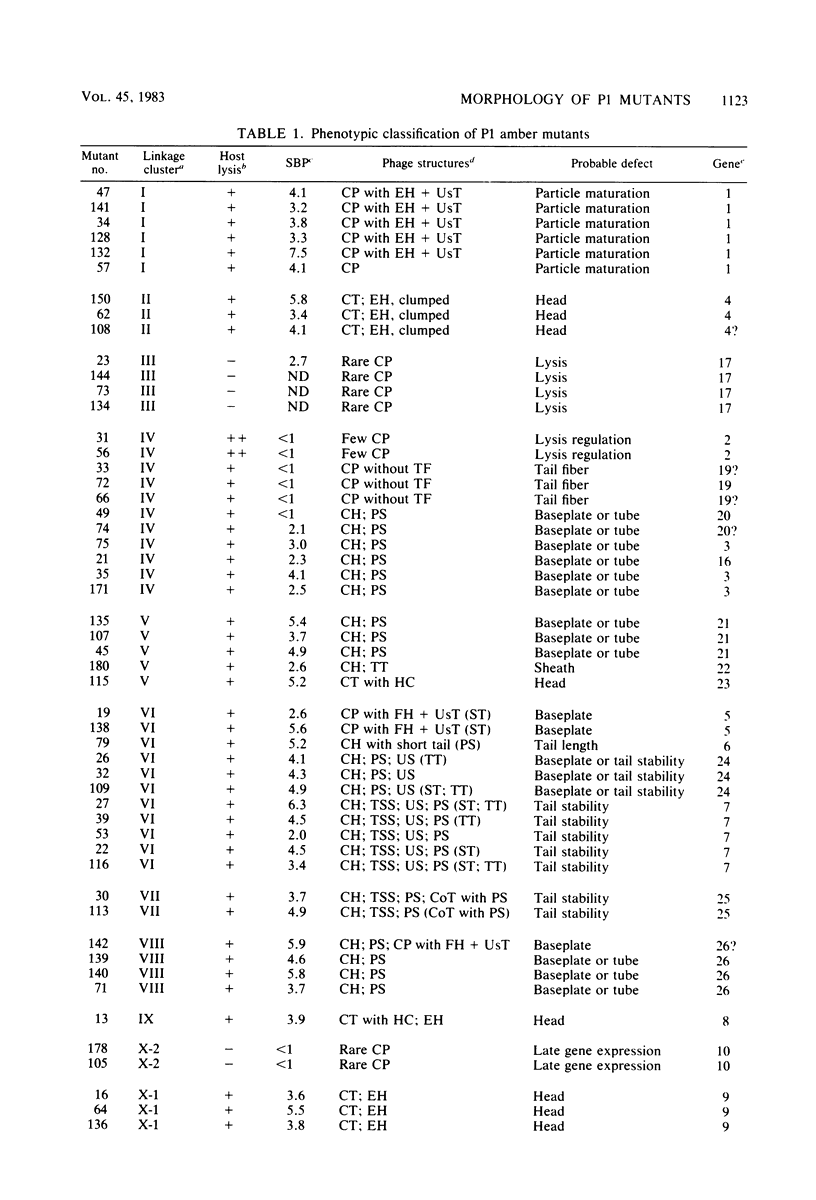
Abstract
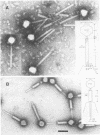
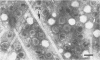
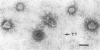
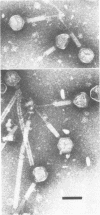
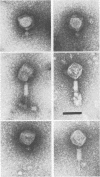
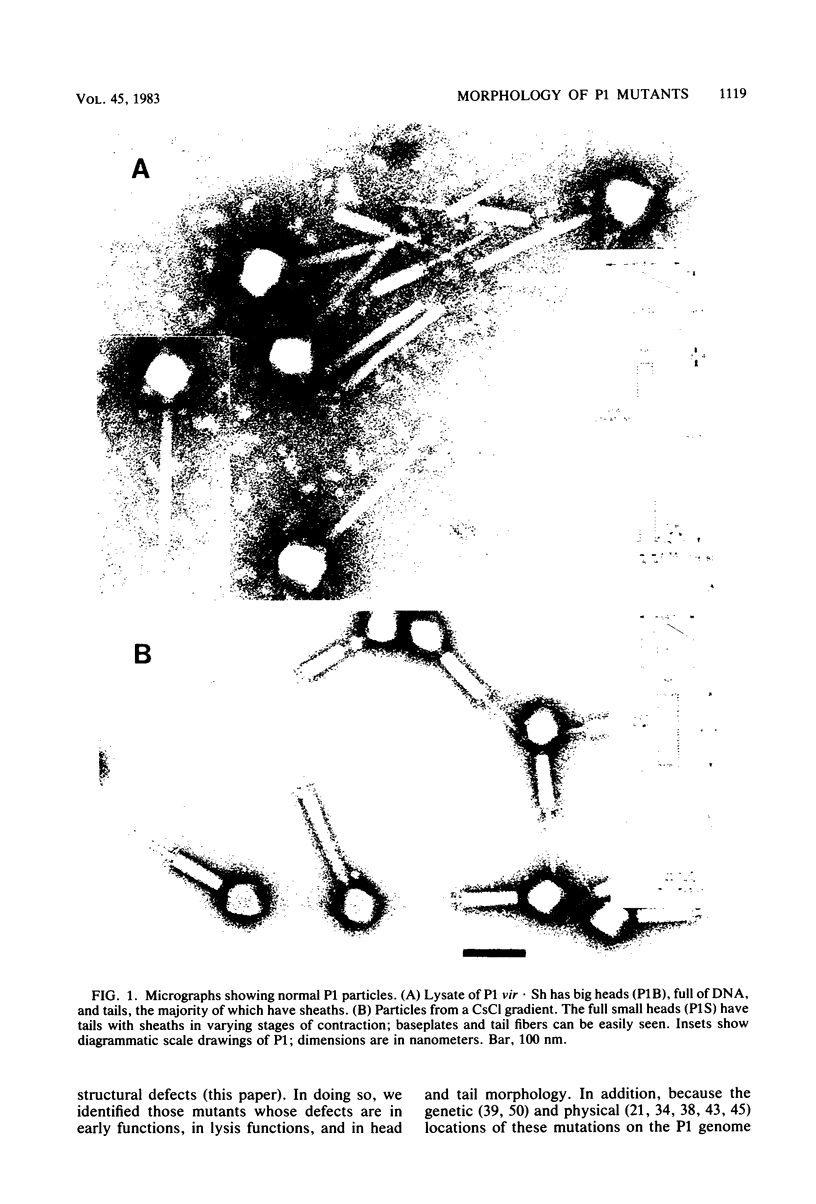
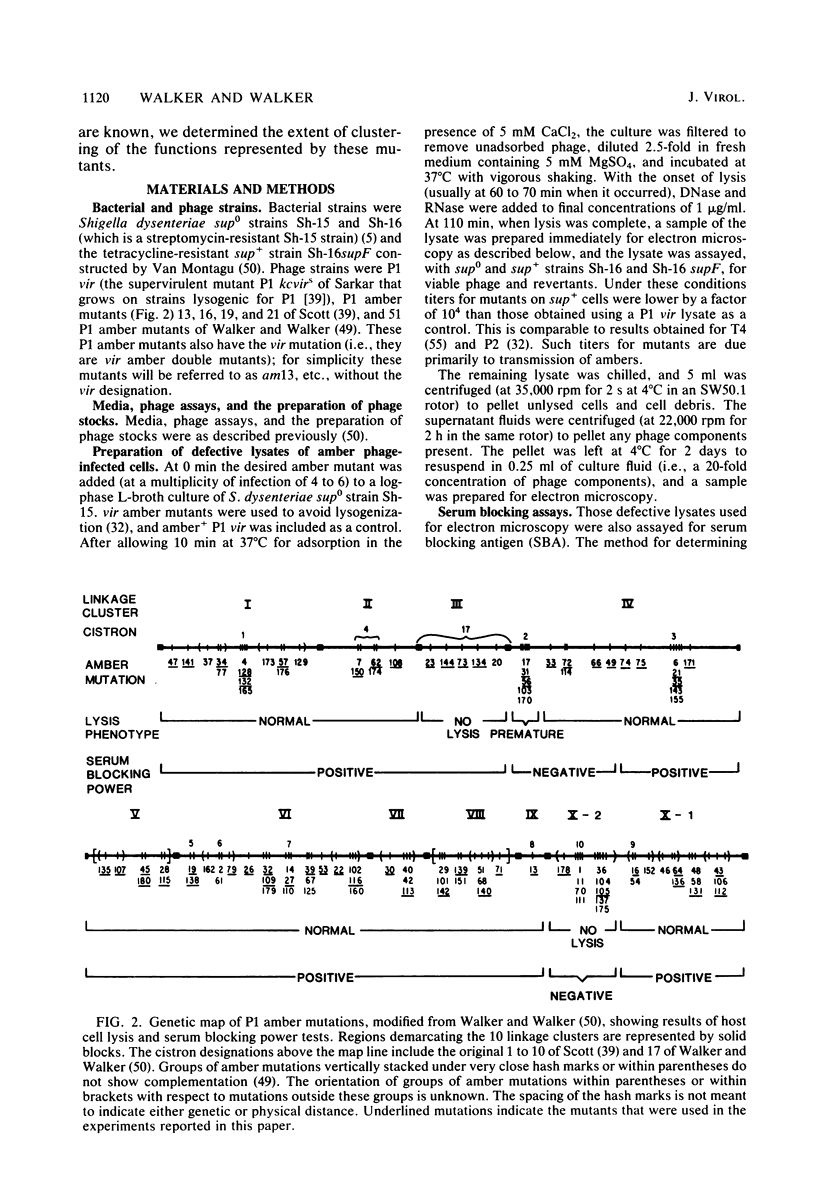
We used electron microscopy and serum blocking power tests to determine the phenotypes of 47 phage P1 amber mutants that have defects in particle morphogenesis. Eleven mutants showed head defects, 30 showed tail defects, and 6 had a defect in particle maturation (which could be either in the head or in the tail). Consideration of previous complementation test results, genetic and physical positions of the mutations, and phenotypes of the mutants allowed assignment of most of the 47 mutations to genes. Thus, a minimum of 12 tail genes, 4 head genes, and 1 particle maturation gene are now known for P1. Of the 12 tail genes, 1 (gene 19, located within the invertible C loop) codes for tail fibers, 6 (genes 3, 5, 16, 20, 21, and 26) code for baseplate components (although one of these genes could code for the tail tube), 1 (gene 22) codes for the sheath, 1 (gene 6) affects tail length, 2 (genes 7 and 25) are involved in tail stability, and 1 (gene 24) either codes for a baseplate component or is involved in tail stability. Of the four head genes, gene 9 codes for a protein required for DNA packaging. The function of head gene 4 is unclear. Head gene 8 probably codes for a minor head protein, whereas head gene 23 could code for either a minor head protein or the major head protein. Excluding the particle maturation gene (gene 1), the 12 tail genes are clustered in three regions of the P1 physical genome. The four head genes are at four separate locations. However, some P1 head genes have not yet been detected and could be located in two regions (for which there are no known genes) adjacent to genes 4 and 8. The P1 morphogenetic gene clusters are interrupted by many genes that are expressed in the prophage.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arisaka F., Tschopp J., Van Driel R., Engel J. Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a co-operative association process. J Mol Biol. 1979 Aug 15;132(3):369–386. doi: 10.1016/0022-2836(79)90266-3. [DOI] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckendorf S. K. Structure of the distal half of the bacteriophage T4 tail fiber. J Mol Biol. 1973 Jan;73(1):37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- Becker A., Gold M. Isolation of the bacteriophage lambda A-gene protein. Proc Natl Acad Sci U S A. 1975 Feb;72(2):581–585. doi: 10.1073/pnas.72.