Abstract
Filamentous phages are thread-shaped bacterial viruses. Their outer coat is a tube formed by thousands equal copies of the major coat protein pVIII. Libraries of random peptides fused to pVIII domains were used for selection of phages probes specific for a panel of test antigens and biological threat agents. Because the viral carrier in the phage borne bio-selective probes is infective, they can be cloned individually and propagated indefinitely without needs of their chemical synthesis or reconstructing. As a new bioselective material, landscape phages combine unique characteristics of affinity reagents and self assembling proteins. Biorecognition layers formed by the phage-derived probes bind biological agents with high affinity and specificity and generate detectable signals in analytical platforms. The performance of phage-derived materials as biorecognition interface was illustrated by detection of Bacillus anthracis spores and Salmonella typhimurium cells. With further refinement, the phage-derived analytical platforms for detecting and monitoring of numerous threat agents may be developed, since phage interface against any bacteria, virus or toxin may be readily selected from the landscape phage libraries. As an interface in the field-use detectors, they may be superior to antibodies, since they are inexpensive, highly specific and strong binders, resistant to high temperatures and environmental stresses.
Keywords: Biosensor, sensor, molecular recognition, interface, bacteriophage, landscape phage, phage display, phage library, Bacillus anthracis, Salmonella typhimurium, nanotechnology, ELISA, quartz crystal microbalance
1. Introduction
Analytical electronic devices that transduce a molecular recognition event into a measurable signal are generally called “sensors” [1]. Their principal components are a sensing interface that interacts with an analyte, and a signal processor that transduces the binding impulse into an observed signal. When a sensor interface is composed of biological entities, or when a sensor is designed to detect a biological agent, they are generically called “biomolecular sensors”, or shortly—“biosensors” [2]. The major desired characteristics of the biosensors—sensitivity, selectivity, robustness and prompt performance—are determined mostly by the nature of the biorecognition interface. A commonly used recognition elements in biosensors are antibodies (reviewed by [3]), although a variety of other bioorganic molecules have been also effectively used as an interface: peptides, enzymes, lectins, carbohydrates, nucleic acids, aptamers, recombinant proteins or molecularly imprinted polymers [4]. In a more recent detection format, the whole cells were explores as the binding entities [5, 6]. No one of these types of recognition interfaces, however, could respond well to the sensor performance requirements. A new biorecognition material—the landscape phage—emerged recently as a result of in the depth study of genetics and structure of the filamentous phage and the development of the phage display technology [7]. This review focuses on the progress made in the development of this new nonomaterial and discusses a prospect of using the landscape phages as bioselectable robust analytical probes and as an interface in biosensors.
2. Phage as bioselective nanomaterial
The filamentous bacteriophages Ff (fd, f1 and M13) are flexible, 800-900 nm long, thin fibers with a diameter of about 6.5 nm (Fig.1). They consist of a single-stranded circular DNA packed in a tube composed of the major coat protein pVIII (98% by mass), and a few copies of the minor coat proteins capping the ends of the phage particle (reviewed in [8, 9]).
Fig. 1.

Electron micrograph of filamentous phage. The aminoterminus of pIII proteins are visible and pointed by arrow (cortesy of Irina Davidovich, Gregory Kishchenko and and Lee Makowski).
A phage display library is an ensemble of up to 10 billion different phage clones, each harboring a unique foreign DNA, and therefore displaying a specified guest peptide on the virion surface [26]. In particular, foreign peptides may be genetically fused to every pVIII subunit and displayed on the phage surface in thousands of copies increasing the virion's total mass by up to 20% [10, 11]. It was shown that the foreign peptides replacing three or four mobile amino acids at the N-terminus of the protein pVIII (shown as white on the Fig. 2), or replacing amino acids 12-19 in its central part (shown as yellow) don't disturb considerably the general architecture of virions. Yet, remarkably, such fusion phages can retain their ability to infect the host bacteria Escherichia coli and form phage progeny up to 1000 identical phage particles per bacterial cell during the doubling period. Such particles were eventually given the name “landscape phage” to emphasize the dramatic change in surface architecture caused by arraying thousands of copies of the guest peptide in a dense, repeating pattern around the tubular capsid, as illustrated by Fig. 2. The foreign peptides decorating the phage body create defined organic surface structures (landscapes) that vary from one phage clone to the next. A landscape library is a huge population of such phages, encompassing billions of clones with different surface structures and biophysical properties [11]. The binding peptide comprising up to 20% of the phage mass and up to 50% of the phage surface may be easily prepared by precipitation from a culture of infected bacteria. Further purification, if necessary, can be readily performed by the large scale hydroxyapatite chromatography [12].
