Abstract
MauG is a novel 42 kDa di-heme protein which is required for the biosynthesis of tryptophan tryptophylquinone, the prosthetic group of methylamine dehydrogenase. The visible absorption and resonance Raman spectroscopic properties of each of the two c-type hemes, and the overall redox properties of MauG are described. The absorption maxima for the Soret peaks of the oxidized and reduced hemes are 403 nm and 418 nm for the low-spin heme, and 389 nm and 427 nm for the high-spin heme, respectively. The resonance Raman spectrum of oxidized MauG exhibits a set of marker bands at 1503 cm−1 and 1588 cm−1 which exhibit similar frequencies to the ν3 and ν2 band of c-type heme proteins with bis-histidine coordination. Another set of marker bands at 1478 cm−1 and 1570 cm−1 are characteristic of a high-spin heme. Two distinct oxidation-reduction midpoint potential (Em) values of −159 mV and −244 mV are obtained from spectrochemical titration of MauG. However, the two ν3 bands located at 1478 and 1503 cm−1 shift together to 1467 and 1492 cm−1 upon reduction, as do the Soret peaks of the low- and high-spin hemes in the absorption spectrum. Thus, the two hemes with distinct spectral properties are reduced and oxidized to approximately the same extent during redox titrations. This indicates that the high- and low-spin hemes have similar intrinsic Em values but exhibit negative redox cooperativity. After the first one-electron reduction of MauG the electron equilibrates between hemes. This makes the second one-electron reduction of MauG more difficult. Thus, the two Em values do not describe redox properties of distinct hemes, but the first and second one-electron reductions of a di-heme system with two equivalent hemes. The structural and mechanistic implications of these findings are discussed.
MauG is a 42 kDa di-heme protein (1) which is required for the biosynthesis of tryptophan tryptophylquinone (TTQ) (2), the prosthetic group of methylamine dehydrogenase (MADH) (3). TTQ is synthesized through post-translational modification of two endogenous tryptophan residues. This modification involves two oxygenation reactions and one covalent cross-linking reaction. It has been shown that the incorporation of the second oxygen into βTrp57 and the covalent cross-linking of βTrp57 to βTrp108 are MauG-dependent processes (Figure 1). These reaction steps are severely compromised in vivo when mauG is mutated or deleted. Mutation of mauG in vivo leads to accumulation of a biosynthetic intermediate of TTQ in which βTrp57 is monohydroxylated and the covalent cross-link between βTrp57 and βTrp108 is absent (4). These steps may then be catalyzed in vitro upon addition of MauG to the isolated biosynthetic intermediate. Incubation of this biosynthetic intermediate in vitro with purified MauG results in the incorporation of the second oxygen into βTrp57 and formation of the cross-link with βTrp108 to form TTQ (5).
Figure 1.
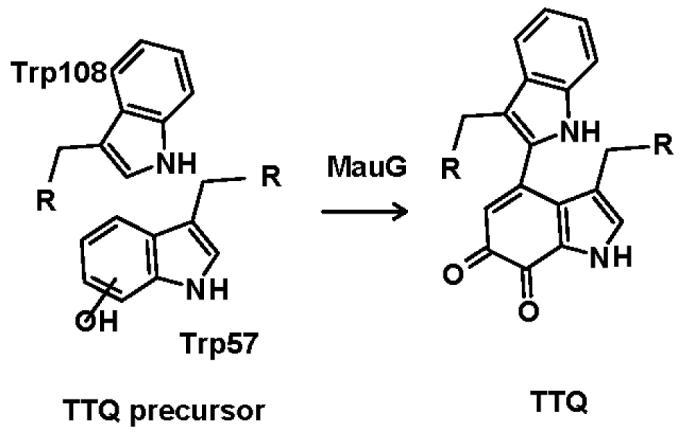
Role of MauG in tryptophan tryptophylquinone (TTQ) biosynthesis. This scheme is based on information presented in references (1, 5).
