Abstract
We generated transgenic mice expressing chimeric receptors, which comprise extracellular domains of the human granulocyte–macrophage colony-stimulating factor (hGM-CSF) receptor and transmembrane and cytoplasmic domains of the mouse leukemia inhibitory factor receptor. In suspension cultures of lineage-negative (Lin−), 5-fluorouracil-resistant bone marrow cells of the transgenic mice, a combination of hGM-CSF and stem cell factor (SCF) induced exponential expansions of mixed colony-forming unit. The combination of hGM-CSF and SCF was effective on enriched, Lin−Sca-1+c-kit+ progenitors and increased either mixed colony-forming unit or cobblestone area–forming cells. In case of stimulation with hGM-CSF and SCF, interleukin-6 (IL-6) and SCF, or IL-11 and SCF, the most efficient expansion was achieved with hGM-CSF and SCF. When Lin−Sca-1+c-kit+CD34− further enriched progenitors were clone sorted and individually incubated in the presence of SCF, hGM-CSF stimulated a larger number of cells than did IL-6, IL-6 and soluble IL-6 receptor (IL-6R), or IL-11. These data suggest the presence of IL-6Rα-, IL-11Rα-, and gp130-low to -negative primitive hematopoietic progenitors. Such primitive progenitors are equipped with signal transduction molecules and can expand when these chimeric receptors are genetically introduced into the cells and stimulated with hGM-CSF in the presence of SCF.
INTRODUCTION
It is widely accepted that multistage developmental processes from multipotential hematopoietic stem cells (HSCs) to terminal differentiation in various lineages are supported by variety of cytokines. Although various combinations of cytokines have also been tested regarding amplification of HSCs (Holyoake et al., 1996; Yonemura et al., 1997), mechanisms governing the amplification of HSCs are not well understood, mostly because receptors and downstream signal transduction pathways in HSCs have remained to be clarified. To investigate related factors, we wanted to introduce into primitive hematopoietic progenitors genetically manipulated signal-transducing molecules to modify their proliferation and differentiation.
New technologies using retrovirus vector, adenovirus vector, or adeno-associated virus vector have been developed to deliver genes into stem cells. However, gene transfer into immature hematopoietic cells with high efficiency has required more study. Several investigators, including our group, have used transgenic mice, and we found that use of related technology is a pertinent approach to introduce genes into hematopoietic cells and to analyze effects of the gene products on their commitment (Nishijima et al., 1995; Takagi et al., 1995; Kirby et al., 1996). To determine functions of introduced molecules, these molecules have to be turned on by extracellular signals, with no untoward effects on endogenous molecules. For this purpose, the human granulocyte–macrophage colony-stimulating factor (hGM-CSF)/hGM-CSF receptor (CSFR) system can serve as a switch, because it functions on target cells in a species-specific manner (Muto et al., 1995; Nishijima et al., 1995).
Although little is known of signal transduction pathways required for expansion of HSCs, signals from gp130, a common component shared with interleukin 6 (IL-6), IL-11, leukemia inhibitory factor (LIF), oncostatin M, ciliary neurotrophic factor, and the cardiotrophin-1 receptor system, are considered to transmit essential signals in hematopoiesis (Kishimoto et al., 1995), because targeted disruption of gp130 leads to serious hematological disorders in embryogenesis (Yoshida et al., 1996). In addition, LIF-deficient mice obtained by gene targeting have decreased numbers of HSCs (Escary et al., 1993), which suggests that LIF receptor (LIFR) signaling is also required for the maintenance of HSCs. Thus, to control expansion of primitive hematopoietic progenitors, use of the intracellular signaling of LIFR was considered to be appropriate.
We generated transgenic mice expressing chimeric receptors, which contained the extracellular domains of hGM-CSFR (α and β) and transmembrane and cytoplasmic domains of mouse LIFR (LIFRβ and gp130). Analysis of hematopoietic cells using fluorescence-activated cell sorting (FACS) or transgenic mouse technology revealed that primitive hematopoietic progenitors are included in the IL-6Rα− gp130+ population (Tajima et al., 1996; Peters et al., 1997). Using our chimeric receptor transgenic mice, we wanted to determine whether there is an IL-6Rα− gp130− more primitive hematopoietic progenitor population which can expand when the hGM-CSF and mouse LIF (mLIF) chimeric receptor genes are genetically introduced and expressed in cells.
MATERIALS AND METHODS
Construction of hGM-CSFR–mLIFR Chimeric Receptor Gene and Generation of Transgenic Mice
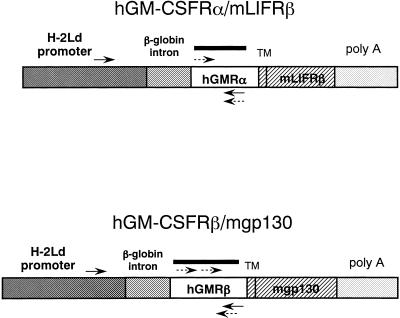
The chimeric receptor constructions, which carry the extracellular domain of hGM-CSFRα or β linked to transmembrane and cytoplasmic regions of either mLIFRβ or mgp130, were generated as described (Nakamura et al., 1998). The chimeric receptor genes were designed to be under transcriptional control of the mouse major histocompatibility complex class I (H-2Ld) promoter (Figure 1) (Suematsu et al., 1992). Both H-2Ld-hGM-CSFRα–mLIFRβ and H-2Ld-hGM-CSFRβ–mgp130 fragments were coinjected into pronuclei of fertilized eggs of C57BL/6 mice as described (Hogan et al., 1994). The chimeric receptor transgenic mice were screened by PCR and Southern blot analysis of tail DNA using hGM-CSFRα and β cDNA fragment as probes (Figure 1). Mice were bred and maintained at our animal facility under specific pathogen-free conditions.
Figure 1.
Construction of chimeric receptor. Chimeric receptor was constructed by linking the extracellular domain of hGM-CSFRα or hGM-CSFRβ to the transmembrane and cytoplasmic region of mLIFRβ or mgp130, respectively. Both transgenes were under transcriptional control of a mouse H-2Ld promoter. Bars represent the probes for genomic Southern blotting. Arrows and hashed arrows represent the primers for genomic PCR and RT-PCR, respectively.
