Abstract
A combination of soluble and extracellular matrix (ECM)-immobilised molecular gradients steer cell migration and location in vivo. Here, gradients of ECM molecules are formed in vitro by the combination of a surface nanopatterning technique called block copolymer nanolithography (BCN) and a biofunctionalisation technique. A modified substrate dip coating process of BCN allows for the formation of precise molecular gradients of cyclic RGDfK peptide patches at interfaces which are presented to cells for testing cell adhesion and polarisation. Surfaces formed by BCN consist of hexagonally ordered gold dot patterns with a gradient in particle spacing. Each dot serves as a chemical anchor for the binding of cyclic RGDfK peptides which are specifically recognised by αvβ3 integrins. Due to steric hindrance only up to one integrin binds to one functionalised gold dot. Applying particle spacing gradients we demonstrate how cell morphology, adhesion area, actin and vinculin distribution as well as polarisation are influenced by the peptide patch spacing gradient. As a consequence, these gradients of adhesive ligands induce cell orientation towards smaller particle spacing when the gradient strength is 15 nm/mm at least. Eventually, this measures a minimal value of ligand patch spacing sensitivity of adhesive cells which is approximately 1 nm across the cell body.
Keywords: Nanotechnology, Peptide gradient, Biofunctionalisation, Cell adhesion, Cell polarisation
Introduction
Cell polarisation and directed cell migration plays a crucial role in many physiological processes such as embryonic development, wound healing, tissue remodelling, or angiogenesis, as well as in pathological processes such as inflammatory diseases or cancer metastasis (Lauffenburger and Horwitz, 1996). Cell polarisation and migration is initiated by several external stimuli such as topography, elasticity, mechanic pressure, soluble or bound chemicals. The last is termed haptotaxis (greek: haptein, to fasten; taxis, arrangement), describing the phenomenon of directed migration along an immobilised ECM gradient (Carter, 1965). Many cells are dependent on contacts with either a substratum like the extracellular environment in a tissue or surrounding cells (Gilmore, 2005). Adhesion-dependent cells which fail to form external contacts initiate apoptosis (Gilmore, 2005). Thus, polarisation and migration of cells is often associated with cells looking for environments which allow their survival and development. In order to initiate migration, tissue cells such as fibroblasts establish a polarised morphology with spatially differentiated adhesion stability. This allows for the generation of a force which contracts the cytoskeleton and enables it to shift the body forward (Mitchison and Cramer, 1996).
The most common haptotaxis assay is performed with Boyden chambers where cells migrate across a porous membrane which is coated with ECM proteins on its lower side and the number of traversed cells is quantified (Boyden, 1962). However this system is not suitable for live-cell imaging and completely lacks any information of the molecular composition of the gradient in the porous mesh. Therefore, it is desirable to mimic transparent substrates with a defined gradient of immobilised biomolecules. Although many concentration gradient systems have already been established (DeLong et al., 2005; Kang et al., 2004), none of those can exclude local aggregation effects or possess a precise spatial control of the bound ligands. The latter, however, is crucial for the study of cooperative effects of receptor clustering and spacing in signal transduction and for understanding the mechanism of gradient sensing looking at small membrane protrusions in the micro- and nanometer size range (Maheshwari et al., 2000).
The first approaches to produce immobilised biomolecule gradients were realised by adsorbing proteins to surfaces with gradients of wettability (Fisher, 1989). Further progress in the development of more elaborated and better defined gradients was, for example, achieved with microfluidic systems (Dertinger et al., 2002), photoimmobilisation of peptides on self-assembled monolayers (Herbert et al., 1997) or by coupling electrochemical potential gradients with electrosorption reactions of organothiols (Plummer and Bohn, 2002; Plummer et al., 2003).
So far, all techniques for gradient fabrication consisted only of proteins or bioactive molecules deposited at an average density on a surface where the concentration continuously varied in average as a function of substrate position. However, controlling the average densities of biomolecules and proteins at interfaces is not sufficient to unfold molecular processes involved in cell signalling (Arnold et al., 2004; Cavalcanti-Adam et al., 2006). Therefore, a profound knowledge of local biomolecule or protein density, respectively presentation, with a resolution of single molecules is necessary for understanding the complex cell-ECM interaction at a nanometer level. The importance of designing model surfaces with nanometer accuracy is underlined by the observation that collagen fibres interact at a spatial periodicity of 67 nm with tissue cells in vivo (Poole et al., 2005). Another example is given by the process of integrin clustering which requires a maximum inter-integrin ligand spacing of 58 nm in order to form focal adhesions (Arnold et al., 2004; Cavalcanti-Adam et al., 2006). Different cellular responses can be triggered not only by varying the average bound concentration of bioactive molecules on the surface but also by their mere position relative to each other.
