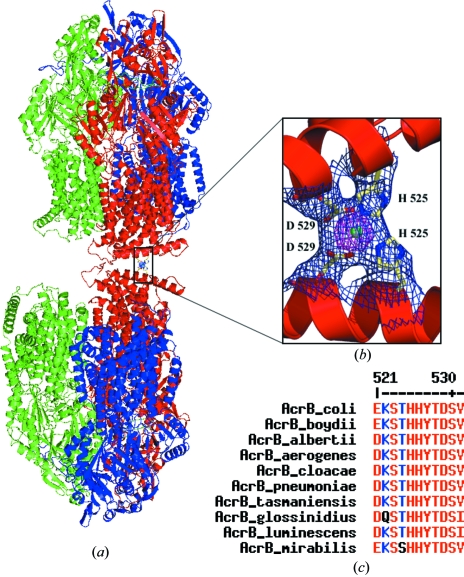
Examination of the crystal packing reveals that the AcrB trimers are arranged in a ‘head-to-neck’ manner (Fig. 4 ▶ a). Each AcrB biological unit interacts with six equivalent trimers in the crystal lattice, three via their cytoplasmic regions and three via their periplasmic domains. We found some residual electron density at each interface between the cytoplasm-protruding regions of two AcrB trimers that corresponds to a unique nonprotein atom coordinated by four residues, two histidines and two aspartates, provided by the two adjacent trimers. As NaCl was the only salt added during purification, we assigned the atom as an Ni2+ ion from leakage of the affinity column used for purification. Indeed, the fortuitous assistance of such an Ni2+ ion in crystal packing has previously been observed in the case of the leukotriene C4 synthase structure (Molina et al., 2007 ▶). Interestingly, in our case this Ni2+ ion mediates a crystal contact between two AcrB trimers (Fig. 4 ▶ b). The same residual electron density, confirmed by Fourier difference maps, is also present in some of the other deposited AcrB structures solved from rhombohedral crystals, but no assignment was reported. A multiple sequence alignment of nine AcrB homologues from different species reveals a high level of conservation of the two residues involved in metal binding (His525 and Asp529; Fig. 4 ▶ c). Thus, if AcrB rhombohedral crystals were obtained from one of these homologues, a similar ion-mediated crystal contact might be expected. Nevertheless, the biological relevance of this metal-ion binding site is certainly low because its formation requires that two AcrB trimers from two different layers interact in a ‘head-to-neck’ manner and this is more than unlikely in vivo.
Figure 4.
AcrB crystal packing. (a) AcrB ‘head-to-neck’ organization in the crystal lattice. The three protomers constituting each functional AcrB trimer are individually coloured (blue, red and green). (b) Enlargement of the Ni2+ ion-mediated crystal contact between two protomers from two different AcrB trimers. Residues His525 and Asp529 from each protomer are shown in stick representation and the Ni2+ ion is depicted as a green sphere. 2F o − F c electron density for these four residues and the Ni2+ ion is contoured at 1σ (blue) and 5σ (pink). (c) Multiple sequence alignment of AcrB homologues, revealing the very high degree of conservation for the residues binding the Ni2+ ion. Homologous AcrB sequences are from Escherichia coli, Shigella boydii, Escherichia alberti, Enterobacter aerogenes, Enterobacter cloacea, Klebsiella pneumoniae, Erwinia tasmaniensis, Sodalis glossinidius, Photorhabdus luminescens and Proteus mirabillis. The alignment was performed with MULTIALIN (Corpet, 1988 ▶).
3.5. Comparison with other AcrB structures
The structure of AcrB solved in this study superimposes very well with previously published symmetric AcrB structures. The root-mean-square deviation (r.m.s.d.) between all matching main-chain atoms in our structure relative to PDB entry 2i6w (our molecular-replacement search model) and PDB entry 2rdd (the most recently solved AcrB symmetric structure) is 0.98 and 1.72 Å, respectively. Our symmetric AcrB structure is very similar to molecules A and B and significantly different from molecule C of the highest resolution asymmetric AcrB structure (PDB code 2j8s; data not shown). This is in agreement with previous observations (Sennhauser et al., 2007 ▶).
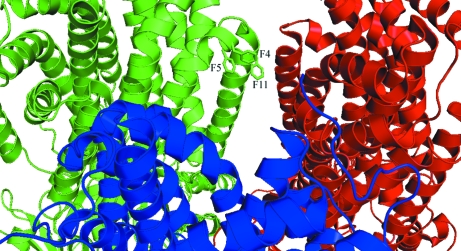
In the RND superfamily the sequence of the N-terminus is highly conserved, with strict conservation of phenylalanine residues 4, 5 and 11 in several AcrB homologues (Das et al., 2006 ▶). This sequence conservation points to a putative functional role for these three residues, which are located at the membrane–cytoplasm interface. Modelling of the six N-terminal residues has only been accomplished in two other symmetric AcrB structures. In one case (PDB code 2rdd) this region is in an α-helical conformation; in the second (PDB code 2i6w) it is in an extended conformation. The calculation of an OMIT map, removing the eight N-terminal residues, allowed us to build these residues in an α-helical conformation, as previously observed in PDB entry 2rdd and in most asymmetric AcrB structures. Consequently, unlike in PDB entry 2i6w, in our model the side chains of phenylalanine residues 4 and 5 are located in close proximity to phenylalanine residue 11 (Fig. 5 ▶). The involvement of this phenylalanine cluster in cytoplasmic substrate transport remains to be investigated.
Figure 5.
The three highly conserved phenylalanine residues at the N-terminus of AcrB. Phe4, Phe5 and Phe11 are shown in stick representations for one monomer. Each protomer is coloured individually (red, blue and green).
