Abstract
Background
White adipose tissue is not only an energy storage organ; it also functions as an endocrine organ. The coordination and integration of numerous gene expression events is required to establish and maintain the adipocyte phenotype.
Findings
We previously observed a 45-fold upregulation for a transcript encoding a novel predicted transmembrane protein, Tmem182, upon brown preadipocyte to adipocyte conversion. Here we use real-time PCR analysis to further characterize Tmem182 transcript expression in the adipocyte lineage. Analysis across a panel of 10 murine tissues revealed highest Tmem182 transcript expression in white adipose tissues (WAT), with 10-fold to 20-fold higher levels than in brown adipose tissue (BAT). Tmem182 transcript expression is ~3-fold upregulated in BAT of genetically obese (ob/ob) mice vs. wild type C57BL/6. Analysis of three in vitro models of white adipogenesis indicates markedly enriched expression of Tmem182 transcript in adipocytes vs. preadipocytes. Compared to 3T3-L1 preadipocytes, a 157-fold higher level of Tmem182 transcript is detected at 3 day post-induction of adipogenesis and an ~2500-fold higher level in mature 3T3-L1 adipocytes. TNFα treatment of 3T3-L1 adipocytes resulted in a ~90% decrease in Tmem182 transcript level. As skeletal muscle and heart were also found to express Tmem182 transcript, we assessed expression in C2C12 myogenesis and observed a ~770-fold upregulation upon conversion of myoblasts to myocytes.
Conclusion
WAT is the most prominent site of Tmem182 transcript expression and levels of transcript for Tmem182 are altered in adipose tissues of ob/ob mice and upon exposure of 3T3-L1 adipocytes to the proinflammatory cytokine TNFα. The dramatic upregulation of Tmem182 transcript during in vitro adipogenesis and myogenesis suggests Tmem182 may function in intracellular pathways important in these two cell types.
Background
White adipose tissue (WAT) is the major site for storage of excess energy and these triglyceride stores are mobilized to meet the energy needs of the organism. Adipose tissue is now also recognized as an endocrine organ with synthesis and secretion of a variety of soluble factors such as leptin, resistin, adiponectin, retinol binding protein-4 and TNFα [1-4]. Adipocytes make up from one-third to two-thirds of the cell population found in adipose tissue, with endothelial cells, nerve cells, macrophages, fibroblast-like interstitial cells and preadipocytes, and perhaps other cell types, comprising the remaining stromal-vascular component [5]. Mature adipocytes form as the result of the differentiation of preadipocyte precursors present in adipose tissue [6-10]. Established preadipocyte cell lines such as 3T3-L1 [11] have been an extensively used in vitro model to define genes central to the adipocyte phenotype [12,13]. Adipogenesis is accompanied by increased transcription of genes that encode proteins key to adipocyte function, for example lipogenesis, lipolysis, lipid transport, and hormone responsiveness [7,14,15]. In vitro and in vivo studies have uncovered a pivotal role for peroxisome proliferator-activated receptor γ (PPARγ), a member of the ligand-activated steroid hormone receptor family, in the adipogenic program [8,10,16-19]. Studies have also illustrated the important contributions of the CCAAT/enhancer-binding proteins (C/EBPs) and other transcriptional signals to adipogenesis [8,10,20].
Methods
Culture of cell lines and adipogenic conversion for 3T3-L1, ScAP-23 and wt-BAT, for the fractionation of adipose tissues, and for culture and differentiation of murine preadipocytes from subcutaneous (SC) WAT was as described [21,22]. C2C12 cells were maintained and passaged as subconfluent cultures in DMEM with10% FBS. For differentiation, cultures at 70% confluence were switched to DMEM with 2% horse serum and 10 μg/ml insulin, and were cultured under these conditions for 7 days. For treatment of 3T3-L1 adipocytes with TNFα and various pharmacological inhibitors the method was as described [23-25]. After serum-starvation for 6 h, 3T3-L1 adipocytes were pretreated with either 50 μM LY294002, 50 μM PD98059, 20 μM SB203580, 100 nM wortmannin, 1 μM rapamycin (Sigma-Aldrich, St. Louis, MO), or DMSO vehicle for 1 h and then cultured in 10 ng/ml of TNFα for 16 h in the presence of inhibitors. RNA was purified using TriZol Reagent (Invitrogen Corp.) according to manufacturer's instruction. For studies of Tmem182 transcript expression in murine tissues, 8 wk old C57BL/6 or ob/ob male mice were utilized, with all animal treatments conducted with approval of the University of Toledo Health Science Campus Institutional Animal Care and Use Committee.