Reverse Transcription (RT)-PCR
Total RNAs were isolated from various tissues and cells of these mice using Trizol reagent (Life Technologies, Grand Island, NY). Polyinosinic acid (10 μg; Clontech Laboratories, Palo Alto, CA) was added as a carrier when RNA was extracted from lineage-negative (Lin−)Sca-1+c-kit+CD34− bone marrow (BM) cells. In all cases, RNA preparations were subjected to a DNase I (Ambion, Austin, TX) digestion step before cDNA synthesis, thus eliminating any remaining genomic DNA. The first-strand cDNA synthesis was performed with d(T)18 or d(N)6 primer using SuperScript II RNaseH− reverse transcriptase (Life Technologies) for 1 h at 37°C. Using oligonucleotides of hGM-CSFRα (sense, nucleotide positions 192–211; antisense, 851–868), hGM-CSFRβ (sense, 785–804; antisense, 958–977), and mouse glyceraldehyde-3-phosphate dehydrogenase (G3PDH, as a positive control; sense, 759–783; antisense, 976–998) as primers, PCR was run for 35 cycles (hGM-CSFRα and mG3PDH: 94°C for 30 s, 60°C for 30 s, and 72°C for 1 min; hGM-CSFRβ: 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min), followed by 7 min at 72°C. We used a DNA thermocycler (Gene Amp PCR system 2400; Perkin-Elmer, Foster City, CA). To check for genomic DNA contamination, a control without reverse transcriptase reaction was always included. In the case of RT-PCR analysis of Lin−Sca-1+c-kit+CD34− BM cells, the first PCR was run for 40 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min), followed by 7 min at 72°C, using oligonucleotides of hGM-CSFRα (sense, 192–211; antisense, 851–868), hGM-CSFRβ (sense, 321–340; antisense, 957–976), mgp130 (sense, 1214–1234; antisense, 1853–1873), mIL-6Rα (sense, 657–677; antisense, 1348–1368), and mIL-11Rα (sense, 381–401; antisense, 1096–1116) as primers. After removal of primers using a QIAquick PCR purification kit (Qiagen, Valencia, CA), the first PCR product was used for the second nested PCR. The second PCR was run for 40 cycles under the same conditions as the first PCR, using oligonucleotides of hGM-CSFRα (sense, 213–232; antisense, 791–810), hGM-CSFRβ (sense, 371–391; antisense, 906–925), mgp130 (sense, 1390–1410; antisense, 1833–1853), mIL-6Rα (sense, 746–766; antisense, 1281–1301), and mIL-11Rα (sense, 427–447; antisense, 835–855) as primers.
Flow Cytometry
BM cells from transgenic mice and their normal littermates were prepared after removing red blood cells with use of ammonium chloride buffer solution. After incubation with 300 ng of anti-mouse CD16/CD32 (Fcγ III/II receptor, 2.4G2; PharMingen, San Diego, CA) mAb, ∼3 × 105 cells in 30 μl of PBS containing 5% fetal bovine serum (FBS; Life Technologies) were incubated with 300 ng of mouse anti-hGM-CSFRα (S-20, immunoglobulin G2a [IgG2a]; Santa Cruz Biotechnology, Santa Cruz, CA) or mouse anti-hGM-CSFRβ (S-16, IgG; Santa Cruz Biotechnology) for 30 min at 4°C. Cells were pelleted, washed, and incubated with FITC-conjugated goat anti-mouse IgG (heavy and light chain [H + L]; Boehringer Mannheim, Indianapolis, IN) for 30 min at 4°C. These cells were washed, resuspended in 1 ml of PBS, and analyzed on a FACScan (Becton Dickinson, Mountain View, CA).
Cell Preparation
BM cells and spleen cells from 8- to 20-wk-old chimeric receptor transgenic mice were used. BM cells were flushed from femurs and tibiae into α-medium (Life Technologies), using a 26-gauge needle. Spleen cells were prepared by teasing the spleen in 3 ml of α-medium in a 35-mm suspension culture dish (171099; Nunc, Naperville, IL) and by repeated pipetting. Red blood cells were lysed with an ammonium chloride buffer. BM cells or spleen cells were passed through a 70-μm nylon cell strainer (2350; Becton Dickinson Labware, Franklin Lakes, NJ). For some experiments, 150 mg/kg body weight 5-fluorouracil (5-FU) (Sigma, St. Louis, MO) was administered through tail veins of mice. BM cells were harvested 2 d after the 5-FU injection. BM cells were enriched for progenitors by negative selection using immunomagnetic beads (Dynabeads M-450, coated with sheep anti-rat IgG; Dynal, Oslo, Norway) and a mixture of mAbs specific for CD45R/B220 (RA3-6B2), CD4 (RM4-5), CD8 (53-6.72), Gr-1 (RB6-8C5), and CD11b (M1/70, Mac-1 α chain) (each purchased from PharMingen). For some experiments, the Lin− cells were then stained with Texas Red (TXR)-conjugated anti-rat IgG (H + L; Caltag, Burlingame, CA), FITC-conjugated anti-mouse Ly6A/E (E13-161.7, Sca-1, rat IgG2a; PharMingen) and phycoerythrin (PE)-conjugated anti-mouse CD117 (2B8, c-kit, rat IgG2b; PharMingen), and the TXR−FITC+PE+ population (Lin−Sca-1+c-kit+) was sorted, based on cells stained with FITC-rat IgG2a (PharMingen) and PE-rat IgG2b (PharMingen) as isotype-matched controls with a FACS Vantage (Becton Dickinson), as described (Okumura et al., 1996).
Clone Sorting and Single-Cell Culture
Lin− cells were stained with TXR-conjugated anti-rat IgG (H + L), PE-conjugated anti-mouse Sca-1 (PharMingen), allophycocyanin (APC)-conjugated anti-mouse c-kit (PharMingen), and FITC-conjugated anti-mouse CD34 (RAM34, rat IgG2a; PharMingen), and the TXR−PE+ APC+ population (Lin−Sca-1+c-kit+) was gated, based on cells stained with PE-rat IgG2a (PharMingen) and APC-rat IgG2b (PharMingen), as isotype-matched controls. Finally, a TXR−PE+APC+ FITC− (Lin−Sca-1+c-kit+CD34−) population was obtained, based on cells stained with FITC-rat IgG2a (PharMingen), PE-anti-mouse Sca-1, and APC-anti-mouse c-kit as isotype-matched controls. Individual Lin−Sca-1+c-kit+CD34− cells were sorted into each well of a 96-well flat-bottom plate (Falcon 3072) with a FACS Vantage equipped with an automatic cell deposition unit (Becton Dickinson). Each well contained 200 μl of serum-free medium. After confirming the presence of a single cell in each well using an inverted microscope, cytokines were added to each well, followed by incubation of the preparation at 37°C in a humidified atmosphere with 5% CO2 in air.
Receptor and Cytokines
Recombinant mIL-3, rat stem cell factor (SCF), hGM-CSF, hIL-6, hIL-11, human megakaryocyte differentiation and growth factor (MDGF), and human erythropoietin (Epo) were kindly provided by Amgen (Thousand Oaks, CA). Recombinant human soluble IL-6R (sIL-6R) was kindly provided by Biotechnology Research Laboratory, Tosoh (Kanagawa, Japan). Concentrations of growth factors used in this study were as follows: IL-3, 10 ng/ml; IL-6, 100 ng/ml; IL-11, 100 ng/ml; GM-CSF, 100 ng/ml; MDGF, 10 ng/ml; Epo, 2 U/ml; SCF, 100 ng/ml; and soluble IL-6R (sIL-6R), 1000 ng/ml.