We chose a cyclic RGDfK peptide selective for the αvβ3 integrin (Haubner et al., 1996) to perform experiments on chemical gradient sensing of Mc3t3 osteoblasts. The formation of gradients depends on hexagonal arrays of gold nanoparticles which were fabricated on the basis of block copolymer micellar self-assembly (Spatz et al., 2000, 2002; Glass et al., 2003a,2003b). By a modified dip-coating procedure we were able to vary the spacing between individual nanoparticles continuously in the range of several mm2 on a single substrate with nanometer precision. The gold particles served as anchor points to which cyclic RGDfK peptides provided with a thiol anchor are covalently bound, called cyclic RGDfK peptide patches. Since each cyclic RGDfK patch forms a binding site for up to one integrin per patch (Wolfram et al., 2006), this model platform enabled us to quantify the response of cells to continuously changing nanometer-defined integrin-integrin spacing.
Materials and methods
Preparation of gradient surfaces
The general preparation method to form gold nanodot patterns (regular arrangement of gold nanodots) from diblock copolymer micellar solutions is described elsewhere (Spatz et al., 2000, 2002; Glass et al., 2003a,2003b; Haupt et al., 2002, 2003). For this study we used a polystyrene-block-poly[2-vinylpyridine-(HgoldCl4)] diblock copolymer (Polymer Source), consisting of a polystyrene block with in average 989 monomers and a poly[2-vinylpyridine] block with in average 385 monomer units. We dissolved 50 mg in 10 ml toluene and added 17.2 mg of tetrachloraurate(III). By providing the exact amount of gold precursor in the micellar solution the gold particles are formed in the required size of approximately 8 nm. For exact gradient formation a custom-made dipping machine was connected via a voltage-controllable power supply unit to a computer and the retraction velocity along the desired gradient length (2 mm for the samples shown here) was decreased from 40 mm/min to 8 mm/min in such a way that equivalent stretches were pulled for each velocity.
Prior to dipping, cover slides were cleaned in piranha solution (3:1 H2SO4/H2O2) for approximately 1 h, rinsed with MilliQ water in an ultra sound bath and dried under nitrogen. To remove all organic components and to reduce the gold precursor, samples were treated with H2 plasma for 40 min (H2 pressure: 0.4 mbar, power: 150 W; machine: TePla 100-E, Plasma System).
To prevent unspecific protein or cell binding, the space between the gold dots was covalently linked to polyethylene glycol (PEG, molecular weight 2000) (Blümmel et al., 2007; Arnold et al., 2004). For cell experiments the gold dots were functionalised by the cyclic RGD-based peptide, i.e. c(-RGDfK-), which consists of the cyclic adhesive peptide linked via the spacer aminohexanoic acid to mercaptopropionic acid (Kantlehner et al., 2000). For immobilisation of c(-RGDfK-)-thiols on gold dots, the PEG-functionalised substrates were immersed for 24 h in a 25-µM c(-RGDfK-)-thiol/water solution to link the molecule via the thiol group to the gold nanodots. The substrates were then rinsed extensively with MilliQ water and shaken for 24 h with several water exchanges to remove non-covalently bound c(-RGDfK-)-thiols.
Cell culture
Mc3t3 osteoblasts were cultured in DMEM (Invitrogen, Germany) supplemented with 10% fetal bovine serum (FBS) and 1 % L-glutamine (Invitrogen) at 37 °C and 10% CO2. Only cells at passage 5– 25 were used. Before plating on the patterned glass substrates, cells were detached with 0.25 % trypsin and 1 mM EDTA in Hanks’ buffered salt solution (HBSS). The samples were sterilised in 70 % ethanol for 15 min and washed with phosphate-buffered saline (PBS) at room temperature. Cells were plated at a density of 40 cells/mm2 in DMEM containing 1 % FBS and 1 % L-glutamine for all the experiments.