Real-time PCR analysis was as previously described [23,24,26]. For this, total RNA was subject to purification with an RNeasy RNA purification kit with DNase I treatment (Qiagen Corp., Valencia, CA) and 5 μg used for first strand cDNA synthesis with SuperScript II RNase H-reverse transcriptase (Invitrogen Corp.) and an oligo(dT)-22 primer. Real time PCR was conducted with an ABI 7500 Real Time PCR System. Target cDNA levels were analyzed by SYBR green-based real-time PCR in 25 μl reactions containing 1× SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 100 nM each forward and reverse primers, and 10 ng of cDNA. Analyses were performed in triplicate and expression of each gene was normalized against Gapdh transcript level. The cycle threshold value was generated using ABI PRISM 7500 SDS software version 1.2 and exported to an Excel spreadsheet to calculate fold differences. Sequence of PCR primers used were: Tmem182 (5'-ACACCAATCAGCCACCATCC-3' and 5'-GCCACGGTAAATAATTGCGGAG-3'); Gapdh (5'-GGCAAATTCAACGGCACAG-3' and 5'-CGGAGATGATGACCCTTTTG-3'); and Myogenin (5'-GCCATCCAGTACATTGAGC-3'and 5'-GTAAGGGAGTGCAGATTGTG-3'). Primers were designed to span introns.
Properties of Tmem182 gene and sequence
During the characterization and utilization of a new in vitro model of brown adipogenesis, mBAP-9, we previously identified ten new genes with increased expression in mBAP-9 adipocytes vs. preadipocytes (manuscript in preparation). Of the ten genes identified, the gene found to exhibit the highest fold upregulation in mBAP-9 adipogenesis (45-fold), encoded an uncharacterized novel predicted transmembrane protein termed Tmem182. Its transcript expression and regulation, however, was not further characterized at the time. Tmem182 is represented by UniGene Mm.334678 [GenBank: NM_001081198]. It encodes a wholly novel 229 amino acid protein with a calculated molecular mass of 25,845. The protein sequence of Tmem182 is shown in Figure 1A. The Kyte-Doolittle analysis of Tmem182 protein sequence identifies four putative membrane-spanning regions (Figure 1B, and underlined and bolded in Figure 1A), indicative of an integral membrane topology. Database analysis (NCBI Homologene) indicates that Tmem182 homologs are found in human (90%), dog (87%), rat (87%) chick (71%), with numbers in parentheses indicating percent amino acid identity to the murine protein. Homologs are also present in pig, horse, monkey and zebrafish.
Figure 1.
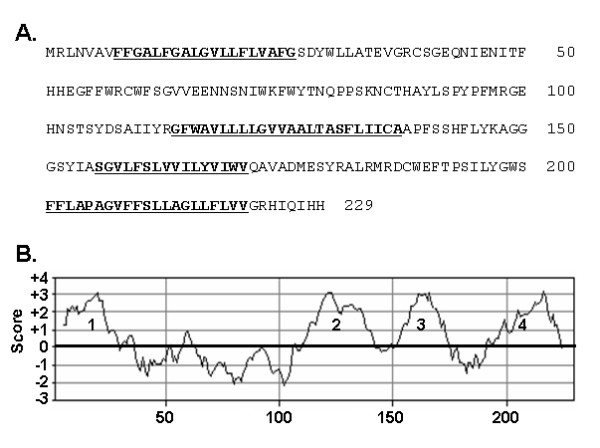
Sequence analysis predicts a transmembrane localization for Tmem182 protein. (A) Amino acid sequence of murine Tmem182. The numbers at right indicate amino acid positions. The four putative membrane-spanning regions are underlined. (B) Results of Kyte-Doolittle analysis with a window size of 10, indicating 4 putative transmembrane regions. Hydrophobicity score is on the y-axis and amino acid residue position on the x-axis.