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Waddell C. H., King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J Mol Biol. 1973 Nov 15;80(4):669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Bächi B., Arber W. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol Gen Genet. 1977 Jun 24;153(3):311–324. doi: 10.1007/BF00431596. [DOI] [PubMed] [Google Scholar]
- Coombs D. H., Eiserling F. A. Studies on the structure, protein composition and aseembly of the neck of bacteriophage T4. J Mol Biol. 1977 Nov 5;116(3):375–405. doi: 10.1016/0022-2836(77)90076-6. [DOI] [PubMed] [Google Scholar]
- DE MARS R. I. The production of phage-related materials when bacteriophage development in interrupted by proflavine. Virology. 1955 May;1(1):83–99. doi: 10.1016/0042-6822(55)90007-6. [DOI] [PubMed] [Google Scholar]
- Echols H., Murialdo H. Genetic map of bacteriophage lambda. Microbiol Rev. 1978 Sep;42(3):577–591. doi: 10.1128/mr.42.3.577-591.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S., Lielausis I. Some steps in the assembly of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):263–276. doi: 10.1016/0022-2836(68)90008-9. [DOI] [PubMed] [Google Scholar]
- Favre R., Boy de la Tour E., Segrè N., Kellenberger E. Studies on the morphopoiesis of the head of phage T-even. I. Morphological, immunological, and genetic characterization of polyheads. J Ultrastruct Res. 1965 Oct;13(3):318–342. doi: 10.1016/s0022-5320(65)80080-6. [DOI] [PubMed] [Google Scholar]
- Frankel F. R., Batcheler M. L., Clark C. K. The role of gene 49 in DNA replication and head morphogenesis in bacteriophage T4. J Mol Biol. 1971 Dec 28;62(3):439–463. doi: 10.1016/0022-2836(71)90147-1. [DOI] [PubMed] [Google Scholar]
- Georgopoulos C. P., Hendrix R. W., Kaiser A. D., Wood W. B. Role of the host cell in bacteriophage morphogenesis: effects of a bacterial mutation on T4 head assembly. Nat New Biol. 1972 Sep 13;239(89):38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Plasterk R. H., van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982 May 27;297(5864):339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- Granboulan P., S echaud J., Kellenberger E. On the fragility of phage T4-related particles. Virology. 1971 Nov;46(2):407–425. doi: 10.1016/0042-6822(71)90042-0. [DOI] [PubMed] [Google Scholar]