Cell data analysis and statistics
Cell images were taken with an Olympus IX inverted microscope placed in an incubation chamber with 5 % CO2. Phase-contrast images were taken with a 10x objective at spacings of 1 to 9 mm every mm over the whole substrate. Projected cell area, eccentricity, angle of orientation and gradient polarisation ratio (GPR) of a large number of cells were analyzed. The GPR is given by the ratio of the maximum x-width (length parallel to the gradient axis) with the maximum y-width (length perpendicular to the gradient axis) of a cell body. Data were analyzed with MatLab 7.0 and Image J 1.37x software. Data sets were compared using Student's t-test; P values less than 0.05 were considered statistically significant.
Critical point drying and scanning electron microscopy
Cells adhering to RGD-nanopatterned glass coverslips were fixed in 2 % glutaraldehyde in PBS for 15 min and then dehydrated in graded ethanol. Samples were then transferred to the chamber of the critical point dryer (CPD 030 critical point dryer; Bal-Tec) to replace ethanol with liquid CO2 at a temperature of 10 °C and a pressure of 50 bar. Then, a change in phase from liquid to gas CO2 was obtained at 40 °C and at a pressure of 74 bar.
Glass samples were sputter-coated with a carbon layer to be imaged with a field-emission scanning electron microscope (FE-SEM, LEO-1530, LEO, Oberkochen, Germany). To visualise the gold nanoparticles on the surface, an acceleration voltage of 3 kV was applied under a pressure of 5310−6 mbar. The geometrical order and nanoparticle spacing were then analyzed using ImageJ software version 1.37×.
Immunofluorescence staining
After 2.5–24 h on the substrates, as indicated, cells were washed with PBS and fixed with 3.7 % paraformaldehyde in PBS for 10 min. The cells were then permeabilised with 0.1 % Triton X-100, blocked with 1 % BSA in PBS for 10 min and incubated with a 1:50 dilution of mouse anti-human vinculin (Sigma) for 1 h at room temperature. Then the cells were labeled with 1:50 dilutions of appropriate Alexa 488-conjugated secondary antibodies (Molecular Probes, Eugene, OR, USA) in PBS for 1 h. Filamentous actin was labeled with TRITC-conjugated phalloidin (Sigma). Cells were visualised with the DeltaVision system (Applied Precision Inc., Issaquah, WA, USA) on an Olympus IX inverted microscope (Olympus, Hamburg, Germany).
Results and discussion
Development of particle spacing gradients by micellar block copolymer nanolithography
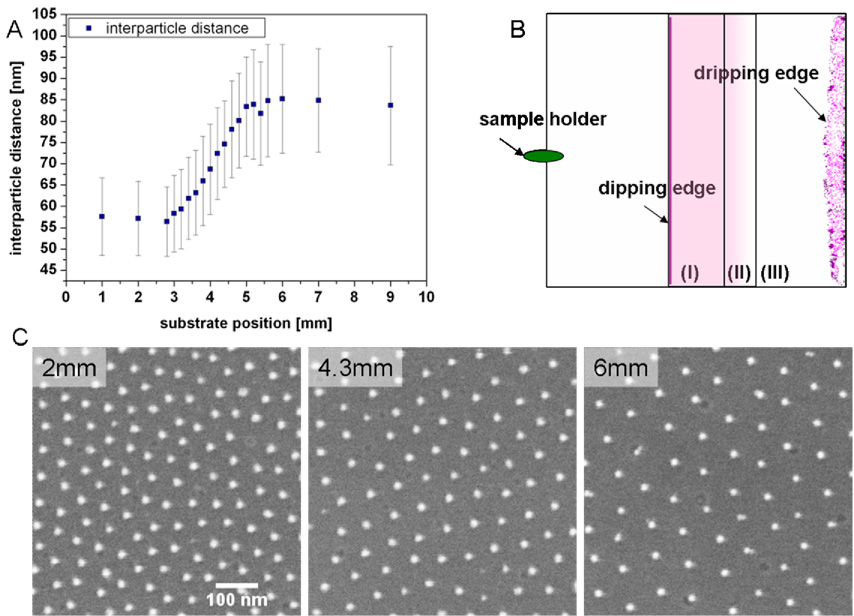
We have developed a dip-coating technique for substrates based on the self-assembly of diblock copolymer micelles resulting in hexagonally ordered gold dot patterns on flat surfaces like glass coverslips (Spatz et al., 2000, 2002; Glass et al., 2003a,2003b). The lateral spacing between the gold dots is controlled by the molecular weight of the diblock copolymers, the polymer concentration and the speed at which the substrate is retracted from the micellar block copolymer solution (Arnold, 2006). The higher the pulling rate, the thicker is the micellar film on the surface and the denser is the packing of the micelles, resulting in less particle spacing. Continuous variations of the substrate retraction speed results in the gradual variation of the spacing between the gold nanoparticles, forming the gold nanoparticle spacing gradient (Fig. 1).