Tissue distribution of Tmem182 transcript expression
We determined transcript expression of Tmem182 in a panel of murine tissues using real-time PCR. As shown in Figure 2A, of the tissues examined, subcutaneous WAT is the most enriched site of transcript expression (p < 0.001), with expression in muscle, heart and lung at levels from ~10% to ~50% of that in WAT. Lower relative levels of Tmem182 transcript are found in kidney, spleen, testis, brain and liver. We also examined Tmem182 transcript level in brown adipose tissue (BAT) and three distinct WAT depots, subcutaneous (SC), epididymal (EP) and retroperitoneal (RP), Figure 2B. We find that each of the three WAT depots have a similar magnitude of expression of Tmem182 transcript with from ~10- to ~20-fold higher levels (p < 0.001) than that found in BAT.
Figure 2.
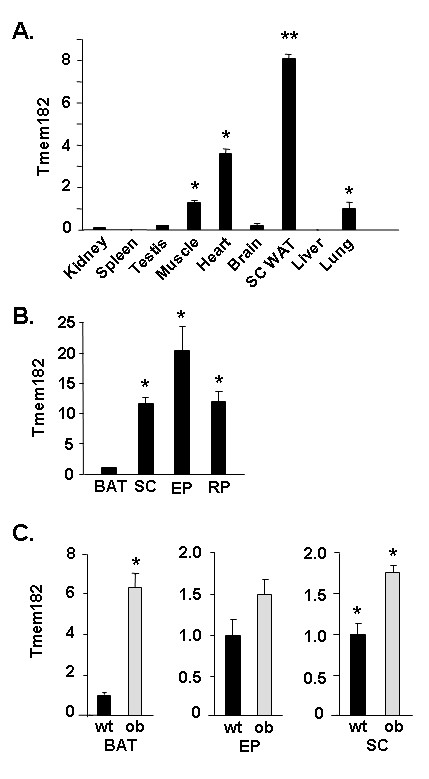
White adipose tissue enrichment of Tmem182 and dysregulation in ob/ob adipose tissues. (A) Tmem182 transcript expression in various murine tissues. RNA was harvested from the indicated murine tissues and Tmem182 transcript level was analyzed by real-time PCR as described in Methods. Values were normalized to Gapdh expression level and transcript expression in lung was set to a value of 1. Statistical analysis was carried out with one-way ANOVA followed by Tukey post-hoc test. ** indicates p < 0.001 for SC WAT vs. all other tissues. * indicates p < 0.01 for muscle, heart, and lung vs. all other tissues. (B) Tmem182 transcript expression in wild type brown and white adipose tissues and (C) Expression of Tmem182 transcript in wild type (wt) vs. ob/ob white and brown adipose tissues. RNA was harvested from subcutaneous (SC) WAT, epididymal (EP) WAT and brown adipose tissue (BAT) of wild type (wt) and ob/ob (ob) mice and real-time PCR for Tmem182 transcript was performed as described in Methods. Values were normalized to Gapdh transcript expression level. Transcript expression in BAT was set to a value of 1 for (B) and wt was set to a value of 1 for (C). Statistical analysis was carried out with single-factor ANOVA. For (B), * indicates p < 0.001 for SC, EP, and RP vs. BAT. For (C), * indicates p < 0.001 for ob BAT vs. wt BAT and for ob SC vs. wt SC.
To begin to address the modulation of Tmem182 transcript levels in regard to the pathophysiology of adipocytes, we compared transcript expression in BAT and WAT from wild type C57BL/6 mice and ob/ob mice, the latter a well-studied murine model of genetic obesity. For Tmem182 transcript, we find a slight increase (~1.7-fold, p < 0.001) in ob/ob WAT for the SC depot (Figure 2C). Furthermore, compared to wt BAT, we find a 6.3-fold upregulation (p < 0.001) of Tmem182 transcript level in ob/ob vs. wt BAT, suggestive of a role for dysregulation of Tmem182 in the obese state (Figure 2C). It would thus appear that in the ob/ob genetic model, BAT shifts to a level of Tmem182 transcript expression that is more similar to that found in WAT.