4. Conclusions
The observation that E. coli AcrB spontaneously binds to IMAC resins was reported 9 y ago (Zgurskaya & Nikaido, 1999 ▶), but several European membrane-protein crystallography laboratories have recently encountered the problem of unintended AcrB crystallization (E-MeP members, personal communication). Furthermore, only two structures of endogenous E. coli AcrB have been deposited in the PDB (Das et al., 2006 ▶; Törnroth-Horsefield et al., 2007 ▶) and no report has made a clear critical analysis of this problem.
In the present case, it was not possible to detect AcrB contamination using classical Coomassie-stained SDS–PAGE. We estimated the AcrB concentration in our CorA purified sample to be about 0.1 mg ml−1. This highlights the fact that AcrB can crystallize from very low concentrations and that it represents a major risk in membrane-protein crystallization when overexpression is carried out in E. coli and purification involves an IMAC step.
To avoid this problem, we propose either introducing a protease-cleavage site that allows removal of the His tag or using another affinity tag such as a Strep tag. Another elegant approach might be to use E. coli strains that are devoid of the acrB gene. A general cautious approach would also be to check, when crystals are obtained, whether this combination of space group and unit-cell parameters are present in the PDB. Finally, an analysis of dissolved crystals should be performed by silver-stained SDS–PAGE and eventually by PMF–MALDI–TOF mass spectrometry.
This paper is the first to be dedicated to description of the AcrB contamination problem and of ways to avoid it. We also report for the first time an Ni2+ ion-mediated crystal contact in AcrB rhombohedral crystals.
Supplementary Material
PDB reference: AcrB, 3d9b, r3d9bsf
Acknowledgments
We gratefully acknowledge fruitful discussions with the partners of the EU programme E-MeP as well as Professor Eric Chabrière. This work was funded in part by the EU E-MeP programme (FP6 Contract No. LSHG-CT2004-504601), by a Marie-Curie (AST-3D Contract No. MEIF-CT-2005-024761) grant to GS, by the Marseille-Nice Genopole and by a PhD grant from the ‘Ministère Français de l’Enseignement Supérieur et de la Recherche’ No. 22976-2006 to DV.
References
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Cámara-Artigas, A., Hirasawa, M., Knaff, D. B., Wang, M. & Allen, J. P. (2006). Acta Cryst. F62, 1087–1092. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Corpet, F. (1988). Nucleic Acids Res.16, 10881–10890. [DOI] [PMC free article] [PubMed]
- Das, D., Xu, Q. S., Lee, J. Y., Ankoudinova, I., Huang, C., Lou, Y., DeGiovanni, A., Kim, R. & Kim, S. (2006). J. Struct. Biol.158, 494–502. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Eshaghi, S., Niegowski, D., Kohl, A., Molina, D. M., Lesley, S. A. & Nordlund, P. (2006). Science, 313, 354–357. [DOI] [PubMed]
- Hao, Q. (2001). Acta Cryst. D57, 1410–1414. [DOI] [PubMed]
- Helling, R. B., Janes, B. K., Kimball, H., Tran, T., Bundesmann, M., Check, P., Phelan, D. & Miller, C. (2002). J. Bacteriol.184, 3699–3703. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- Kehres, D. G. & Maguire, M. E. (2002). Biometals, 15, 261–270. [DOI] [PubMed]
- Knoop, V., Groth-Malonek, M., Gebert, M., Eifler, K. & Weyand, K. (2005). Mol. Genet. Genomics, 274, 205–216. [DOI] [PubMed]
- Lohkamp, B. & Dobritzsch, D. (2008). Acta Cryst. D64, 407–415. [DOI] [PubMed]
- Lunin, V. V., Dobrovetsky, E., Khutoreskaya, G., Zhang, R., Joachimiak, A., Doyle, D. A., Bochkarev, A., Maguire, M. E., Edwards, A. M. & Koth, C. M. (2006). Nature (London), 440, 833–837. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Molina, D. M., Wetterholm, A., Kohl, A., McCarthy, A. A., Niegowski, D., Ohlson, E., Hammarberg, T., Eshaghi, S., Haeggström, J. Z. & Nordlund, P. (2007). Nature (London), 448, 613–617. [DOI] [PubMed]
- Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T. & Yamaguchi, A. (2006). Nature (London), 443, 173–179. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Payandeh, J., Canhui, L., Ramjeesingh, M., Poduch, E., Bear, C. E. & Pai, E. F. (2008). J. Biol. Chem.283, 11721–11733. [DOI] [PubMed]
- Payandeh, J. & Pai, E. F. (2006). EMBO J.25, 3762–3773. [DOI] [PMC free article] [PubMed]
- Seeger, M. A., Schiefner, A., Eicher, T., Verrey, F., Diederichs, K. & Pos, K. M. (2006). Science, 313, 1295–1298. [DOI] [PubMed]
- Sennhauser, G., Amstutz, P., Briand, C., Storchenegger, O. & Grütter, M. G. (2007). PLoS Biol.5, 106–113. [DOI] [PMC free article] [PubMed]
- Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996). Anal. Chem.68, 850–858. [DOI] [PubMed]
- Suhre, K. & Sanejouand, Y.-H. (2004). Acta Cryst. D60, 796–799. [DOI] [PubMed]
- Törnroth-Horsefield, S., Gourdon, P., Horsefield, R., Brive, L., Yamamoto, N., Mori, H., Snijder, N. & Neutze, R. (2007). Structure, 12, 1663–1673. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Xu, B., Jahic, M. & Enfors, S. (1999). Biotechnol. Prog.15, 81–90. [DOI] [PubMed]
- Zgurskaya, H. I. & Nikaido, H. (1999). Proc. Natl Acad. Sci. USA, 96, 7190–7196. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AcrB, 3d9b, r3d9bsf