The genes that encode the MADH subunits, together with nine other genes that relate to MADH expression and function, are clustered in the methylamine utilization (mau) locus (6, 7). Four genes, mauFEDG, were shown to be essential for MADH maturation 6, 7). The MauG protein is the only one of these gene products that has been expressed and characterized (1). It contains two c-type hemes as predicted from its primary sequence, which contains two CXXCH motifs. In contrast to typical c-type cytochromes, the reduced form of MauG binds CO and is oxidized by O2. The electron paramagnetic resonance spectrum of oxidized MauG exhibits signals corresponding to one high-spin heme and one low-spin heme. The g values exhibited by these hemes are atypical of c-type cytochromes and much more similar to those of hemes that bind and activate oxygen, such as ligand complexes of cytochrome P450CAM and the complex of heme oxygenase with heme.
The description of MauG-mediated TTQ biosynthesis in vitro demonstrated a novel function for this c-type heme protein, which is consistent with its physical properties that are atypical for a c-type heme protein. In this paper we describe in detail the visible absorption and resonance Raman (RR) spectroscopic properties of each of the two c-type hemes, and the redox properties of MauG. The results indicate the high-spin heme and low-spin heme have very similar oxidation-reduction midpoint potential (Em) values but exhibit negative cooperativity such that two distinct Em values are obtained during the two-electron reduction of MauG. The structural and mechanistic implications of this unusual redox behavior are discussed.
METHODS
Materials
Homologous expression of MauG in P. denitrificans and methods for its isolation and purification have been described previously (1). All reagents were obtained from commercial sources and used without further purification.
Redox Titrations of MauG
Stoichiometric reduction and oxidation of MauG were performed anaerobically using sodium dithionite and potassium ferricyanide, respectively. Stock solutions of titrants were freshly prepared in 50 mM potassium phosphate, pH 7.4, which had been previously de-gassed and purged with nitrogen gas. The concentrations of titrants were standardized before and after MauG titrations by anaerobic titration against amicyanin, a protein standard with established redox properties (8) that is routinely used in our lab. The MauG sample was placed in a sealed cuvette and made anaerobic by repeated cycles of evacuation and argon replacement. Standardized solutions of titrants were quantitatively transferred with a gas-tight syringe and the MauG sample was mixed with a stir bar at the bottom of cuvette. Absorption spectra were recorded with a Multispec-1501 Shimadzu spectrophotometer.
Spectrochemical Titrations
The Em values of MauG were determined by spectrochemical titration. The ambient potential was measured directly with a Corning combination redox electrode which was calibrated using quinhydrone (a 1:1 mixture of hydroquinone and benzoquinone) as a standard with an Em value of +286 mV at pH 7.0 (9). The reaction mixture contained 5.0 μM MauG in 50 mM potassium phosphate buffer at pH 7.4 at 25°C. Flavin mononucleotide and safranin T were present as mediators. The mixture was titrated by addition of incremental amounts of dithionite, which was used as a reductant. The reaction was fully reversible. In the oxidative direction, titration by addition of potassium ferricyanide was performed. The absorption spectrum of MauG and the ambient potential were recorded after each incremental addition of reductant or oxidant. The concentrations of the oxidized and reduced MauG were determined by comparison with the spectra of the completely oxidized and reduced forms of MauG after the absorbance of mediators was subtracted from the recorded spectrum. Em values were obtained by fitting the experimental data to the following equations which describe the redox behavior of a system with either a single redox active component (eq 1) or two distinct redox-active components (eq 2), where a is the fraction of the total absorbance change that is attributable to one heme and (1-a) is the fraction of the total absorbance change that is attributable to the other heme. Em values are reported versus the normal hydrogen electrode (NHE).
| (1) |
| (2) |
Direct Electrochemical Analysis
A CH Instruments model CHI 660A electrochemical workstation was used for recording cyclic voltammograms (CVs) of MauG with a three-electrode cell consisting of a saturated Ag/AgCl reference electrode (Ag/AgCl), a Pt wire auxiliary electrode, and a single-walled carbon nanotube (SWNT) coated glassy carbon disk working electrode (CH Instruments, 3mm diameter). The working electrode was modified with SWNT which are pre-treated with 3:1/v/v solution of concentrated sulfuric acid (98%) and concentrated nitric acid (70%) as reported in the literature (10). Em values are reported versus the NHE.