Fig. 1.
Nanoparticle spacing gradient surfaces. (A) Gradient sample with a 2 mm length from substrate position 3 mm to 5 mm which covers nanodot spacings ranging from ca. 55 nm to 85 nm. Each gold particle has a diameter of approximately 6 nm. Origin of the graph indicates the dipping edge as explained in (B). Error bars represent standard deviations (standard errors ~1 %). (B) Schematic drawing of the dip-coated substrates illustrating the differently patterned areas. (I) 3 mm homogenously nanopatterned area, small constant particle spacing; (II) 2 mm particle spacing gradient; (III) homogenously nanopatterned area, large constant particle spacing, including the dripping edge which displays an area of uncontrolled nanoparticle aggregation forming upon solvent drying. (C) Scanning electron microscopy pictures taken at different positions of the substrate: area (I) at position 2 mm, area (II) at position 4.3 mm and area (III) at position 6 mm.
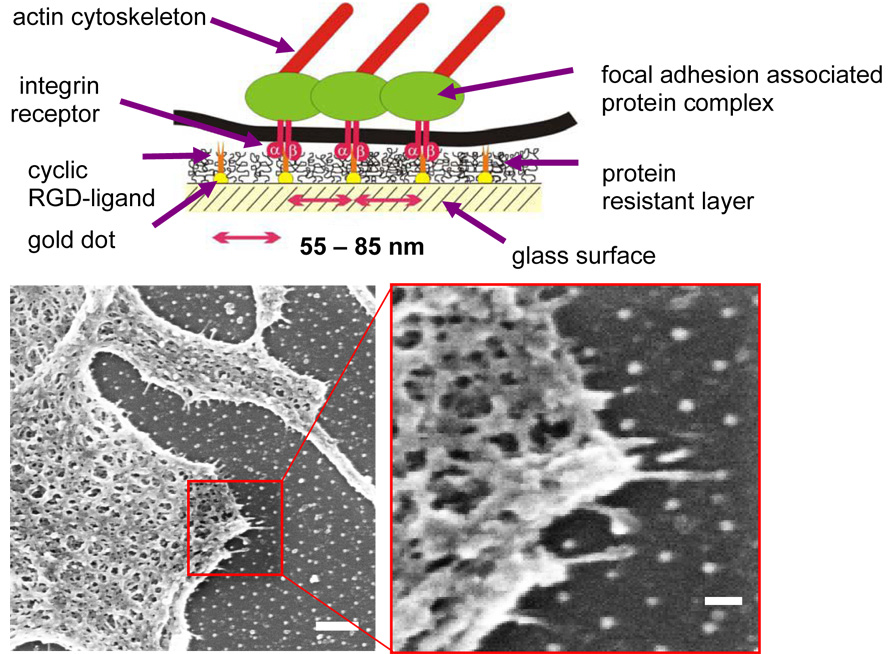
Depending on the above mentioned parameters, spacing between dots from 20 to 300 nm and gradual variations in spacing of Δ=30–40 nm per sample could be obtained (Arnold, 2006). A single dot had a diameter of approximately 6 nm. Bearing in mind that the diameter of one integrin molecule is in the range of 8–12 nm (Xiong et al., 2001), only one integrin can potentially bind to one c(-RGDfK-) peptide patch which is presented by a nanodot after biofunctionalisation. Before functionalisation of the nanodot the surrounding glass substrate was blocked with PEG which prevents adsorption of cell-derived adhesive proteins or others in the media and consequently prevents a possible non-specific adhesion of cells onto the glass surface (Fig. 2). PEG-based substrates are widely used as biologically inert interfaces (Blümmel et al., 2007). Topographic effects of the nanodots are minimised since the PEG layer when swollen by water is designed such that the same height as the height of gold dots is reached. This was confirmed by scanning force microscopy and X-ray photoelectron spectroscopy data (Blümmel et al., 2007).