- Heilmann H., Reeve J. N., Pühler A. Identification of the repressor and repressor bypass (antirepressor) polypeptides of bacteriophage P1 synthesized in infected minicells. Mol Gen Genet. 1980 Apr;178(1):149–154. doi: 10.1007/BF00267223. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Schumm J. W., Taylor A. L. The S and U genes of bacteriophage mu are located in the invertible G segment of mu DNA. Virology. 1979 Jan 15;92(1):108–124. doi: 10.1016/0042-6822(79)90218-6. [DOI] [PubMed] [Google Scholar]
- Iida S., Arber W. Multiple physical differences in the genome structure of functionally related bacteriophages P1 and P7. Mol Gen Genet. 1979 Jun 20;173(3):249–261. doi: 10.1007/BF00268635. [DOI] [PubMed] [Google Scholar]
- Iida S., Arber W. Plaque forming specialized transducing phage P1: isolation of P1CmSmSu, a precursor of P1Cm. Mol Gen Genet. 1977 Jun 24;153(3):259–269. doi: 10.1007/BF00431591. [DOI] [PubMed] [Google Scholar]
- Israel J. V., Anderson T. F., Levine M. in vitro MORPHOGENESIS OF PHAGE P22 FROM HEADS AND BASE-PLATE PARTS. Proc Natl Acad Sci U S A. 1967 Feb;57(2):284–291. doi: 10.1073/pnas.57.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D., Syvanen M., Masuda T. DNA packaging steps in bacteriophage lambda head assembly. J Mol Biol. 1975 Jan 15;91(2):175–186. doi: 10.1016/0022-2836(75)90158-8. [DOI] [PubMed] [Google Scholar]
- Kemp C. L., Howatson A. F., Siminovitch L. Electron microscopy studies of mutants of lambada bacteriophage. I. General description and quantitation of viral products. Virology. 1968 Nov;36(3):490–502. doi: 10.1016/0042-6822(68)90174-8. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975 Dec 25;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- King J. Bacteriophage T4 tail assembly: four steps in core formation. J Mol Biol. 1971 Jun 28;58(3):693–709. doi: 10.1016/0022-2836(71)90034-9. [DOI] [PubMed] [Google Scholar]
- King J., Lenk E. V., Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J Mol Biol. 1973 Nov 15;80(4):697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- King J., Mykolajewycz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J Mol Biol. 1973 Apr 5;75(2):339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Mölbert E., Showe M., Kellenberger E. Form-determining function of the genes required for the assembly of the head of bacteriophage T4. J Mol Biol. 1970 Apr 14;49(1):99–113. doi: 10.1016/0022-2836(70)90379-7. [DOI] [PubMed] [Google Scholar]