Fig. 2.


A. Alpha-helical domain of the fusion major coat protein pVIII. B. Space-filling model of the phage (about 1% of its length). White area in A and B depicts the foreign random peptides attached to amino acid Asp-4 or Asp-5 of pVIII in the landscape libraries [11] (for simplicity they are presented in alpha-helical conformation, although they can adapt any conformation); yellow area pictures amino acids 12-19 randomized in the α-library [14]. Numbers 1-4 point different areas of the same pVIII subunit starting from the N-terminal foreign peptide. About half of amino acids are exposed, while another half is buried in the capsid (shown as red in A). The model was built by Dr. Alexey M. Eroshkin using Marvin's phage model [8].
3. Landscape phages as substitute antibodies
Affinity selection is a characteristic aspect of the phage display technology that allows obtaining ligands against any receptor, including biopolymers, organic compounds or inorganic materials [29]. Therefore, the landscape phage is unique micro-fibrous material that can be selected with desired properties by the affinity binding protocol. To obtain the specific phage ligand, an immobilized target molecule or a corpuscular particle, called here “selector,” is exposed to a phage display library. Phage particles whose displayed peptides bind the selector are captured, while all other phages are washed away. The captured phage, generally a 1/108-1/107 fraction of the initial library population, can be then eluted from the support with mild acid, alkaline or detergent solutions without affecting phage infectivity, and propagated by infecting bacterial host cells. A single round of affinity selection is able to enrich for selector-binding clones by many orders of magnitude; a few rounds suffice to survey a library with billions or even trillions of initial clones for exceedingly rare guest peptides with particularly high affinities for the selector. After several rounds of affinity selection, individual phage clones are propagated and their ability to bind the selector is confirmed.
Phages can be selected from landscape phage display libraries with affinities for a wide range of simple targets such as dioxin, Cibacron blue, β-galactosidase, streptavidin, neutravidin and fibrinogen [11, 13, 14, 27], as well as for more complex targets such as prostate cancer cells [15, 16, 28], malignant glial cells [17, 25, 30], or serum antibodies [31]. Landscape phages have been shown to serve as substitutes for antibodies and detection probes in enzyme-linked immuno assay (ELISA) and biosensors [13-23], as immunogens [24], gene-delivery vehicles [25] and affinity matrices [30, 32].
4. Landscape phages as detection probes
It was demonstrated that phage landscape libraries contain many potential probes for surface markers of cells, spores and bacteria. Phage probes against biological threat agents, such as Bacillus anthracis spores and Salmonella typhimurium were isolated in a nonbiased multistage selection procedure using immobilized spores or bacteria as a selector [18-20]. Binding of the selected phages to their respective targets was characterized by a precipitation test, fluorescence-activated cell sorting, enzyme-linked immunosorbent assay (ELISA), and fluorescent, optical and electron microscopy. These tests demonstrated specific dose-dependent binding of each antigen to the phage it has selected. Inhibition ELISA verified that non-immobilized synthetic peptides and peptide-bearing phages compete with immobilized phage for binding to their respective antigens [13]. These experiments with different antigens have shown that landscape phages may be used as a new type of substitute antibodies—filaments that can bind protein and glycoprotein antigens with nanomolar affinities and high specificity.
It is interesting to note that representative landscape phage selected with B. anthracis spores in a non-biased selection scheme can bind the selector strain at a higher level than other species of Bacillus spores [18]. Similarly, the phage selected with S. typhimurium demonstrated higher affinity to the selector strain in comparison with nine other gram-negative bacteria, predominately Enterobacteriaceae [20]. A small amount of cross reactivity of this phage was noted with Yersinia enterocolitica and Citrobacter freundii. The complex of phage with bacteria was visualized by fluorescence microscopy and transmission electron microscopy (TEM) (Fig. 3) and showed a multivalent character of the phage-bacteria binding.