Resonance Raman Spectroscopy
Resonance Raman (RR) spectra were obtained using a Raman spectrometer consisting of a Spex model 1877 triple spectrograph and a CCD detector as reported previously (11). The protein solution was placed in a homemade flow system and maintained at a constant flow during the experiment to avoid laser-induced damage. A 406.7-nm line from a argon-krypton ion laser (Spectra-Physics BeamLokTM model 2080-KV) was used as the excitation source and the Raman signal was collected in a 120° geometry. The laser power was adjusted to ∼6 mW at the sample. Each spectrum was recorded with a 30-s accumulation time, and 15 repetitively measured spectra were averaged to improve the quality of the final spectrum. The frequencies of the Raman bands were calibrated using a standard argon lamp.
Resonance Raman Spectroscopy of Different Redox States of MauG
The RR spectra of oxidized MauG, cytochrome c and myoglobin are shown and compared in Fig. 3. The RR spectrum of MauG much more closely resembles that of cytochrome c since most of the major marker bands have similar frequencies and intensities. This result further confirms that MauG is a c-type heme protein despite physical properties that are atypical of cytochromes c and more typical of oxygen-binding hemes (1). With 406.7 nm excitation, the spectrum of MauG obtained at pH 7.5 shows two ν3 bands at 1503 and 1478 cm−1, two ν2 bands at 1570 cm−1 and 1588 cm−1, and a ν4 band at 1374 cm−1. These marker bands, which are sensitive to the spin and coordination states of the heme iron, along with the slightly broad ν10 band at ∼1636 cm−1 reveal that MauG has two heme centers of different structure. The set of marker bands at 1503 cm−1 and 1588 cm−1 exhibit very similar frequencies to the ν3 and ν2 band of c-type heme proteins with bis-histidine coordination (12-16) and strongly suggests that one heme center is a six coordinated low-spin species with histidines as the proximal and distal axial ligands. The set of marker bands at 1478 cm−1 and 1570 cm−1 are suggestive of a high-spin species. However, these bands are significantly lower in frequency than that of most high-spin c-type heme centers, which are normally 5-coordinate (17). Since the ν3 band from 6-coordinate high-spin species normally appears at a lower frequency than that from 5-coordinated one (18, 19), the ν3 band at 1478 cm−1 may indicate that the high-spin heme is a 6-coordinated species when oxidized.
Figure 3.
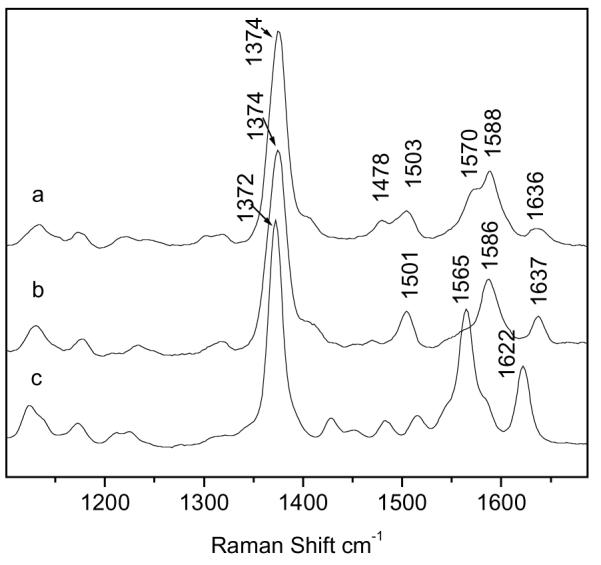
Resonance Raman spectra of MauG (a), cytochrome c (b) and myoglobin (c). Protein concentrations were 0.25 mM.
When fully reduced by sodium dithionite, the RR spectrum of MauG (Fig. 4A) shows a ν4 band at 1359 cm−1 and two ν3 bands at 1467 cm−1 and 1492 cm−1. The Raman band at 1492 cm−1 is coming from the reduced form of the low-spin heme center. The 1467 cm−1 band arises from the high-spin heme center, however, the frequency of this band now suggests a 5-coordinate heme center (17). This result suggests that, upon reduction, the high-spin heme center in MauG undergoes a structural change that may involve displacement of a weak exogenous ligand such as water. Such a structural change will result in a vacant distal pocket for the high-spin heme in reduced MauG which may be important for substrate binding.
Figure 4.