Differentiation-dependent expression of Tmem182 transcript in adipogenesis
We originally identified Tmem182 during the characterization of a new brown adipocyte in vitro cell culture model, termed mBAP-9, wherein we found Tmem182 transcript to be upregulated 45-fold during the adipogenic conversion of these cells from preadipocytes to mature adipocytes (manuscript in preparation). Our tissue expression studies, however, revealed that although BAT is a site of expression of Tmem182 transcript, the most dominant site of expression in vivo is WAT. We therefore determined if Tmem182 transcript evidenced differentiation-dependent expression in various in vitro models of white adipogenesis. The best characterized cell culture model of adipogenesis is 3T3-L1, these cells were developed ~30 years ago [11] and undergo differentiation from preadipocytes to mature white adipocytes following treatment with the adipogenic agents Dex and MIX. Real-time PCR analysis indicates that 3T3-L1 adipocytes express ~2500-fold higher levels of Tmem182 transcript than 3T3-L1 preadipocytes (Figure 3A). The increase in transcript expression is first noted at day 3, with a 157-fold increase noted. In addition to the 3T3-L1 model, we examined Tmem182 transcript expression in two other models of white adipogenesis. ScAP-23 cells are a cell line established in this laboratory derived from preadipocytes present in murine SC WAT, in contrast to the embryonic derivation of 3T3-L1 cells. As shown in Figure 3B, adipogenesis of ScAP-23 cells leads to a 75-fold increase in levels of Tmem182 transcript. Figure 3C shows that adipocyte conversion of primary cultures of murine stromal vascular fraction cells from SC WAT, wherein preadipocytes are found, is accompanied by an ~22-fold increase in Tmem182 transcript. Although we find differing magnitudes of upregulation of Tmem182 transcript across the adipogenesis models that we assessed, in all cases a significant increase in Tmem182 transcript level accompanies adipocyte conversion.
Figure 3.
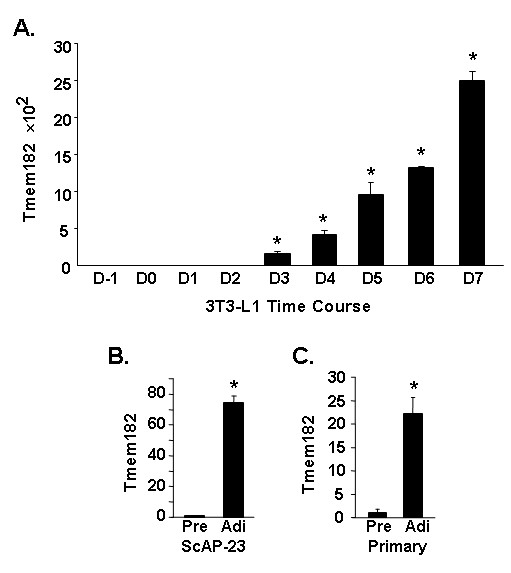
Tmem182 is a differentiation-dependent gene in white adipogenesis. (A) Tmem182 transcript expression during a time course of 3T3-L1 differentiation. 3T3-L1 RNA was harvested daily at the indicated days preceding (D-1) and during differentiation. Tmem182 transcript level was analyzed by real-time PCR as described in Methods. Values were normalized to Gapdh transcript expression level. For D-1, D0, D1, and D2, a value of 40 cycles was assigned to calculate a delta Ct value and transcript expression in D-1 sample was set to a value of 1. Statistical analysis was carried out using one-way ANOVA followed by Tukey post-hoc test; * indicates p < 0.001 for D3, D4, D5, D6 and D7 vs. all other samples. (B) Tmem182 transcript expression in ScAP-23 preadipocytes (Pre) and adipocytes (Adi) and (C) Tmem182 transcript expression in primary murine subcutaneous preadipocytes (Pre) and adipocytes (Adi). RNA was harvested from the indicated cultures before and after differentiation and Tmem182 transcript level was analyzed by real-time PCR as described in Methods. Values were normalized to Gapdh transcript expression level and transcript expression in Pre set to a value of 1. For (B) and (C), statistical analysis was carried out with single-factor ANOVA; * indicates p < 0.001 for Pre vs. Adi.