In Vitro Colony Assay
Methylcellulose clonal culture was carried out in 35-mm suspension culture dishes (171099; Nunc) as described (Nakahata and Ogawa, 1982). One milliliter of culture mixture consisted of α-medium, 0.9% 4000 centipoise methylcellulose (Sigma), 30% FBS (Hyclone, Logan, UT), 1% deionized fraction V BSA (Sigma), 100 μM 2-mercaptoethanol (Sigma), and hematopoietic growth factors. Dishes were incubated at 37°C in a humidified atmosphere with 5% CO2 in air. Colony types were determined on days 7–16 of culture by in situ observation using an inverted microscope, according to the criteria described elsewhere (Nakahata et al., 1982). To assess the accuracy of in situ identification of the colonies, individual colonies were lifted using an Eppendorf micropipette under direct microscopic visualization, spread on glass slides using a cytocentrifuge (Cytospin 2; Shandon Southern Instruments, Sewickly, PA), and then stained for May-Grünwald-Giemsa and acetylcholinesterase. Abbreviations of colony types are Meg, megakaryocyte colonies; and Mix, colonies consisting of more than trilineages including granulocyte–macrophage–megakaryocyte colonies, granulocyte–erythrocyte–macrophage colonies, and granulocyte–erythrocyte–macrophage–megakaryocyte colonies.
Serum-free Suspension Culture
Serum-free suspension culture was carried out in 24-well plates (3047; Becton Dickinson) as described (Iscove and Melchers, 1978; Koike et al., 1988). Cells were incubated in 1 ml of α-media containing 1% deionized, crystallized BSA (Sigma), 300 μg/ml 30% iron-saturated human transferrin (Boehringer Mannheim), 160 μg/ml soybean lecithin (Sigma), 96 μg/ml cholesterol (Sigma), and 100 nM sodium selenite (Sigma).
Cobblestone Area Formation on OP9 Stromal Cell
Cobblestone area forming cell (CAFC) development was measured using a mouse OP9 stromal cell line (Kodama et al., 1994). This cell line was provided by RIKEN Cell Bank (Tsukuba, Japan) with permission of Dr. H. Kodama (Bayer Yakuhin, Nara, Japan). OP9 stromal cells were maintained in α-medium containing 10% FBS in a 24-well plate previously coated with type I-A collagen (Nitta Gelatin, Osaka, Japan). One hundred Lin−Sca-1+c-kit+ cells were preincubated with cytokine combinations in serum-free suspension cultures for 4 d. After three washes with α-medium, the proliferated cells were added to a well of OP9 stromal cell culture and then incubated at 37°C with 5% CO2 without adding cytokines. On the other hand, with no preincubation, the same number of Lin−Sca-1+c-kit+ cells was directly cocultured with OP9 stromal cells in the same manner. On day 7, numbers of cobblestone areas seen under an inverted phase-contrast microscope were counted. On days 12, 20, and 28, nonadherent cells were harvested by three gentle washes of the well with α-medium then analyzed for colony formation in the presence of IL-3, IL-6, SCF, Epo, and MDGF. After harvesting the nonadherent cells, freshly prepared media were added, and the coculture was continued up to day 28.
RESULTS
Generation of Transgenic Mice
To ubiquitously express the hGM-CSFR–mLIFR chimeric receptor, we used the H-2Ld promoter (Figure 1). We found that the chimeric receptor could transduce self-renewal signals in embryonic stem cells in response to exogenous hGM-CSF (Nakamura et al., 1998). To make use of Ly6A/E (Sca-1) as a marker of HSCs, we used the C57BL/6 strain to generate the transgenic mice. Both H-2Ld-hGM-CSFRα–mLIFRβ and H-2Ld-hGM-CSFRβ–mgp130 fragments were coinjected into pronuclei of fertilized eggs of C57BL/6 mice. Among 343 offspring, 14 founder mice carried both α and β transgenes. Finally, expression of both α and β chains of chimeric receptor were confirmed by FACS analysis for both BM cells and thymus cells in 11 mice. The results of in vitro colony formation, in response to hGM-CSF, showed the same tendency; therefore, one representative chimeric receptor transgenic mouse line, maintained by crossing with C57BL/6 mice, was used for further analysis. We examined the heterozygote chimeric receptor transgenic mice in this study. In all experiments, we used as negative controls normal littermates with genetic backgrounds identical to those of the chimeric receptor transgenic mice.
Expression of Chimeric Receptors in the Transgenic Mice
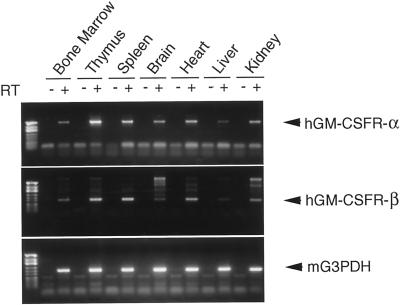
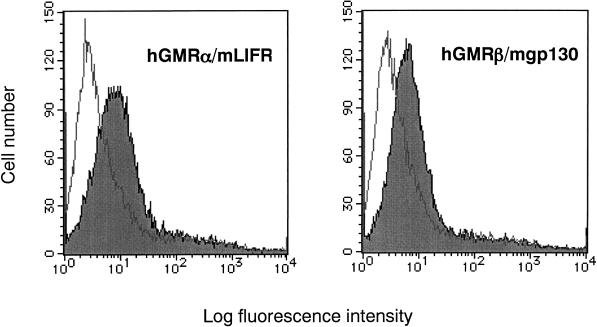
RT-PCR revealed that the transgenes were ubiquitously expressed in BM, thymus, spleen, brain, liver, and kidney (Figure 2). Expression of the both α and β chains in BM cells was also confirmed, using FACS analysis (Figure 3). The chimeric receptor transgenic mice appeared normal and healthy, currently up to 2 y of age, and bred well. There were no significant differences in blood cell count and the total blood picture between chimeric receptor transgenic mice and their normal littermates (our unpublished results).
Figure 2.
RT-PCR analysis of transgene expression. RNA was prepared from various tissues and cells of the adult chimeric receptor transgenic mice. The lane marked − is the PCR product of a mock cDNA (no reverse transcriptase included in the cDNA synthesis reaction). First lane is DNA size marker (HinfI-digested φX174). The expected sizes of the PCR products were α, 677 bp; β, 193 bp; and G3PDH, 240 bp.