Fig. 3.

TEM micrograph of bacteria-phage complex. Phage is labeled with gold nanoparticles (arrows). Adapted from [19].
The performance of the probes in detection of biological agents was illustrated by quartz crystal microbalance (QCM), in which the phages immobilized on a gold electrode of the QCM unit reacted with their analytes in solution phase [22]. Phage were immobilized onto the sensor surfaces by phage self-assemblage on Langmuir-Blodgett (LB) phospholipid by biotin/streptavidin coupling [22], or by direct physical adsorption of phage to the sensor surface [21]. In the LB method monolayers containing biotinylated phospholipids were transferred onto the gold surface of acoustic wave sensors and treated with streptavidin and biotinylated phage. The phage-loaded sensor demonstrated specific dose-dependent binding of β-galactosidase from E.coli. It was observed that the affinity of the complex depends on the mode of phage immobilization and type of analytical platform: 0.6 nM by acoustic wave sensor versus 30 nM by enzyme-linked immunosorbent assay (ELISA). The difference in affinities was attributed to the monovalent (ELISA) and divalent (sensor) interaction of the phage with β-galactosidase, as was indicated by the analysis of binding curves using the Hill presentation. It was hypothesized that one or another mode of interaction depends on the conformational freedom of the phage immobilized to the solid surface. Binding of the phage is quite specific because the response was reduced by 85% if β–galactosidase is preincubated with 4 nM phage. It was shown that binding of the phage to β–galactosidase is selective: presence of 1000-fold excess of bovine serum albumin in mixture with β–galactosidase did not affect the ELISA signal and reduces the biosensor signal only by 4%.
It was found that phages readily adsorb onto the gold surfaces [21], although the possible mechanism of the adsorption is still obscure [33]. When the acoustic wave sensor (Maxtek) with gold electrodes was exposed to the β-galactosidase-binding phage in suspension and then tested with gradually increasing concentrations of β–galactosidase, the sensor showed the value of EC50 of approximately 2 nM, comparable with results obtained by the self-assembling LB method [34]. Another biosensors specific for S. typhimurium demonstrated a linear dose-response relationship over six decades of bacterial concentration [21]. Bacterial binding to the sensor was confirmed by scanning electron microscopy (SEM) (Fig. 4). The sensitivity of the biosensor (-10.9 Hz) was vastly greater than the established background. The lower limit of detection based on the dose-response curve was estimated at 100 cells/ml.
Fig. 4.
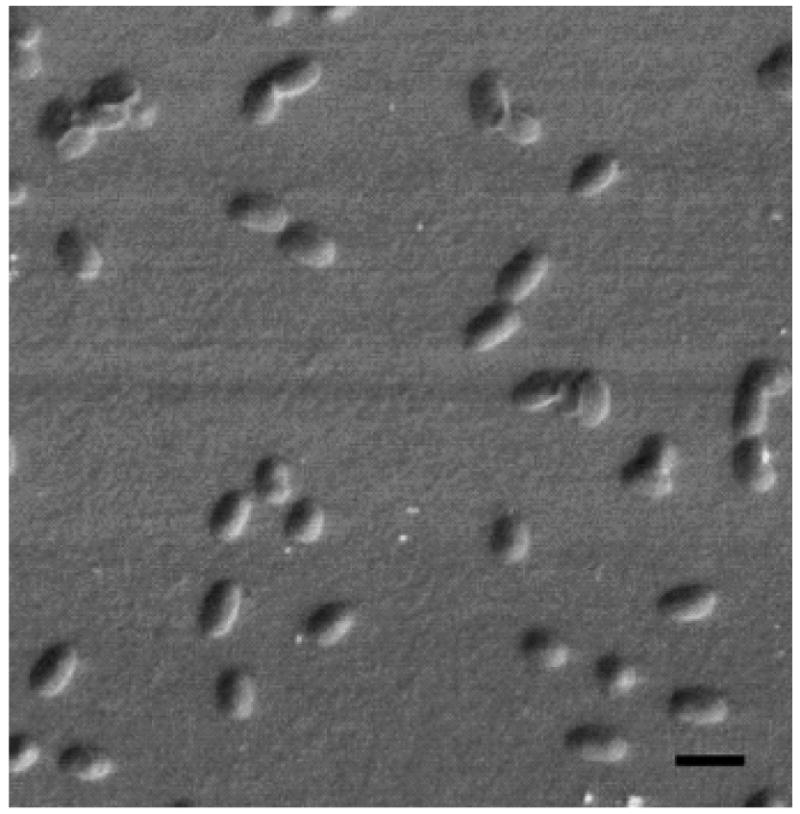
Scanning electron micrograph of S. typhimurium binding to phage immobilized to the surface of a sensor by physical adsorption. Magnification 3000×; bar = 5μm. Adapted from [21].