Fig. 2.
(Upper panel) Scheme of biofunctionalised nanopatterns to control integrin clustering (Arnold et al., 2004): gold dots are functionalised by c(-RGDfK-) thiols; glass areas between cell-adhesive gold dots are covalently bound to polyethyleneglycol to prevent unspecific protein binding. Therefore, cell adhesion is only mediated via c(-RGDfK-)-covered gold nanodots. (Bottom panels) Mc3t3 osteoblast in contact with a biofunctionalised 80-nm pattern and exhibiting cell protrusions sensing the pattern. Bars: 20 µm (left); 200 nm (right).
Effect of c(-RGDfK-) patch spacing gradient on the substrate-projected cell area
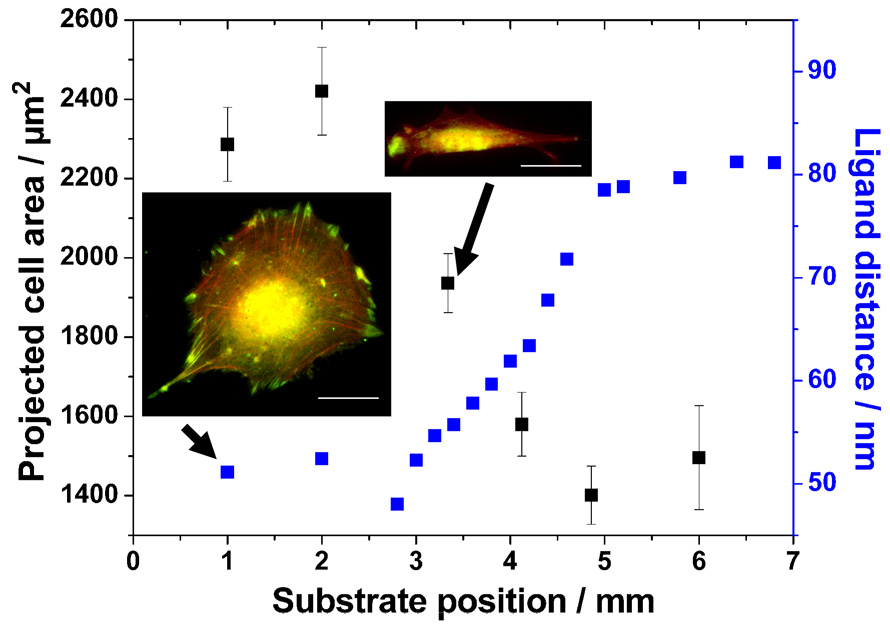
The cell adhesion area depends strongly on the strength of adhesion to the surrounding environment (DiMilla et al., 1993). Mc3t3 osteoblasts were plated onto a c(-RGDfK-) patch spacing gradient surface for approximately 23 hours and the projected cell adhesion area was evaluated. The patch spacing covered a range from ~50 nm to 80 nm along 2 mm effective length of the substrate. Thus, the gradient strength is Δ15 nm/mm. The graph of the projected cell area as a function of substrate position is shown in Figure 3. Cells on the dense part formed an average adhesion area of ~2400 µm2 while the ones attaching to c(-RGDfK-) patch spacing of ≥70 nm decreased their area to ~1500 µm2.
Fig. 3.
Projected cell area as a function of substrate position. Projected cell area along a 2-mm c(-RGDfK-) patch spacing gradient on a sample covering a spacing from 50–80 nm after 23 h cell culture. Insets: Mc3t3 osteoblasts after 23 h adherence on a homogeneously nanopatterned area with 50 nm c(-RGDfK-) patch spacing and along the spacing gradient, respectively. The latter one displays a section of the gradient which represents approximately 70 nm c(-RGDfK-) patch spacing. Cells were immunostained for vinculin (green), and actin was visualised using TRITC-phalloidin (red). Bars: 20 µm.