Figure 3.
Expression of chimeric receptors on BM cells of transgenic mice. Expression of chimeric receptors was confirmed by flow cytometry with FACScan, using mAbs directed against the α and β subunits of the hGM-CSF receptor.
hGM-CSF Stimulates In Vivo Hematopoiesis of Chimeric Receptor Transgenic Mice
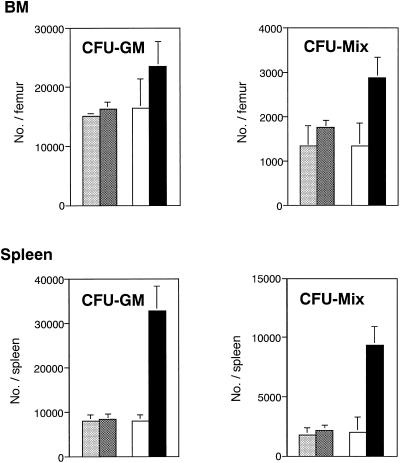
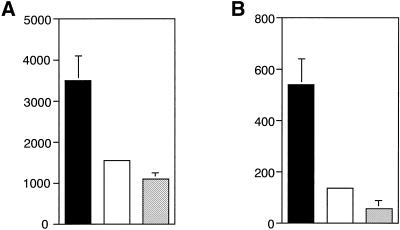
The in vivo effects of hGM-CSF were first examined. After injection of 10 μg/kg hGM-CSF per day subcutaneously for 2 wk, white blood cell and platelet counts in peripheral blood of the chimeric receptor transgenic mice increased 1.5 and 1.3 times, respectively, although no change in differential cell count of white blood cells was noted (our unpublished results), and change in red cell counts was nil (our unpublished results). Next, hematopoietic cells in spleen, BM, and peripheral blood were examined. The number of spleen cells increased to approximately double in the chimeric receptor transgenic mice, and both CFU-GM and CFU-Mix in spleens increased dramatically (Figure 4). Although no change was found in BM cell counts, CFU-Mix in BM also increased. In contrast, no hematopoietic change was observed in normal littermates after injection of hGM-CSF. These data suggest that hGM-CSF stimulated in vivo hematopoiesis of the chimeric receptor transgenic mice through receptors introduced into the hematopoietic cells. However, one would need to rule out the possibility that factors induced by hGM-CSF from nonhematopoietic cells stimulated the hematopoietic cells in vivo.
Figure 4.
hGM-CSF effects on in vivo hematopoiesis of in chimeric receptor transgenic mice. Three hundred nanograms (10 μg/kg) of hGM-CSF were injected subcutaneously once per day for 14 d to three chimeric receptor transgenic mice (▪) and three normal littermates (▩), which were then killed. BM cells and spleen cells were harvested from a femur and a whole spleen, respectively. After counting the total number, the cells were analyzed for in vitro colony assay in the presence of IL-3, IL-6, SCF, Epo, and MDGF. Separately, three chimeric receptor transgenic mice (□) and 3 normal littermates (░⃞) were killed without giving hGM-CSF injection, and clonogenic cells in BM and spleen were evaluated in the same manner. All of the mice used in this experiment were 20 wk old.
In Vitro Colony Formation and Expansion of Clonogenic Cells in Response to hGM-CSF
To examine the direct effects of hGM-CSF on proliferation and differentiation of hematopoietic progenitors of chimeric receptor transgenic mice, fresh BM cells were analyzed for in vitro colony formation. Although hGM-CSF had no effects on BM cells of normal littermates, it did stimulate developments of GM colonies, erythroid bursts, and mixed colonies from BM cells of the chimeric receptor transgenic mice in the presence of Epo (Table 1). The numbers of colonies reached a maximum with 100 ng/ml hGM-CSF (our unpublished results). In the absence of Epo, hGM-CSF supported only nonerythroid GM and granulocyte–macrophage–megakaryocyte colonies (our unpublished results). The lack of erythroid colony formation in the presence of hGM-CSF alone indicates the inability of hGM-CSF to substitute for Epo. In the culture of Lin− BM cells from 5-FU-treated chimeric receptor transgenic mice, hGM-CSF and SCF synergistically enhanced the mixed colony formations (Table 1) in the presence of Epo, although there were no significant differences among cultures stimulated with hGM-CSF and SCF, IL-6 and SCF, or IL-11 and SCF.
Table 1.
Colony formation from BM cells of the chimeric receptor transgenic mice
| Cytokines | E | GM | Mix | B | Mast | |
|---|---|---|---|---|---|---|
| Mice | ||||||
| Transgenic | hGM-CSF + Epo | 94.7 ± 11.4 | 36.7 ± 0.6 | 6.3 ± 1.5 | 4.0 ± 1.7 | 0.0 ± 0.0 |
| IL-3 + Epo | 90.7 ± 7.6 | 73.3 ± 1.5 | 4.0 ± 1.7 | 2.0 ± 0.0 | 7.0 ± 1.0 | |
| Epo | 110.0 ± 13.1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Normal | hGM-CSF + Epo | 102.0 ± 7.2 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| IL-3 + Epo | 92.7 ± 9.2 | 76.0 ± 3.6 | 4.0 ± 0.0 | 1.3 ± 1.5 | 4.3 ± 1.5 | |
| Epo | 100.7 ± 21.4 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| Cytokines | ||||||
| hGM-CSF + SCF + Epo | 26.3 ± 4.9 | 6.7 ± 2.1 | ||||
| IL-6 + SCF + Epo | 22.3 ± 4.2 | 5.0 ± 1.7 | ||||
| IL-11 + SCF + Epo | 25.0 ± 1.0 | 4.0 ± 1.0 | ||||
| hGM-CSF + Epo | 4.3 ± 0.6 | 2.0 ± 2.0 | ||||
| IL-6 + Epo | 3.0 ± 2.0 | 0.3 ± 0.6 | ||||
| IL-11 + Epo | 1.3 ± 1.5 | 0.0 ± 0.0 | ||||
| SCF + Epo | 1.7 ± 1.1 | 0.0 ± 0.0 | ||||
| Epo | 0.0 ± 0.0 | 0.0 ± 0.0 |
For mice, 20,000 BM cells from the chimeric receptor transgenic mice or from their normal littermates were cultured in the presence of the designated cytokine(s). E, erythroid colonies; B, erythroid bursts. For cytokines, 2000 Lin− BM cells from 5-FU-treated chimeric receptor transgenic mice were cultured in the presence of the designated cytokine(s). The number of colonies indicates mean ± SD of triplicate cultures.