5. Robustness of the phage probes
Most detection devices traditionally rely on antibodies as diagnostic probes (reviewed recently in [3]). Their use outside of a laboratory, however, may be problematic because antibodies are often unstable in severe environmental conditions. Environmental monitoring requires thermostable probes, such as preselected phages that are superior to antibodies and can operate in non-controlled conditions. For example, when thermostability of a landscape phage probe and a monoclonal antibody specific for β-galactosidase was examined in parallel in the ELISA format they were both stable for greater than 6 months at room temperature, but at higher temperatures the antibody degraded more rapidly than the phage probe [18]. At 37 °C, phage degraded only slightly (half-life of phage as a probe at this temperature was 950 days), while monoclonal antibodies lost virtually all of their activity for 30-week study (half-life 107 days). At 50 °C, both phage and monoclonal antibodies progressively degraded, but monoclonal antibody activity was undetectable after five weeks, while phage still retained more than 50% of its activity at the same time point. At 63 °C, monoclonal antibodies were found to be completely inactivated after just 24 h. The phage probe was significantly more stable at this temperature, maintaining detectable activity for six weeks. Phage was shown to retain binding activity even after short incubations at 76 °C. While a small amount of degradation was detectable after only 4 h, phage had a half-life of 2.4 days at 76 °C. These results confirm that the phage probes are highly thermostable and can function even after exposure to high temperatures during shipping, storage and operation.
6. Conclusion
The presented data show that the phage engineering, which grounds on the natural mechanisms of selection, amplification and self assemblage, is a powerful and very precise technique that allows directed nanofabrication of bioselective materials, with possible applications to biosensors, nanoelectronics, biosorbents, and other areas of medicine, technology, and environmental monitoring. In particular, the genetically driven “phage landscaping” allows the generation of libraries possessing diverse nanostructures accommodated on the phage's surface – a huge resource of diagnostic and detection probes. Biorecognition interface fabricated from the selected phage-derived probes bind biological agents, and as a part of analytical platforms, generate detectable signals. They may be suitable as robust and inexpensive antibody substitutes for field-use detectors and real time monitoring devices for biological and chemical threat agents.
Acknowledgments
This work was supported by ARO/DARPA grant # DAAD 19-01-10454 (to VAP), NIH grant # NIH-1 R21 AI055645 (to VAP), and Auburn University Animal Health & Disease Research Funds grant #354 (to VAP). The author is grateful to Director of Auburn University Detection and Food Safety Center Professor Bryan A. Chin for support and fruitful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cunningham AJ. Introduction to Bioanalytical Sensors. New york: John Wiley & Sons, Inc.; 1998. p. 418. [Google Scholar]
- 2.Gizeli E, Lowe CR, editors. Biomolecular sensors. Taylor & Francis; London, New York: 2002. p. 322. [Google Scholar]
- 3.Goodchild S, Love T, Hopkins N, Mayers C. Engineering antibodies for biosensor technologies. Adv Appl Microbiol. 2006;58:185–226. [PubMed] [Google Scholar]
- 4.Cooper J, Cass T, editors. Biosensors. Second. University Press; Oxford, New York: 2004. p. 251. [Google Scholar]