Formation of actin fibres and focal adhesion clusters
Focal adhesions are macromolecular assemblies linking the ECM with the intracellular cytoskeleton for mechanical anchoring, cell shape control and biochemical signalling. When integrins bind to matrix ligands, clustering of integrins is initiated and actin filaments are coupled via anchor proteins to the intracellular domain of the receptor which together form the focal adhesion. In Figure 3 representative cells on a homogenous 50-nm pattern and along the gradient at a position where the c(-RGDfK-) patch spacing is ~70 nm are shown. Cells were stained for vinculin (green) and actin (red). The cell on the small, homogeneous spacing pattern presents well developed, major vinculin clusters symmetrically distributed over the periphery of the cell. Well-established actin stress fibres connect mature focal adhesions. Along the gradient we found highly elongated cells with fine, aligned actin stress fibres and rather unorganised vinculin aggregates mainly at the front and the rear of the cell body. Remarkably, the size of the vinculin clusters is considerably larger towards the gradient direction which offers a smaller c(-RGDfK-) patch spacing. Actin stress fibres are more dominant towards smaller dimensions indicating an asymmetry in actin distribution. Still, focal adhesions of both cellular sides are connected via actin stress fibres. Mechanical tension generated between these focal adhesions by myosin-activated actin stress fibre contraction might enable the cell to test the differential stability of connected focal adhesions. This might result in deleting weak focal adhesions and indicate a mechanism by which cells may orient towards more stable ECM adhesion areas. These results are in agreement with earlier findings (Arnold et al., 2004; Cavalcanti-Adam et al., 2006, 2007) where an increase of the spacing between the adhesive dots to more than 73 nm resulted in limited cell adhesion, cell spreading and formation of focal adhesions.
Cell orientation
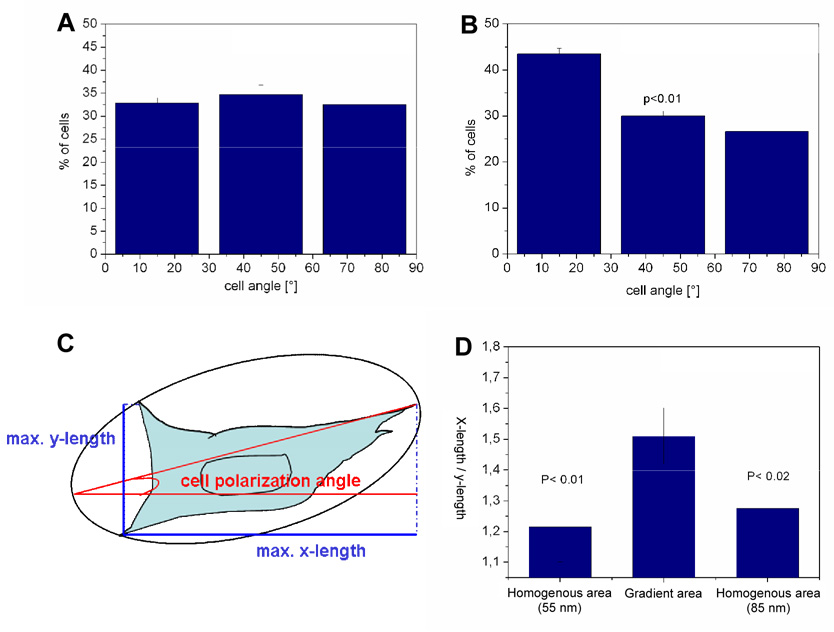
Cell orientation was determined for each cell by fitting an ellipse to the cell body and analysing the angle of the major axis, called cell body axis, relative to the direction of the c(-RGDfK-) gradient (Fig. 4C). The cell orientation angle measurements after 23 hours cell culture indicated a significantly higher amount of cells to be aligned in the direction of the gradient compared to cells adhering to homogenously nanopatterned areas (Fig. 4A, B). The analysis of the gradient polarisation ratio (GPR) which is the ratio of the maximal width of the cell shape parallel and vertical to the gradient axis (scheme in Fig. 4C) presents a significantly larger GPR value along the gradient than for cells on small (50 nm) or large (80 nm) homogenously c(-RGDfK-) patterned areas (Fig. 4D). Therefore, the cell cortex must be polarised in response to the anisotropy of the cell-adhesive environment which is also retrieved from the distribution of actin and vinculin in Figure 3. The length of a cell adhering along a gradient with a strength of Δ15 nm/mm after 23 hours (Fig. 3) equals ca. 70 µm. Calculating the particle spacing between the front and back of such a cell gives ~ 1 nm. This is the minimum average change in c(-RGDfK-) patch spacing a cell requires to sense between its front and back for 23 h in order to interpret the environment and to orient itself along the gradient direction.