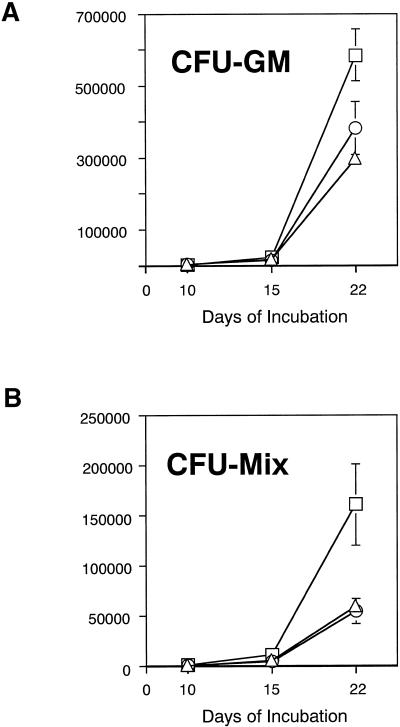
Effects of hGM-CSF on in vitro expansion of hematopoietic cells were then studied using Lin− BM cells from 5FU-treated chimeric receptor transgenic mice in serum-free suspension cultures. Accumulated expansion rates of CFU-GM and CFU-Mix during three sequential cultures were calculated. Although no single agent, hGM-CSF, IL-6, IL-11, or SCF, could support proliferation of the cells (our unpublished results), a combination of hGM-CSF and SCF led to proliferation of enriched cells and exponentially expanded both CFU-GM and CFU-Mix (Figure 5, A and B). When compared with combinations of IL-6 and SCF and IL-11 and SCF, the highest expansion on day 22 was achieved in culture containing hGM-CSF and SCF. These observations are interpreted to mean that hGM-CSF stimulated 5-FU-resistant, dormant hematopoietic progenitors of the chimeric receptor transgenic mice and expanded the cells more efficiently than did IL-6 or IL-11, but the presence of SCF was required.
Figure 5.
Time course analysis of expansion of clonogenic cells in serum-free suspension culture. Ten thousand Lin− cells obtained from 5-FU-treated chimeric receptor transgenic mice were incubated with hGM-CSF and SCF (□), IL-6 and SCF (○), or IL-11 and SCF (▵) in serum-free conditions. The starting 10,000 cells contained 73.3 CFU-GM and 16.7 CFU-Mix. On days 10, 15, and 22, total cell numbers were counted. Ten thousand cells were transferred to freshly prepared media containing the same combination of cytokines as for the previous culture, and the culture was continued. At each time of replating, 2000 cells were analyzed for CFU-GM and CFU-Mix by in vitro colony assay in the presence of IL-3, IL-6, SCF, Epo, and MDGF. The y-axis represents the accumulated expansion rate of CFU-GM or CFU-Mix during the serum-free suspension cultures. Five cultures for each cytokine combination were prepared and then repeated in two independent experiments.
hGM-CSF Stimulates Enriched, Lin−Sca-1+c-kit+ BM Cells and Expands Multipotent Hematopoietic Progenitors
Lin−Sca-1+c-kit+ cells were obtained from BM of the chimeric receptor transgenic mice by FACS sorting and were then analyzed for expansion of multipotent hematopoietic cells in response to hGM-CSF. One hundred cells that contained 50.7 CFU-Mix and 33.3 CFU-GM and 13.3 CFU-Meg before incubation were incubated for 8 d with hGM-CSF and SCF, IL-6 and SCF, or IL-11 and SCF in serum-free suspension cultures. In cultures stimulated with hGM-CSF and SCF, Sca-1, c-kit double-positive cells achieved twice the expansion as those stimulated with IL-6 and SCF or IL-11 and SCF (Figure 6A). The most efficient expansion of CFU-Mix was observed in cultures stimulated with hGM-CSF and SCF (Figure 6B).
Figure 6.
hGM-CSF effects on Lin−Sca-1+c-kit+ BM cells. Lin−Sca-1+c-kit+ cells were purified from BM of the chimeric receptor transgenic mice by FACS sorting. One hundred Lin−Sca-1+c-kit+ cells were incubated with hGM-CSF and SCF (▪), IL-6 and SCF (□), or IL-11 and SCF (░⃞) in serum-free suspension cultures. The starting 100 Lin−Sca-1+c-kit+ cells contained 50.7 CFU-Mix, 33.3 CFU-GM, and 13.3 CFU-Meg. On day 8 of incubation, total cell number, percent of Sca-1+c-kit+ cells, and number of CFU-Mix were examined. The y-axis represents expansion rates of Sca-1+c-kit+ cells (A) and CFU-Mix (B). Assays were done in triplicate cultures and repeated in two independent experiments.
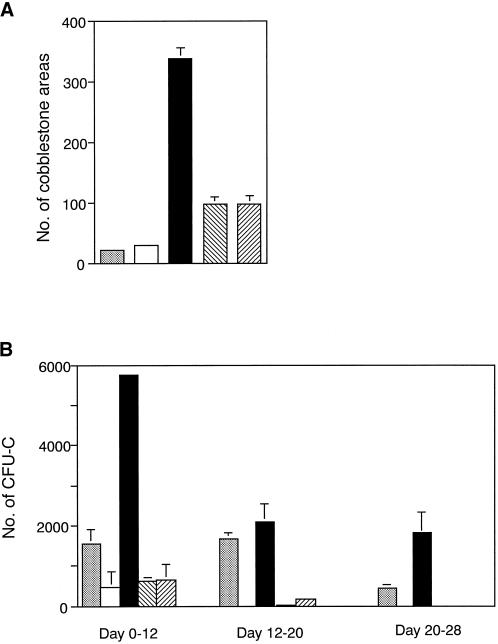
Using the same enriched cells, the effects of hGM-CSF on expansion of CAFCs were examined. With no preincubation, 100 Lin−Sca-1+c-kit+ cells were cultured on an OP9 stromal cell layer without cytokines. The same numbers of enriched cells were preincubated for 4 d with hGM-CSF and SCF, IL-6 and SCF, or IL-11 and SCF in serum-free suspension culture and then washed well with α-medium and cultured on an OP9 stromal cell layer without cytokines. On day 7 of coculture, the cobblestone areas were counted. Preincubation with hGM-CSF and SCF for 4 d dramatically increased the number of CAFCs on day 7 of coculture (Figure 7A) compared with the case of no preincubation. After day 7 of coculture, these cobblestone areas differed in size, and their numbers did not seem to represent hematopoietic activity. Thus, instead of the number of cobblestone areas, we adopted the number of clonogenic cells produced on the stromal cell layer during days 0–12, 12–20, and 20–28 of coculture to precisely evaluate the hematopoietic potential of the cobblestone areas. Throughout the coculture period, the cobblestone areas formed by hGM-CSF and SCF–treated CAFCs showed more active hematopoiesis than did cobblestone areas formed by nontreated CAFCs (Figure 7B). Preincubation with IL-6 and SCF or IL-11 and SCF also increased the number of CAFCs on day 7. However, the CAFCs seemed to lose potency during the preincubation, because their potential to produce progenitors gradually declined and produced no progenitors after day 20 of coculture. These data suggest that Lin−Sca-1+c-kit+ primitive hematopoietic progenitors expand most effectively by stimulation through the chimeric receptor in the presence of SCF when compared with IL-6R or IL-11R.