- 5.Rider TH, Petrovick MS, Nargi FE, Harper JD, Schwoebel ED, Mathews RH, Blanchard DJ, Bortolin LT, Young AM, Chen J, Hollis MA. A B cell-based sensor for rapid identification of pathogens. Science. 2003;301(5630):213–215. doi: 10.1126/science.1084920. [DOI] [PubMed] [Google Scholar]
- 6.Kuang Y, Walt DR. Monitoring “promiscuous” drug effects on single cells of multiple cell types. Anal Biochem. 2005;345(2):320–325. doi: 10.1016/j.ab.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Sidhu SS, editor; Carmen A, editor. Phage Display in Biotechnology and Drug Discovery. Vol. 3. CRC Press, Taylor & Francis Group; Boca Raton, London, New York, Singapore: 2005. p. 748. (Drug Discovery). [Google Scholar]
- 8.Marvin DA, Welsh LC, Symmons MF, Scott WR, Straus SK. Molecular structure of fd (f1, M13) filamentous bacteriophage refined with respect to X-ray fibre diffraction and solid-state NMR data supports specific models of phage assembly at the bacterial membrane. J Mol Biol. 2006;355(2):294–309. doi: 10.1016/j.jmb.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 9.Marvin DA. Filamentous phage structure, infection and assembly. Curr Opin Struct Biol. 1998;8(2):150–158. doi: 10.1016/s0959-440x(98)80032-8. [DOI] [PubMed] [Google Scholar]
- 10.Ilyichev AA, Minenkova OO, Tatkov SI, Karpyshev NN, Eroshkin AM, Petrenko VA, Sandakhchiev LS. Construction of M13 viable bacteriophage with the insert of foreign peptides into the major coat protein. Doklady Biochemistry (Proc Acad Sci Ussr)-Engl Tr. 1989;307:196–198. [Google Scholar]
- 11.Petrenko VA, Smith GP, Gong X, Quinn T. A library of organic landscapes on filamentous phage. Protein Eng. 1996;9(9):797–801. doi: 10.1093/protein/9.9.797. [DOI] [PubMed] [Google Scholar]
- 12.Smith GP, Gingrich TR. Hydroxyapatite chromatography of phage-display virions. Biotechniques. 2005;39(6):879–884. doi: 10.2144/000112032. [DOI] [PubMed] [Google Scholar]
- 13.Petrenko VA, Smith GP. Phages from landscape libraries as substitute antibodies. Protein Eng. 2000;13(8):589–592. doi: 10.1093/protein/13.8.589. [DOI] [PubMed] [Google Scholar]
- 14.Petrenko VA, Smith GP, Mazooji MM, Quinn T. Alpha-helically constrained phage display library. Protein Eng. 2002;15(11):943–950. doi: 10.1093/protein/15.11.943. [DOI] [PubMed] [Google Scholar]
- 15.Romanov VI, Durand DB, Petrenko VA. Phage display selection of peptides that affect prostate carcinoma cells attachment and invasion. Prostate. 2001;47(4):239–251. doi: 10.1002/pros.1068. [DOI] [PubMed] [Google Scholar]
- 16.Romanov VI, Whyard T, Adler HL, Waltzer WC, Zucker S. Prostate cancer cell adhesion to bone marrow endothelium: the role of prostate-specific antigen. Cancer Res. 2004;64(6):2083–2089. doi: 10.1158/0008-5472.can-03-3487. [DOI] [PubMed] [Google Scholar]
- 17.Samoylova TI, Petrenko VA, Morrison NE, Globa LP, Baker HJ, Cox NR. Phage probes for molecular profiling of malignant glial cells. Molecular Cancer Therapeutic. 2003;2:1129–1137. [PubMed] [Google Scholar]
- 18.Brigati J, Williams DD, Sorokulova IB, Nanduri W, Chen IH, Turnbough CL, Jr, Petrenko VA. Diagnostic probes for Bacillus anthracis spores selected from a landscape phage library. Clinical Chemistry. 2004;50(11):1899–1906. doi: 10.1373/clinchem.2004.038018. [DOI] [PubMed] [Google Scholar]