Fig. 4.
Cell polarisation along c(-RGDfK-) patch spacing gradients. Angles of the main cell body axis with the gradient direction after 23 h on 55 nm homogenously nanopatterned samples (A) and on the c(-RGDfK-) patch spacing gradient ranging from 50 to 80 nm spacing along a 2 mm substrate length (B). Data were acquired from three separate experiments. For graph (A) 347 cells and graph (B) 428 cells were evaluated, error bars present standard error, p<0.01. (C) Describes the analysis of the main cell body angle with the direction of the gradient and the gradient polarisation ratio (GPR). The GPR is given by the ratio of the maximum x-width (length parallel to the gradient axis) with the maximum y-width (length perpendicular to the gradient axis) of a cell body. GPR of a cell adhering on homogeneously nanopatterned areas (50 nm, 80 nm) and on the c(-RGDfK-) patch spacing gradient area with spacing ranging from 50 to 80 nm along 2 mm substrate length after 23 h are given in (D).
Conclusion
The c(-RGDfK-) patch spacing gradient fabrication method is of great ease and highly adjustable with respect to gradient strength and ligand patch spacing. The projected cell area and the cell shape give clear indications of the sensitivity of the cell to ligand patch spacing gradients. Mc3t3 osteoblasts polarised their cell body towards smaller c(-RGDfK-) interligand spacings if an average ligand patch spacing difference of at least 1 nm between the back and the front of a cell is presented to cells. Differential strength of oppositely placed focal adhesions might be tested by coupling focal adhesion through myosin-activated, contracting actins stress fibres. This results in deleting weak focal adhesions and indicates a mechanism by which cells may orient towards more stable ECM adhesion areas.
Acknowledgement
We are thankful to Prof. Benjamin Geiger for general and detailed discussions. Thanks go to Dr. Jacques Blümmel, Nicole Plath and Nadine Perschmann for synthesising mPEG-triethoxysilanes, Thorsten Brach and Ann-Marie Michalski for technical assistance, Christian Hökl, Dr. Patrick Heil, Dr. Philippe Girard, and Dr. Jennifer Curtis for writing the Image Analysis program. The work was supported by the Landesstiftung Baden-Württemberg within the framework of the programme “Spitzenforschung Baden-Württemberg” and partly by the National Institutes of Health through the NIH Roadmap for Medical Research (PN2 EY016586). M. López-García thanks the Humboldt foundation for a grant. This work was also supported by the Max Planck Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold M. PhD thesis. Heidelberg, Germany: University of Heidelberg; 2006. Molecularly defined nanostructured interfaces as tools for the regulation and measurement of functional length scales in cell adhesion mediating protein clusters. [Google Scholar]
- Arnold M, Cavalcanti-Adam EA, Glass R, Blümmel J, Eck W, Kanthlener M, Kessler H, Spatz JP. Activation of integrin function by nanopatterned adhesive interfaces. Chem. Phys. Chem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- Blümmel J, Perschmann N, Aydin D, Drinjakovic J, Surrey T, López-García, M, Kessler H, Spatz JP. Protein repellent properties of covalently attached PEG coatings on nanostructured SiO2 based interfaces. Biomaterials. 2007;28:4739–4747. doi: 10.1016/j.biomaterials.2007.07.038. [DOI] [PubMed] [Google Scholar]
- Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J. Exp. Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SB. Principles of cell motility: the direction of cell movement and cancer invasion. Nature. 1965;208:1183–1187. doi: 10.1038/2081183a0. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam EA, Micoulet A, Blümmel J, Goldernheimer J, Kessler H, Spatz JP. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell Biol. 2006;85:219–224. doi: 10.1016/j.ejcb.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong SA, Moon JJ, West JL. Covalently immobilized gradients of bFGF on hydrogel scaffolds for directed cell migration. Biomaterials. 2005;26:3227–3234. doi: 10.1016/j.biomaterials.