Figure 7.
Expansion of CAFCs. Lin−Sca-1+c-kit+ cells were purified from BM of the chimeric receptor transgenic mice by FACS sorting. Without preincubation, 100 Lin−Sca-1+c-kit+ cells were incubated on an OP9 stromal cell layer without adding cytokines (░⃞). On the other hand, 100 Lin−Sca-1+c-kit+ cells were incubated with SCF (□), hGM-CSF and SCF (▪), IL-6 and SCF (▧), or IL-11 and SCF (▨) in serum-free suspension culture for 4 d and then washed well with α-medium and incubated on an OP9 stromal cell layer without addition of cytokines. (A) On day 7 of coculture, numbers of cobblestone area were counted. (B) To evaluate the hematopoietic capacity of the cobblestone area, on days 12, 20, and 28, clonogenic cells produced from the cobblestone area during the coculture were analyzed by in vitro colony assay. Assays were performed in triplicate cultures and repeated in two independent experiments.
hGM-CSF Stimulates IL-6Rα-, IL-11Rα-, and gp130-low to -negative Primitive Hematopoietic Progenitors and Expands Multipotent Progenitors
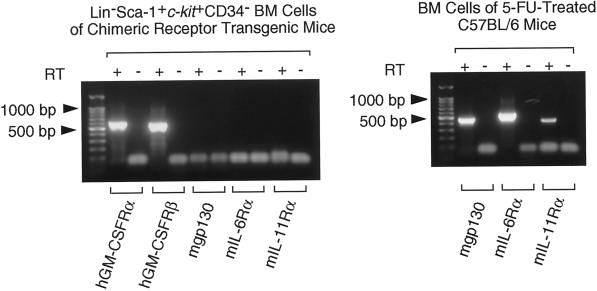
Osawa et al. (1996) reported that HSCs of adult mouse BM were detected in Lin−Sca-1+c-kit+CD34− fractions. Next, we examined expression of the chimeric receptor IL6Rα and gp130 on Lin−Sca-1+c-kit+CD34− cells by RT-PCR. Using cDNA from Lin−Sca-1+c-kit+CD34− cells as a template, genes for the extracellular domain of both hGM-CSFRα and β subunit were amplified. The IL-6Rα subunit gene, the IL-11Rα subunit gene, and the gene for extracellular domain of gp130 were not detected (Figure 8). These data suggest that genetically introduced chimeric receptor genes are expressed on IL-6Rα-, IL-11Rα-, and gp130-low to -negative (low/negative) progenitors.
Figure 8.
RT-PCR analysis of transgene and endogenous cytokine receptor gene expression in primitive hematopoietic progenitors. RNA was prepared from 10 Lin−Sca-1+c-kit+CD34− cells obtained by clone sorting of BM cells of the chimeric receptor transgenic mice with 10 μg of polyinosinic acid as an RNA carrier. Using one quadrant of the RNA, cDNA was synthesized. PCR was run for 40 cycles. After removal of the first PCR primers, 0.1 of the first PCR product was used for the second nested PCR. The second PCR was run for 40 cycles. The expected sizes of the final PCR products were hGM-CSFRα, 598 bp; hGM-CSFRβ, 554 bp; mgp130 (extracellular domain), 464 bp; mIL-6Rα, 556 bp; and mIL-11Rα, 429 bp. The suitability of the primers for mgp130, mIL-6Rα, and mIL-11Rα was confirmed by PCR using cDNA of 10 BM cells of 5-FU-treated C57BL/6 mice as a template. These data are from a single experiment, and similar results were obtained in two additional experiments.
To determine whether chimeric receptors can function in Lin−Sca-1+c-kit+CD34− cells, we then carried out single-cell cultures of cells sorted from the BM of the chimeric receptor transgenic mice by a FACS Vantage automatic cell deposition unit. As shown in Table 2, in the presence of SCF, 61.3% of Lin−Sca-1+c-kit+CD34− cells responded to hGM-CSF, whereas only 9.4 and 14.3% of the cells responded to IL-6 and IL-11, respectively. It is well established that sIL-6R can activate various kinds of cells expressing gp130 signal transducer but lacking the specific IL-6Rα subunit (Hirota et al., 1996; Koshimizu et al., 1996). When IL-6, sIL-6R, and SCF were added to the culture, the number of responding cells increased slightly but never reached the level of stimulation seen with hGM-CSF and SCF. Although all of the proliferated cells contained CFU-Mix regardless of cytokines added, stimulation with hGM-CSF and SCF produced the highest number of CFU-Mix. These observations suggest that the chimeric receptors can transduce signals and markedly expand multipotential progenitors in the presence of SCF. The marked difference in expansion rate of primitive hematopoietic progenitors among hGM-CSF, IL-6, and IL-11 stimuli is probably due to functional expression of the chimeric receptors in IL-6Rα-, IL-11Rα-, and gp130-low/negative progenitor population.
Table 2.
Effect of hGM-CSF on single primitive hematopoietic progenitors
| Cytokines | Positive for proliferation | Total CFU-Mix produced |
|---|---|---|
| SCF | 0 /8 | 0 |
| IL-6 | 0 /8 | 0 |
| IL-6 + sIL-6R | 0 /8 | 0 |
| IL-11 | 0 /8 | 0 |
| hGM-CSF | 0 /8 | 0 |
| IL-6 + SCF | 3 /32 (9.4%) | 30 |
| IL-6 + sIL-6R + SCF | 7 /31 (22.6%) | 80 |
| IL-11 + SCF | 3 /21 (14.3%) | 18 |
| hGM-CSF + SCF | 19 /31 (61.3%) | 378 |
Single Lin−Sca-1+c-kit+CD34− cells were obtained by clone-sorting of BM cells of the chimeric receptor transgenic mice and individually incubated with cytokines in a serum-free suspension culture. On day 6, each well was checked for the presence of proliferated cells. After three washes with α-medium, the proliferated cells in each well were individually replated into methylcellulose media containing IL-3, IL-6, SCF, Epo, and MDGF to examine the number of CFU-Mix. Data are from a single experiment, and similar results were obtained in two additional experiments.