Regulation of Tmem182 transcript by TNFα
We determined the regulation of Tmem182 transcript following incubation of in vitro differentiated 3T3-L1 white adipocytes with TNFα. TNFα is a proinflammatory cytokine central to adipose tissue pathophysiology and acts to suppress expression of many adipocyte genes; elevated TNFα levels are associated with obesity [27-31]. TNFα treatment of adipocytes promotes lipolysis and dedifferentiation [32-34], with the latter ascribed TNFα-mediated transcriptional downregulation of the key adipocyte transcription factors PPARγ [35] and C/EBPα [36,37]. Studies of TNFα effects on Tmem182 transcript expression were carried out using treatment with TNFα alone, or by treating cells with TNFα after adipocytes had been pretreated with pharmacological inhibitors including those for intracellular signaling pathways with a role in adipocyte gene expression and/or function. For example, reports indicate a role for p38 MAP kinase in the effects of TNFα on adipocyte gene expression [38,39] and the TNFα effects on the transcript expression of the novel adipocyte lipase ATGL are attenuated by pretreatment with PD-98059, LY-294002, or rapamycin, suggesting involvement of the p44/42 MAP kinase, PI 3-kinase, and p70 ribosomal protein S6 kinase signals [40]. As shown in Figure 4, TNFα treatment results in an ~90% decrease (p < 0.005) in transcript level for Tmem182 transcript. We postulated that identification of the intracellular signaling pathway(s) involved in the TNFα-mediated decrease might be one step towards gaining insights into Tmem182 function. For example, TNFα might exert its inhibitory effects by the same intracellular signaling mechanism(s) for a particular subset of genes that share similar and/or related functions in adipocytes. However, the data in Figure 4 reveal that none of the tested pharmacological inhibitors blocked the effects of TNFα, suggesting that TNFα-mediated diminution of Tmem182 transcript expression occurs via a signaling pathway(s) that is not reliant on p38 MAP kinase, p44/42 MAP kinase, p70 S6 or PI 3-kinase.
Figure 4.
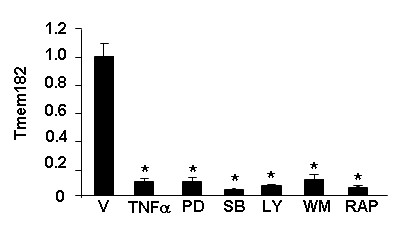
TNFα downregulates Tmem182 transcript expression in 3T3-L1 adipocytes. After serum-starvation for 6 h, 3T3-L1 adipocytes were incubated with either DMSO vehicle (V), 50 μM PD98059 (PD), 20 μM SB20358 (SB), 50 μM LY294002 (LY), 100 nM wortmannin (WM), or 1 μM rapamycin (Rap) for 1 h and then either treated or untreated with 10 ng/ml of TNFα for 16 h. RNA samples were harvested and Tmem182 transcript level was analyzed by real-time PCR as described in Methods. Values were normalized to Gapdh transcript expression level and transcript expression in vehicle alone was set to a value of 1. Statistical analysis was carried out using one-way ANOVA followed by Tukey post-hoc test; * indicates p < 0.001 for indicated samples vs. V alone (first column). Treatment with inhibitors alone failed to appreciably alter expression for Tmem182 transcript (data not shown).
Differentiation-dependent expression of Tmem182 transcript in myogenesis
The tissue profiling study in Figure 2 indicates that muscle is also a site of enriched expression of Tmem182 transcript. Adipocytes and myocytes are thought to arise in development from a shared mesenchymal stem cell precursor [8,41]. The annotation for the GenBank entry [GenBank:NM_001081198] for Tmem182 indicates it was among the transcripts identified by Kuninger and coworkers in a study of novel genes induced during growth factor-mediated muscle cell survival and differentiation [42]. However, neither the regulation of Tmem182 transcript expression during myogenesis, nor any mention of Tmem182, was in their published report [42]. To determine if Tmem182 was induced during muscle differentiation we utilized the C2C12 model of in vitro myogenesis [43]. As is shown in the left panel of Figure 5, Tmem182 transcript is markedly upregulated during the conversion of C2C12 myoblasts to myotubes. A 10-fold increase (p < 0.001) in transcript level is noted one day post induction of differentiation and cultures at 7 days post myogenic induction express ~770 times higher (p < 0.001) levels of Tmem182 transcript compared to the level in C2C12 day 0 myoblasts. The right panel of Figure 5 shows the expression of transcript for myogenin, a muscle regulatory gene that serves as a marker for myogenesis [43,44].
Figure 5.