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Dertinger SKW, Jiang X, Li Z, Murthy VN, Whitesides GM. Gradients of substrate-bound laminin orient axonal specification of neurons. Proc. Natl. Acad. Sci. USA. 2002;99:12542–12547. doi: 10.1073/pnas.192457199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J. Cell Biol. 1993;122:729–737. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher PR, Merkl R, Gerisch G. Quantitative analysis of cell motility and chemotaxis in Dictyostelium discoideum by using an image processing system and a novel chemotaxis chamber providing stationary chemical gradients. J. Cell Biol. 1989;108:973–984. doi: 10.1083/jcb.108.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP. Anoikis. Cell Death Differ. 2005;12:1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- Glass R, Arnold M, Blümmel J, Küller A, Möller M, Spatz JP. Micronanostructured interfaces fabricated by the use of inorganic block copolymer micellar monolayers as negative resist for electron beam lithography. Adv. Functional Materials. 2003a;13:569–575. [Google Scholar]
- Glass R, Möller M, Spatz JP. Block copolymer micelle nanolithography. Nanotechnology. 2003b;14:1153–1160. [Google Scholar]
- Haubner R, Gratias R, Diefenbach B, Goodman SL, Jonczyk A, Kessler H. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αvβ3 antagonists. J. Am. Chem. Soc. 1996;118:7461–7472. [Google Scholar]
- Haupt M, Miller S, Ladenburg A, Sauer R, Thonke K, Spatz JP, Riethmüller S, Möller M, Banhart F. Semiconductor nanostructures defined with self-organized polymers. J. Appl. Phys. 2002;91:6057–6059. [Google Scholar]
- Haupt M, Glass R, Ladenburger A, Rauscher H, Roos W, Riethmüller S, Möller M, Sauer R, Thonke K, Spatz JP. Ultraviolet-emitting ZnO nanowhiskers of highest crystal quality. J. Appl. Phys. 2003;93:6252–6257. [Google Scholar]
- Herbert CB, McLernon TL, Hypolite CL, Adams DN, Pikus L, Huang CC, Fields GB, Letourneau PC, Distefano MD, Hu WS. Micropatterning gradients and controlling surface densities of photoactivatable biomolecules on self-assembled monolayers of oligo (ethylene glycol) alkanethiolates. Chem. Biol. 1997;4:731–737. doi: 10.1016/s1074-5521(97)90311-2. [DOI] [PubMed] [Google Scholar]
- Kang CE, Gemeinhart EJ, Gemeinhart RA. Cellular alignment by grafted adhesion peptide surface density gradients. J. Biomed. Mater. Res. A. 2004;71:403–411. doi: 10.1002/jbm.a.30137. [DOI] [PubMed] [Google Scholar]
- Kantlehner M, Schaffner P, Finsinger MJ, Jonczyk A, Diefenbach B, Nies B, Holzemann G, Goodman SL, Kessler H. Surface coating with cyclic RGD peptides stimulates osteoblast adhesion and proliferation as well as bone formation. Chem. Biochem. 2000;1:107–114. doi: 10.1002/1439-7633(20000818)1:2<107::AID-CBIC107>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Plummer ST, Bohn PW. Spatial dispersion in electrochemically generated surface composition gradients visualized with covalently bound fluorescent nanospheres. Langmuir. 2002;18:4142–4149. [Google Scholar]
- Plummer ST, Wang Q, Bohn PW, Stockton R, Schwartz MA. Electrochemically derived gradients of the extracellular matrix protein fibronectin on gold. Langmuir. 2003;19:7528–7536. [Google Scholar]
- Poole K, Khairy K, Friedrichs J, Franz C, Cisneros DA, Howard J, Mueller D. Molecular-scale topographic cues induce the orientation and directional movement of fibroblasts on two-dimensional collagen surfaces. J. Mol. Biol. 2005;349:380–386. doi: 10.1016/j.jmb.2005.03.064. [DOI] [PubMed] [Google Scholar]
- Spatz JP, Möβmer S, Hartmann C, Möller M, Herzog T, Krieger M, Boyen H-G, Ziemann P, Kabius B. Ordered deposition of inorganic clusters from micellar block copolymer films. Langmuir. 2000;16:407–415. [Google Scholar]
- Spatz JP, Chan VZH, Möβmer S, Kamm FM, Plettl A, Ziemann P, Möller M. A combined top down/bottom up approach for nanolithography. Adv. Materials. 2002;14:1827–1832. [Google Scholar]
- Wolfram T, Belz F, Schön T, Spatz JP. Site-specific presentation of single recombinant proteins in defined nanoarrays. Biointerphases. 2006;2:44–48. doi: 10.1116/1.2713991. [DOI] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αvβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]