- Laski F., Jackson E. N. Maturation cleavage of bacteriophage P22 DNA in the absence of DNA packaging. J Mol Biol. 1982 Feb 5;154(4):565–579. doi: 10.1016/s0022-2836(82)80015-6. [DOI] [PubMed] [Google Scholar]
- Lindahl G. Characterization of conditional lethal mutants of bacteriophage P2. Mol Gen Genet. 1974 Feb 6;128(3):249–260. doi: 10.1007/BF00267114. [DOI] [PubMed] [Google Scholar]
- Luftig R. B., Wood W. B., Okinaka R. Bacteriophage T4 head morphogenesis. On the nature of gene 49-defective heads and their role as intermediates. J Mol Biol. 1971 May 14;57(3):555–573. doi: 10.1016/0022-2836(71)90109-4. [DOI] [PubMed] [Google Scholar]
- Mural R. J., Chesney R. H., Vapnek D., Kropf M. M., Scott J. R. Isolation and characterization of cloned fragments of bacteriophage P1 DNA. Virology. 1979 Mar;93(2):387–397. doi: 10.1016/0042-6822(79)90243-5. [DOI] [PubMed] [Google Scholar]
- Onorato L., Showe M. K. Gene 21 protein-dependent proteolysis in vitro of purified gene 22 product of bacteriophage T4. J Mol Biol. 1975 Mar 5;92(3):395–412. doi: 10.1016/0022-2836(75)90288-0. [DOI] [PubMed] [Google Scholar]
- Poteete A. R., King J. Functions of two new genes in Salmonella phage P22 assembly. Virology. 1977 Feb;76(2):725–739. doi: 10.1016/0042-6822(77)90254-9. [DOI] [PubMed] [Google Scholar]
- Ray P., Murialdo H. The role of gene Nu3 in bacteriophage lambda head morphogenesis. Virology. 1975 Mar;64(1):247–263. doi: 10.1016/0042-6822(75)90096-3. [DOI] [PubMed] [Google Scholar]
- Razza J. B., Watkins C. A., Scott J. R. Phage P1 temperature-sensitive mutants with defects in the lytic pathway. Virology. 1980 Aug;105(1):52–59. doi: 10.1016/0042-6822(80)90155-5. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Genetic studies on bacteriophage P1. Virology. 1968 Dec;36(4):564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- Showe M. K., Black L. W. Assembly core of bacteriophage T4: an intermediate in head formation. Nat New Biol. 1973 Mar 21;242(116):70–75. doi: 10.1038/newbio242070a0. [DOI] [PubMed] [Google Scholar]
- Simon L. D. Infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope: T4 head morphogenesis. Proc Natl Acad Sci U S A. 1972 Apr;69(4):907–911. doi: 10.1073/pnas.69.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. A characterization of bacteriophage P1 DNA fragments cloned in a lambda vector. Virology. 