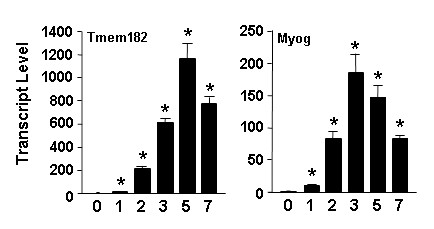
Tmem182 is a differentiation-dependent gene in C2C12 myogenesis. (A) Tmem182 transcript expression during a time course of C2C12 myogenic differentiation. C2C12 RNA was harvested at the time of induction of differentiation (0) or at indicated time points thereafter; numbers below graphs indicate days post-induction. Tmem182 (left panel) and myogenin (right panel) transcript levels were analyzed by real-time PCR as described in Methods. Values were normalized to Gapdh transcript expression level and the value in the day 0 sample set to 1. Statistical analysis was carried out using one-way ANOVA followed by Tukey post-hoc test; * indicates p < 0.001 for indicated values compared to day 0.
Summary
The primary amino acid sequence of Tmem182 predicts an evolutionarily conserved novel transmembrane protein. Tmem182 protein sequence lacks homologies with previously defined protein families and Tmem182 function is currently unknown. Enrichment of Tmem182 transcript in WAT, alteration in obesity, differentiation-dependent upregulation in adipogenesis and regulation by TNFα suggests that expression of Tmem182 may be integral to the adipocyte phenotype. Interestingly, Tmem182 transcript is also enriched in muscle tissue and it is markedly upregulated during in vitro myogenesis of C2C12 myoblasts to myocytes. This suggests Tmem182 may function in cellular pathways shared by adipocytes and myocytes but not by their respective precursor cell types. Future studies will further examine the in vitro and in vivo regulation and the function of Tmem182 in adipocytes and muscle cells.
Abbreviations
PCR: Polymerase chain reaction; FBS: Fetal bovine serum; WAT: White adipose tissue; BAT: Brown adipose tissue; PPAR: Peroxisome proliferator-activated receptor; C/EBP: CCAAT/Enhancer binding protein; Dex: Dexamethasone; Mix: Methylisobutylxanthine; PD: PD98059; SB: SB203580; LY: LY294002; WM: Wortmannin; Rap: Rapamycin
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YW conducted all real-time PCR experiments. CMS carried out the analysis in Figure 1, wrote the manuscript and conceived of the study design. Both authors contributed to data analysis and interpretation, and have read and approved of the manuscript.
Acknowledgments
Acknowledgements
We thank Dr. J.Y. Kim for kindly providing adipocyte RNA samples.
Contributor Information
Yu Wu, Email: yu.wu@utoledo.edu.
Cynthia M Smas, Email: cynthia.smas@utoledo.edu.
References
- Coppack SW. Pro-inflammatory cytokines and adipose tissue. Proc Nutr Soc. 2001;60:349–356. doi: 10.1079/pns2001110. [DOI] [PubMed] [Google Scholar]
- Scherer PE. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes. 2006;55:1537–1545. doi: 10.2337/db06-0263. [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Scherer PE. Adipose tissue-derived factors: impact on health and disease. Endocr Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- Ailhaud G, Grimaldi P, Negrel R. Cellular and molecular aspects of adipose tissue development. Annu Rev Nutr. 1992;12:207–233. doi: 10.1146/annurev.nu.12.070192.001231. [DOI] [PubMed] [Google Scholar]
- MacDougald OA, Mandrup S. Adipogenesis: forces that tip the scales. Trends Endocrinol Metab. 2002;13:5–11. doi: 10.1016/S1043-2760(01)00517-3. [DOI] [PubMed] [Google Scholar]