- 19.Petrenko VA, Sorokulova IB. Detection of biological threats. A challenge for directed molecular evolution. J Microbiol Methods. 2004;58(2):147–168. doi: 10.1016/j.mimet.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Sorokulova IB, Olsen EV, Chen I, Fiebor B, Barbaree JM, Vodyanoy VJ, Chin BA, Petrenko VA. Landscape phage probes for Salmonella typhimurium. J Microbiol Methods. 2005;63(1):55–72. doi: 10.1016/j.mimet.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Olsen EV, Sorokulova IB, Petrenko VA, Chen IH, Barbaree JM, Vodyanoy VJ. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella typhimurium. Biosens Bioelectron. 2006;21:1434–1442. doi: 10.1016/j.bios.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Petrenko VA, Vodyanoy VJ. Phage display for detection of biological threat agents. J Microbiol Methods. 2003;53(2):253–262. doi: 10.1016/s0167-7012(03)00029-0. [DOI] [PubMed] [Google Scholar]
- 23.Petrenko VA, Olsen EV, Sykora JC, Kouzmitcheva GA, Sorokulova IB, Brigati JR, Chen IH, Chin BA, Vodyanoy VJ. Nanofabrication of Bioselective Materials Using Diverse Nanolandscapes Displayed on Live Viruses. 2005 NSTI Nanotechnology Conference and Trade Show; Anaheim, California, U.S.A.. 2005. [Google Scholar]
- 24.Minenkova OO, Ilyichev AA, Kishchenko GP, Petrenko VA. Design of specific immunogens using filamentous phage as the carrier. Gene. 1993;128(1):85–88. doi: 10.1016/0378-1119(93)90157-x. [DOI] [PubMed] [Google Scholar]
- 25.Mount JD, Samoylova TI, Morrison NE, Cox NR, Baker HJ, Petrenko VA. Cell Targeted Phagemid Rescued by Pre-Selected Landscape Phage. Gene. 2004;341:59–65. doi: 10.1016/j.gene.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Petrenko VA, Smith GP. Vectors and Modes of Display. In: Sidhu SS, editor. Phage Display in Biotechnology and Drug Discovery. CRC Pressw, Taylor & Francis Group; Bo Raton, FL, U.S.A.: 2005. p. 714. [Google Scholar]
- 27.Iannolo G, Minenkova O, Gonfloni S, Castagnoli L, Cesareni G. Construction, exploitation and evolution of a new peptide library displayed at high density by fusion to the major coat protein of filamentous phage. Biol Chem. 1997;378(6):517–521. doi: 10.1515/bchm.1997.378.6.517. [DOI] [PubMed] [Google Scholar]
- 28.Legendre D, Fastrez J. Construction and exploitation in model experiments of functional selection of a landscape library expressed from a phagemid. Gene. 2002;290:203–215. doi: 10.1016/s0378-1119(02)00562-0. [DOI] [PubMed] [Google Scholar]
- 29.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem Rev. 2005;105(11):4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 30.Samoylova TI, Cox NR, Morrison NE, Globa LP, Romanov V, Baker HJ, Petrenko VA. Phage matrix for isolation of glioma cell membrane proteins. Biotechniques. 2004;37(2):254–260. doi: 10.2144/04372RR02. [DOI] [PubMed] [Google Scholar]
- 31.Kouzmitcheva GA, Petrenko VA, Smith GP. Identifying diagnostic peptides for lyme disease through epitope discovery. Clin Diagn Lab Immunol. 2001;8(1):150–160. doi: 10.1128/CDLI.8.1.150-160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GP, Petrenko VA, Matthews LJ. Cross-linked filamentous phage as an affinity matrix. J Immunol Methods. 1998;215(12):151–161. doi: 10.1016/s0022-1759(98)00087-8. [DOI] [PubMed] [Google Scholar]
- 33.Souza GR, Christianson DR, Staquicini FI, Ozawa MG, Snyder EY, Sidman RL, Miller JH, Arap W, Pasqualini R. Networks of gold nanoparticles and bacteriophage as biological sensors and cell-targeting agents. Proc Natl Acad Sci U S A. 2006;103(5):1215–1220. doi: 10.1073/pnas.0509739103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanduri V, Samoylov AM, Petrenko VA, Vodyanoy VJ, Simonian AL. Comparison of Optical and Acoustic Wave Phage Biosensors. The Electrochemical Society, Inc. 2004 Joint International Meeting; Honolulu, Hawaii, Hilton Hawaiian Village. 2004. [Google Scholar]