1979 Jul 15;96(1):129–142. doi: 10.1016/0042-6822(79)90179-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D., Austin S., Yarmolinsky M., Hoess R. Site-specific recombination and its role in the life cycle of bacteriophage P1. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- Toussaint A., Lefebvre N., Scott J. R., Cowan J. A., de Bruijn F., Bukhari A. I. Relationships between temperate phages Mu and P1. Virology. 1978 Aug;89(1):146–161. doi: 10.1016/0042-6822(78)90048-x. [DOI] [PubMed] [Google Scholar]
- Walker D. H., Jr, Anderson T. F. Morphological variants of coliphage P1. J Virol. 1970 Jun;5(6):765–782. doi: 10.1128/jvi.5.6.765-782.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. H., Jr, Walker J. T. Genetic studies of coliphage P1. I. Mapping by use of prophage deletions. J Virol. 1975 Sep;16(3):525–534. doi: 10.1128/jvi.16.3.525-534.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker D. H., Jr, Walker J. T. Genetic studies of coliphage P1. III. Extended genetic map. J Virol. 1976 Oct;20(1):177–187. doi: 10.1128/jvi.20.1.177-187.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. T., Walker D. H., Jr Structural proteins of coliphage P1. Prog Clin Biol Res. 1981;64:69–77. [PubMed] [Google Scholar]
- Walker J. T., Walker D. H. Mutations in coliphage p1 affecting host cell lysis. J Virol. 1980 Aug;35(2):519–530. doi: 10.1128/jvi.35.2.519-530.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Luftig R. B., Wilson J. H., Eddleman H., Lyle H., Wood W. B. Assembly of bacteriophage T4 tail fibers. II. Isolation and characterization of tail fiber precursors. J Mol Biol. 1970 Nov 28;54(1):15–31. doi: 10.1016/0022-2836(70)90443-2. [DOI] [PubMed] [Google Scholar]
- Wood W. B., Revel H. R. The genome of bacteriophage T4. Bacteriol Rev. 1976 Dec;40(4):847–868. doi: 10.1128/br.40.4.847-868.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M., Ahmad-Zadeh C. Determination of gene product positions in bacteriophage T4 by specific antibody association. J Mol Biol. 1970 Jul 28;51(2):411–421. doi: 10.1016/0022-2836(70)90151-8. [DOI] [PubMed] [Google Scholar]
- Yanagida M., Boy de la Tour E., Alff-Steinberger C., Kellenberger E. Studies on the morphopoiesis of the head of bacteriophage T-even. 8. Multilayered polyheads. J Mol Biol. 1970 May 28;50(1):35–58. doi: 10.1016/0022-2836(70)90102-6. [DOI] [PubMed] [Google Scholar]
- van de Putte P., Cramer S., Giphart-Gassler M. Invertible DNA determines host specificity of bacteriophage mu. Nature. 1980 Jul 17;286(5770):218–222. doi: 10.1038/286218a0. [DOI] [PubMed] [Google Scholar]