- Gregoire FM. Adipocyte differentiation: from fibroblast to endocrine cell. Exp Biol Med (Maywood) 2001;226:997–1002. doi: 10.1177/153537020122601106. [DOI] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Ailhaud G. Adipose tissue as a secretory organ: from adipogenesis to the metabolic syndrome. C R Biol. 2006;329:570–577. doi: 10.1016/j.crvi.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Rosen ED. The transcriptional basis of adipocyte development. Prostaglandins Leukot Essent Fatty Acids. 2005;73:31–34. doi: 10.1016/j.plefa.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Ross SE, Erickson RL, Gerin I, DeRose PM, Bajnok L, Longo KA, Misek DE, Kuick R, Hanash SM, Atkins KB, Andresen SM, Nebb HI, Madsen L, Kristiansen K, MacDougald OA. Microarray analyses during adipogenesis: understanding the effects of Wnt signaling on adipogenesis and the roles of liver X receptor alpha in adipocyte metabolism. Mol Cell Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- Gregoire FM, Smas CM, Sul HS. Understanding adipocyte differentiation. Physiol Rev. 1998;78:783–809. doi: 10.1152/physrev.1998.78.3.783. [DOI] [PubMed] [Google Scholar]
- Rajala MW, Scherer PE. Minireview: The adipocyte – at the crossroads of energy homeostasis, inflammation, and atherosclerosis. Endocrinology. 2003;144:3765–3773. doi: 10.1210/en.2003-0580. [DOI] [PubMed] [Google Scholar]
- Mueller E, Drori S, Aiyer A, Yie J, Sarraf P, Chen H, Hauser S, Rosen ED, Ge K, Roeder RG, Spiegelman BM. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J Biol Chem. 2002;277:41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- Lazar MA. PPAR gamma, 10 years later. Biochimie. 2005;87:9–13. doi: 10.1016/j.biochi.2004.10.021. [DOI] [PubMed] [Google Scholar]
- Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE, McKeon C, Darlington GJ, Spiegelman BM. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/S1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Tillison K, Smas CM. Cloning, expression, and differentiation-dependent regulation of SMAF1 in adipogenesis. Biochem Biophys Res Commun. 2005;326:36–44. doi: 10.1016/j.bbrc.2004.10.200. [DOI] [PubMed] [Google Scholar]
- Kim JY, Tillison K, Lee JH, Rearick DA, Smas C. The Adipose Tissue Triglyceride Lipase ATGL/PNPLA2 is Downregulated by Insulin and TNFalpha in 3T3-L1 Adipocytes and is a Target for Transactivation by PPARgamma. Am J Physiol Endocrinol Metab. 2006;16:16. doi: 10.1152/ajpendo.00317.2005. [DOI] [PubMed] [Google Scholar]
- Kim JY, Liu K, Zhou S, Tillison K, Wu Y, Smas CM. Assessment of fat-specific protein 27 in the adipocyte lineage suggests a dual role for FSP27 in adipocyte metabolism and cell death. Am J Physiol Endocrinol Metab. 2008;294:E654–667. doi: 10.1152/ajpendo.00104.2007. [DOI] [PubMed] [Google Scholar]
- Wu Y, Smas CM. Wdnm1-like, a new adipokine with a role in MMP-2 activation. Am J Physiol Endocrinol Metab. 2008;295:E205–215. doi: 10.1152/ajpendo.90316.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Liu K, Zhou S, Tillison K, Wu Y, Smas CM. Assessment of Fat Specific Protein 27 (Fsp27) in the Adipocyte Lineage Suggests a Dual Role for Fsp27 in Adipocyte Metabolism and Cell Death. Am J Physiol Endocrinol Metab. 2008;294 doi: 10.1152/ajpendo.00104.2007. [DOI] [PubMed] [Google Scholar]
- Kim JY, Wu Y, Smas CM. Characterization of ScAP-23, a new cell line from murine subcutaneous adipose tissue, identifies genes for the molecular definition of preadipocytes. Physiol Genomics. 2007;31:328–342. doi: 10.1152/physiolgenomics.00206.2006. [DOI] [PubMed] [Google Scholar]
- Ruan H, Hacohen N, Golub TR, Van Parijs L, Lodish HF. Tumor necrosis factor-alpha suppresses adipocyte-specific genes and activates expression of preadipocyte genes in 3T3-L1 adipocytes: nuclear factor-kappaB activation by TNF-alpha is obligatory. Diabetes. 2002;51:1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- Ruan H, Miles PD, Ladd CM, Ross K, Golub TR, Olefsky JM, Lodish HF. Profiling gene transcription in vivo reveals adipose tissue as an immediate target of tumor necrosis factor-alpha: implications for insulin resistance. Diabetes. 2002;51:3176–3188. doi: 10.2337/diabetes.51.11.3176. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Molecular mechanisms of insulin resistance and the role of the adipocyte. Int J Obes Relat Metab Disord. 2000;24:S23–27. doi: 10.1038/sj.ijo.0801497. [DOI] [PubMed] [Google Scholar]