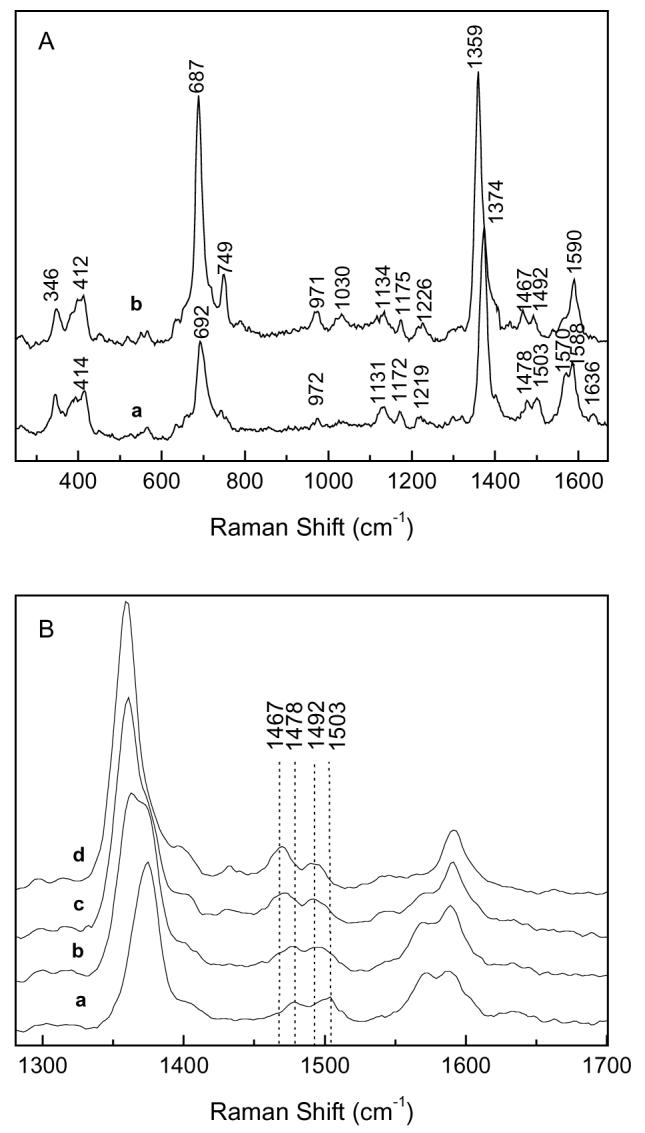
Resonance Raman Spectra of different redox states of MauG. A. Spectra of fully oxidized (a) and fully reduced (b) MauG. B. Spectral changes occurring during reduction of MauG by incremental additions of sodium dithionite: fully oxidized MauG (a), partially reduced MauG (b and c), and fully reduced MauG (d). Protein concentrations were 0.25 mM.
The results of the redox titration that was monitored by visible absorption spectroscopy (Fig. 2) indicated that the two hemes of MauG were reduced to approximately the same extent during the reductive titration. This phenomenon was further investigated using another independent spectroscopic technique, RR spectroscopy, to monitor the reduction of MauG. RR may be performed in solution at ambient temperature so that the results can be directly compared and correlated with those of the redox titrations described above. It was not possible to directly monitor the potential and the RR spectrum simultaneously with our experimental setup. To address the question of whether the two hemes titrate individually and sequentially, or together, the overall redox state of MauG was referenced the changes in the ν4 band which shifts from 1374 cm−1 in oxidized MauG to 1359 cm−1 in fully reduced MauG. Figure 4B shows the RR spectral changes which occur during a reductive titration with sodium dithionite. When MauG is in intermediate stages of reduction, as judged by the shift and broadening in the ν4 band, the ν3 bands from both heme centers at 1478 cm−1 and 1503 cm−1 shift to 1467 cm−1 and 1492 cm−1 at the same time during the reduction process. These data confirm that the two hemes of MauG are reduced in concert rather than sequentially during the reductive titration.
Figure 2.