DISCUSSION
In vitro experiments using either 5-FU-resistant, dormant cells or Lin−Sca-1+c-kit+, enriched cells of the hGM-CSFR-mLIFR chimeric receptor transgenic mice showed that the most effective expansion of primitive hematopoietic progenitors in the presence of SCF was achieved by hGM-CSF, when compared with natural ligands for the gp130 receptor family, such as IL-6 and IL-11. The marked difference of expansion rate of primitive hematopoietic progenitors among hGM-CSF, IL-6, and IL-11 stimuli may relate to the different expression pattern of the receptors. Therefore, we purified Lin−Sca-1+c-kit+CD34− cells from BM of the chimeric receptor transgenic mice by FACS clone sorting and incubated the cells individually in serum-free culture with hGM-CSF, IL-6, IL-6 and sIL-6R, IL-11, SCF, and their combinations. In the presence of SCF, hGM-CSF stimulated to a greater extent Lin−Sca-1+c-kit+CD34− progenitors than did IL-6, IL-6 and sIL-6R, or IL-11 and markedly expanded multipotential progenitors, suggesting that the marked difference in progenitor expansion is mainly due to the different expression pattern of the receptors. RT-PCR analysis of the receptor gene expression in the Lin−Sca-1+c-kit+CD34− progenitors supports this notion. In humans, significant expansion of multipotent hematopoietic progenitors was seen in the case of IL-6Rα− cord blood cells in response to IL-6, sIL-6R, and SCF but not from IL-6Rα+ cells, suggesting the presence of IL-6Rα−gp130+ progenitors that have more hematopoietic capacities than do IL-6Rα+ progenitors (Sui et al., 1995; Tajima et al., 1996). Peters et al. (1997) reported that IL-6–sIL-6R double transgenic mice had a dramatic increase of extramedullary hematopoietic progenitors in liver and spleen, although IL-6 single transgenic mice or sIL-6R single transgenic mice showed no such phenotype. Their results suggest that IL-6Rα−gp130+ progenitors also exist in mice, although the possibility that nonhematopoietic cells produce factor(s) in response to IL-6 and sIL-6R and in turn the factor(s) stimulate the hematopoietic progenitors would need to be ruled out. Our data clearly show that IL-6Rα-, IL-11Rα-, and gp130-low/negative primitive hematopoietic progenitors are present in mice, and that these cells are equipped with signal transduction molecules, which can expand when chimeric receptors are genetically introduced and stimulated by hGM-CSF in the presence of SCF.
When Lin−Sca-1+c-kit+CD34− cells were incubated with IL-6, IL-6 and sIL-6R, and IL-11 in the presence of SCF, some of the cells did proliferate, but their receptors were not detected by RT-PCR. Therefore, the Lin−Sca-1+c-kit+CD34− population may be heterogeneous. A small proportion of the cells may express functional IL-6Rα, IL-11Rα and gp130, whereas the greater number of the cells completely lacks the receptors. Alternatively, it may be that a large part of the population expresses IL-6Rα, IL-11Rα, and gp130. However, the expression level is too low to detect their mRNAs by RT-PCR, and a small number of cells express the receptors at minimum level for required signaling. In any case, the results of Table 2 suggest that a considerable portion of Lin−Sca-1+c-kit+CD34− cells are silent on stimulation by IL-6, sIL-6R, and SCF but are ready to respond to stimulation by hGM-CSF and SCF when the hGM-CSFR–mLIFR chimeric receptors are expressed on the cells. Therefore, hGM-CSF stimulated a large number of Lin−Sca-1+c-kit+CD34− cells and supported the development of CFU-Mix most effectively when compared with IL-6, IL-6 and sIL-6R, or IL-11 in the presence of SCF.
In our transgenic mice, the chimeric receptors were expressed in Lin−Sca-1+c-kit+CD34− cells, regardless of the presence of SCF. However, hGM-CSF alone failed to stimulate proliferation of the cells, suggesting that gp130 signaling alone is insufficient to initiate expansion of primitive hematopoietic progenitors, and that efficient expansion of the cells requires both gp130 and c-kit signaling. Important molecules in the signaling of gp130 are Janus kinase 1 (JAK1), JAK2, and Tyk2, signal transducer and activator of transcription 1 (STAT1) and STAT3, the src-related kinase Tec, and the tyrosine phosphatase SHP2 (Hirano, 1998). Although the signaling events activated by SCF and the receptor tyrosine kinase c-kit require further study, JAK2, STAT1, and Tec were found to be activated (Tang et al., 1994; Weiler et al., 1996; Deberry et al., 1997). Thus, investigations of cross-talk between gp130 and c-kit signaling pathways may elucidate pathways responsible for expansion of primitive hematopoietic progenitors.
In vitro expansion of HSCs is important for analysis of molecular mechanisms related to stem cell self-renewal. Although transgenic mouse technology cannot be directly applied to clinical therapy, the knowledge acquired using transgenic mice can be useful in related work on stem cell transplantation or gene therapy.
ACKNOWLEDGMENTS
We thank Drs. K.-i. Arai, R. Nishinakamura, G. Morstyn, F. Fletcher, and I. McNiece for discussion and critical comments and M. Ohara for comments on the manuscript. The Department of Stem Cell Regulation is supported by Amgen.