- Sethi JK, Hotamisligil GS. The role of TNF alpha in adipocyte metabolism. Semin Cell Dev Biol. 1999;10:19–29. doi: 10.1006/scdb.1998.0273. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Uysal KT, Wiesbrock S, Yang Q, Chen H, Hotamisligil GS. Interaction of tumor necrosis factor-alpha- and thiazolidinedione-regulated pathways in obesity. Endocrinology. 2004;145:2214–2220. doi: 10.1210/en.2003-1580. [DOI] [PubMed] [Google Scholar]
- Souza SC, Palmer HJ, Kang YH, Yamamoto MT, Muliro KV, Paulson KE, Greenberg AS. TNF-alpha induction of lipolysis is mediated through activation of the extracellular signal related kinase pathway in 3T3-L1 adipocytes. J Cell Biochem. 2003;89:1077–1086. doi: 10.1002/jcb.10565. [DOI] [PubMed] [Google Scholar]
- Beutler B, Mahoney J, Le Trang N, Pekala P, Cerami A. Purification of cachectin, a lipoprotein lipase-suppressing hormone secreted by endotoxin-induced RAW 264.7 cells. J Exp Med. 1985;161:984–995. doi: 10.1084/jem.161.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti FM, Dieckmann B, Beutler B, Cerami A, Ringold GM. A macrophage factor inhibits adipocyte gene expression: an in vitro model of cachexia. Science. 1985;229:867–869. doi: 10.1126/science.3839597. [DOI] [PubMed] [Google Scholar]
- Xing H, Northrop JP, Grove JR, Kilpatrick KE, Su JL, Ringold GM. TNF alpha-mediated inhibition and reversal of adipocyte differentiation is accompanied by suppressed expression of PPARgamma without effects on Pref-1 expression. Endocrinology. 1997;138:2776–2783. doi: 10.1210/en.138.7.2776. [DOI] [PubMed] [Google Scholar]
- Jain R, Police S, Phelps K, Pekala PH. Tumour necrosis factor-alpha regulates expression of the CCAAT-enhancer-binding proteins (C/EBPs) alpha and beta and determines the occupation of the C/EBP site in the promoter of the insulin-responsive glucose-transporter gene in 3T3-L1 adipocytes. Biochem J. 1999;338:737–743. doi: 10.1042/0264-6021:3380737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JM, Pekala PH. Transcriptional repression of the C/EBP-alpha and GLUT4 genes in 3T3-L1 adipocytes by tumor necrosis factor-alpha. Regulations is coordinate and independent of protein synthesis. J Biol Chem. 1992;267:13580–13584. [PubMed] [Google Scholar]
- Pandey M, Loskutoff DJ, Samad F. Molecular mechanisms of tumor necrosis factor-alpha-mediated plasminogen activator inhibitor-1 expression in adipocytes. Faseb J. 2005;19:1317–1319. doi: 10.1096/fj.04-3459fje. [DOI] [PubMed] [Google Scholar]
- Trujillo ME, Lee MJ, Sullivan S, Feng J, Schneider SH, Greenberg AS, Fried SK. Tumor necrosis factor alpha and glucocorticoid synergistically increase leptin production in human adipose tissue: role for p38 mitogen-activated protein kinase. J Clin Endocrinol Metab. 2006;91:1484–1490. doi: 10.1210/jc.2005-1901. [DOI] [PubMed] [Google Scholar]
- Kim JY, Tillison K, Lee JH, Rearick DA, Smas CM. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3-L1 adipocytes and is a target for transactivation by PPARgamma. Am J Physiol Endocrinol Metab. 2006;291:E115–127. doi: 10.1152/ajpendo.00317.2005. [DOI] [PubMed] [Google Scholar]
- Otto TC, Lane MD. Adipose development: from stem cell to adipocyte. Crit Rev Biochem Mol Biol. 2005;40:229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- Kuninger D, Kuzmickas R, Peng B, Pintar JE, Rotwein P. Gene discovery by microarray: identification of novel genes induced during growth factor-mediated muscle cell survival and differentiation. Genomics. 2004;84:876–889. doi: 10.1016/j.ygeno.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]