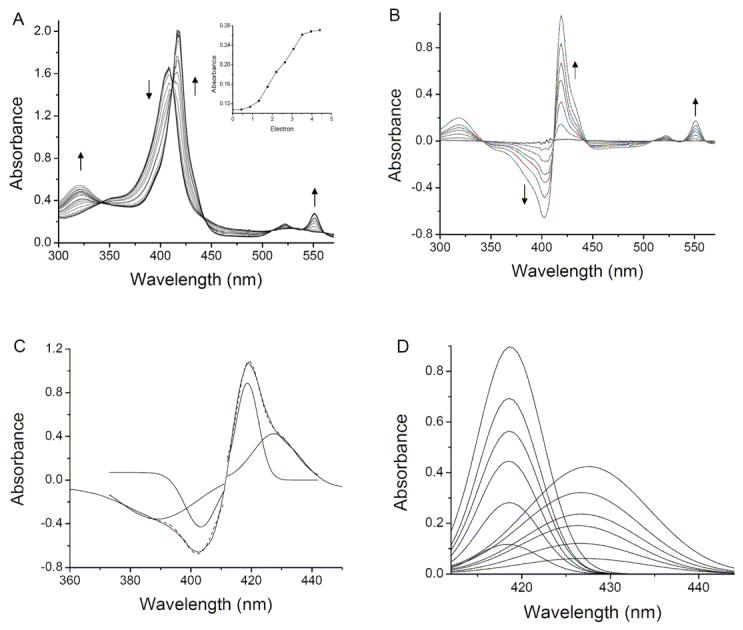
Anaerobic reductive titration of MauG with sodium dithionite. A. Complete reduction by dithionite. The sample contained 7.9 μM MauG in 50 mM potassium phosphate pH 7.4. Spectra were recorded after incremental additions of sodium dithionite. The arrows indicate the direction of the spectral changes during the titration. The change in absorbance at 550 nm on addition of electron equivalents in the form of added dithionite are shown in the inset. B. Reduced minus oxidized difference spectra. In this representation of the data shown in A, the spectrum of the fully oxidized MauG was subtracted from each spectrum that was obtained after each addition of dithionite. C. The individual components of the Soret peaks in the fully reduced minus oxidized spectrum were deconvoluted by Gaussian fitting with Origin 7.0. Each of the two fitted components is drawn under the original spectrum and the overall fit of the sum of the two is drawn as a dashed line which overlays the original spectrum. D. Deconvoluted spectra of the Soret peaks in reduced minus oxidized difference spectra shown in B.
Spectrochemical Redox Titrations
The results described above indicate that the two hemes of MauG have distinct absorption and RR spectra but are reduced and oxidized together during the redox titrations. Intuitively, this suggests that the Em values for the two hemes must be identical. To investigate this further, spectrochemical titrations of MauG were performed to precisely determine the Em values associated with the different redox couples of MauG. Titrations were performed using FMN and safranine T as mediators (Fig. 5). The fraction of reduced MauG was determined by the absorbance at either 550 nm or 406 nm.
Figure 5.
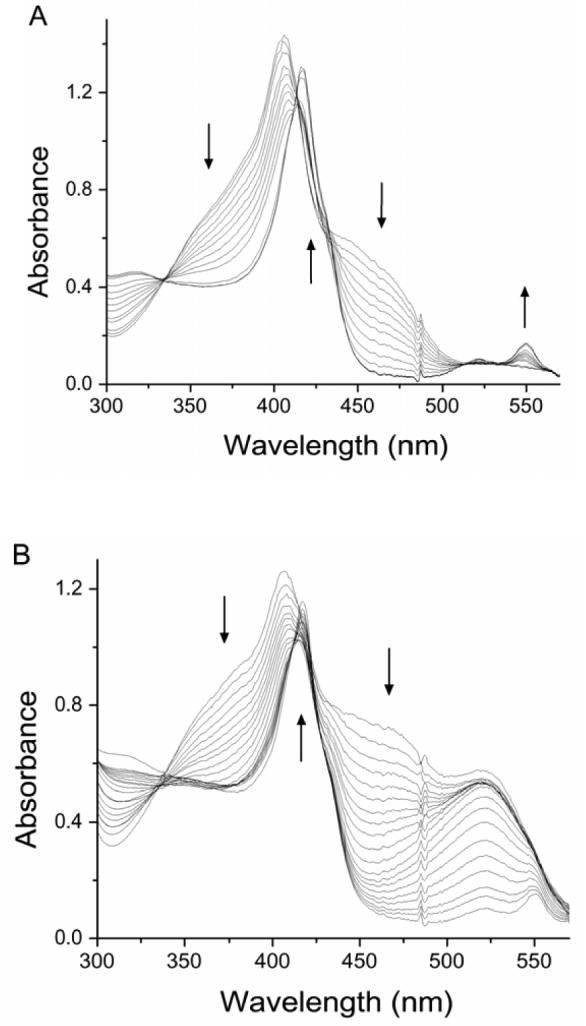
Spectrochemical titrations of MauG (5.0 μM) in the presence of FMN (40 μM) as a mediator (A) and in the presence of FMN (50 μM) plus safranin T (16 μM) as mediators (B). The arrows indicate the direction of the spectral changes during the titration in the reductive direction.