Abbreviations used:
- APC
allophycocyanin
- BM
bone marrow
- CAFC
cobblestone area–forming cell
- CFU
colony-forming unit
- CFU-Mix
mixed CFU
- CSFR
colony-stimulating factor receptor
- Epo
erythropoietin
- FACS
fluorescence-activated cell sorting
- FBS
fetal bovine serum
- 5-FU
5-fluorouracil
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
- hGM-CSF
human granulocyte–macrophage colony-stimulating factor
- H + L
heavy and light chain
- HSC
hematopoietic stem cell
- IgG
immunoglobulin G
- IL
interleukin
- IL-6Rα-
IL-11Rα-, and gp130-low/negative, IL-6Rα-, IL-11Rα-, and gp130-low to -negative
- JAK
Janus kinase
- LIF
leukemia inhibitory factor
- LIFR
LIF receptor
- Lin −
lineage-negative
- MDGF
megakaryocyte differentiation and growth factor
- Meg
megakaryocyte colonies
- mLIF
mouse LIF
- RT
reverse transcription
- SCF
stem cell factor
- sIL-6R
soluble IL-6R
- STAT
signal transducer and activator of transcription
- TXR
Texas Red
REFERENCES
- Deberry C, Mou S, Linnekin D. Stat1 associates with c-kit and is activated in response to stem cell factor. Biochem J. 1997;327:73–80. doi: 10.1042/bj3270073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escary JL, Perreau J, Dumenil D, Ezine S, Brulet P. Leukemia inhibitory factor is necessary for maintenance of hematopoietic stem cells and thymocyte stimulation. Nature. 1993;363:361–364. doi: 10.1038/363361a0. [DOI] [PubMed] [Google Scholar]
- Hirano T. Interleukin 6 and its receptor: ten years later. Int Rev Immunol. 1998;16:249–284. doi: 10.3109/08830189809042997. [DOI] [PubMed] [Google Scholar]
- Hirota H, Kiyama H, Kishimoto T, Taga T. Accelerated nerve regeneration in mice by up-regulated expression of interleukin (IL) 6 and IL-6 receptor after trauma [see comments] J Exp Med. 1996;183:2627–2634. doi: 10.1084/jem.183.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Holyoake TL, Freshney MG, McNair L, Parker AN, McKay PJ, Steward WP, Fitzsimons E, Graham GJ, Pragnell IB. Ex vivo expansion with stem cell factor and interleukin-11 augments both short-term recovery posttransplant and the ability to serially transplant marrow. Blood. 1996;87:4589–4595. [PubMed] [Google Scholar]
- Iscove NN, Melchers F. Complete replacement of serum by albumin, transferrin, and soybean lipid in cultures of lipopolysaccharide-reactive B lymphocytes. J Exp Med. 1978;147:923–933. doi: 10.1084/jem.147.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby SL, Cook DN, Walton W, Smithies O. Proliferation of multipotent hematopoietic cells controlled by a truncated erythropoietin receptor transgene. Proc Natl Acad Sci USA. 1996;93:9402–9407. doi: 10.1073/pnas.93.18.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto T, Akira S, Narazaki M, Taga T. Interleukin-6 family of cytokines and gp130. Blood. 1995;86:1243–1254. [PubMed] [Google Scholar]
- Kodama H, Nose M, Niida S, Nishikawa S. Involvement of the c-kit receptor in the adhesion of hematopoietic stem cells to stromal cells. Exp Hematol. 1994;22:979–984. [PubMed] [Google Scholar]
- Koike K, Nakahata T, Takagi M, Kobayashi T, Ishiguro A, Tsuji K, Naganuma K, Okano A, Akiyama Y, Akabane T. Synergism of BSF-2/interleukin 6 and interleukin 3 on development of multipotential hemopoietic progenitors in serum-free culture. J Exp Med. 1988;168:879–890. doi: 10.1084/jem.168.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshimizu U, Taga T, Watanabe M, Saito M, Shirayoshi Y, Kishimoto T, Nakatsuji N. Functional requirement of gp130-mediated signaling for growth and survival of mouse primordial germ cells in vitro and derivation of embryonic germ (EG) cells. Development. 1996;122:1235–1242. doi: 10.1242/dev.122.4.1235. [DOI] [PubMed] [Google Scholar]
- Muto A, Watanabe S, Miyajima A, Yokota T, Arai K. High affinity chimeric human granulocyte-macrophage colony-stimulating factor receptor carrying the cytoplasmic domain of the beta subunit but not the alpha subunit transduces growth promoting signals in Ba/F3 cells. Biochem Biophys Res Commun. 1995;208:368–375. doi: 10.1006/bbrc.1995.1347. [DOI] [PubMed] [Google Scholar]
- Nakahata T, Gross AJ, Ogawa M. A stochastic model of self-renewal and commitment to differentiation of the primitive hemopoietic stem cells in culture. J Cell Physiol. 1982;113:455–458. doi: 10.1002/jcp.1041130314. [DOI] [PubMed] [Google Scholar]
- Nakahata T, Ogawa M. Identification in culture of a class of hemopoietic colony-forming units with extensive capability to self-renew and generate multipotential hemopoietic colonies. Proc Natl Acad Sci USA. 1982;79:3843–3847. doi: 10.1073/pnas.79.12.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Arai T, Takagi M, Sawada T, Matsuda T, Yokota T, Heike T. A selective switch-on system for self-renewal of embryonic stem cells using chimeric cytokine receptors. Biochem Biophys Res Commun. 1998;248:22–27. doi: 10.1006/bbrc.1998.8900. [DOI] [PubMed] [Google Scholar]
- Nishijima I, Nakahata T, Hirabayashi Y, Inoue T, Kurata H, Miyajima A, Hayashi N, Iwakura Y, Arai K, Yokota T. A human GM-CSF receptor expressed in transgenic mice stimulates proliferation and differentiation of hemopoietic progenitors to all lineages in response to human GM-CSF. Mol Biol Cell. 1995;6:497–508. doi: 10.1091/mbc.6.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura N, Tsuji K, Ebihara Y, Tanaka I, Sawai N, Koike K, Komiyama A, Nakahata T. Chemotactic and chemokinetic activities of stem cell factor on murine hematopoietic progenitor cells. Blood. 1996;87:4100–4108. [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Peters M, Schirmacher P, Goldschmitt J, Odenthal M, Peschel C, Fattori E, Ciliberto G, Dienes HP, Meyer zum Buschenfelde KH, Rose-John S. Extramedullary expansion of hematopoietic progenitor cells in interleukin (IL)-6-sIL-6R double transgenic mice. J Exp Med. 1997;185:755–766. doi: 10.1084/jem.185.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suematsu S, Matsusaka T, Matsuda T, Ohno S, Miyazaki J, Yamamura K, Hirano T, Kishimoto T. Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Proc Natl Acad Sci USA. 1992;89:232–235. doi: 10.1073/pnas.89.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, et al. gp130 and c-Kit signalings synergize for ex vivo expansion of human primitive hemopoietic progenitor cells. Proc Natl Acad Sci USA. 1995;92:2859–2863. doi: 10.1073/pnas.92.7.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima S, et al. Analysis of interleukin 6 receptor and gp130 expressions and proliferative capability of human CD34+ cells. J Exp Med. 1996;184:1357–1364. doi: 10.1084/jem.184.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Hara T, Ichihara M, Takatsu K, Miyajima A. Multi-colony stimulating activity of interleukin 5 (IL-5) on hematopoietic progenitors from transgenic mice that express IL-5 receptor alpha subunit constitutively. J Exp Med. 1995;181:889–899. doi: 10.1084/jem.181.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Mano H, Yi T, Ihle JN. Tec kinase associates with c-kit and is tyrosine phosphorylated and activated following stem cell factor binding. Mol Cell Biol. 1994;14:8432–8437. doi: 10.1128/mcb.14.12.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler SR, Mou S, DeBerry CS, Keller JR, Ruscetti FW, Ferris DK, Longo DL, Linnekin D. JAK2 is associated with the c-kit proto-oncogene product and is phosphorylated in response to stem cell factor. Blood. 1996;87:3688–3693. [PubMed] [Google Scholar]
- Yonemura Y, Ku H, Lyman SD, Ogawa M. In vitro expansion of hematopoietic progenitors and maintenance of stem cells: comparison between FLT3/FLK-2 ligand and KIT ligand. Blood. 1997;89i:1915–1921. [PubMed] [Google Scholar]
- Yoshida K, et al. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc Natl Acad Sci USA. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]