Initial attempts to fit the data from spectrochemical titrations to the standard linear Nernst plot according to (eq 3) consistently yielded plots which were slightly curved with slopes of
| (3) |
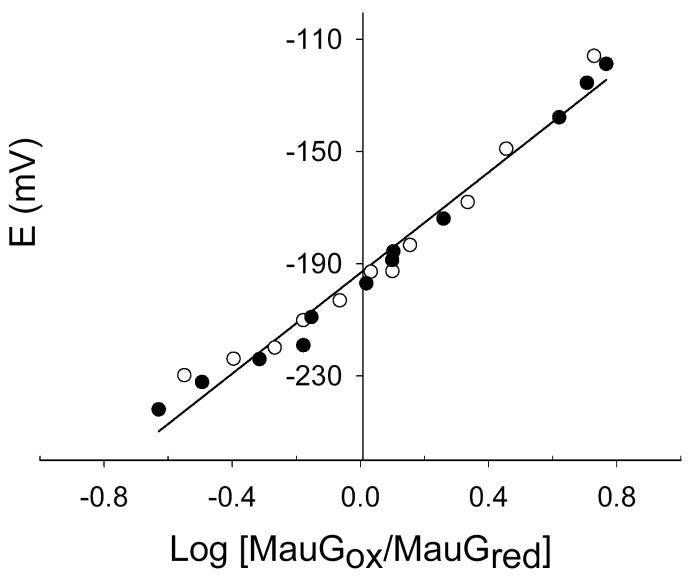
approximately 90 mV (i.e., n=0.67) (Fig. 6). This curvature, and a fitted n value much smaller than the known value of n=1.0 for heme, suggested the possibility of negative cooperativity between the hemes. As such the data were fit to eqs 1 and 2 which allow better visualization of the validity of alternative fits to a system with a single component or two distinct components. If the two hemes were equivalent and did not exhibit cooperativity, then the data would fit well to eq 1 and a fit to eq 2 would yield identical values for Em1 and Em2. This was not the case. As indicated in Table 1, essentially identical results were obtained from analysis of the absorbance changes at either 550 nm or 406 nm, and in the presence or absence of safranine T in addition to FMN. This shows that the experimentally determined Em values were not influenced by the redox dyes or interference in the analysis from spectral overlap. Data consistently yielded better fits to eq 2, which assumes an n value of 1.0 for each heme, with Em values of −159 ± 10 mV and −244 ± 5 mV vs NHE (Fig. 7).
Figure 6.
Nernst plot for the reductive titration of MauG in the presence of FMN as a mediator. Experimental points from the forward and reverse reductive (closed circles) and oxidative (open circles) titrations are shown. Values of log MauGox/MauGred were obtained from the absorbance changes at 550 nm. The straight line shows the fit of these data to eq 3 which yields a slope 90 mV (n=0.67) and intercept of −193 mV.
Table 1.
Em values obtained from spectrochemical titrations of MauGa
| Mediator present | Wavelength (nm)b | Em1 (mV) | Em2 (mV) |
|---|---|---|---|
| FMN | 550 | −240 ± 4 | −159 ± 9 |
| FMN | 406 | −247 ± 5 | −159 ± 14 |
| FMN + safranine T | 406 | −246 ± 5 | −159 ± 4 |
Em values were obtained by fitting the experimental data from titrations shown in Figure 7 and similar experiments to eq 2.
The concentrations of the oxidized and reduced MauG were determined by comparison with the spectra of the completely oxidized and reduced forms of MauG at the indicated wavelength.
Figure 7.
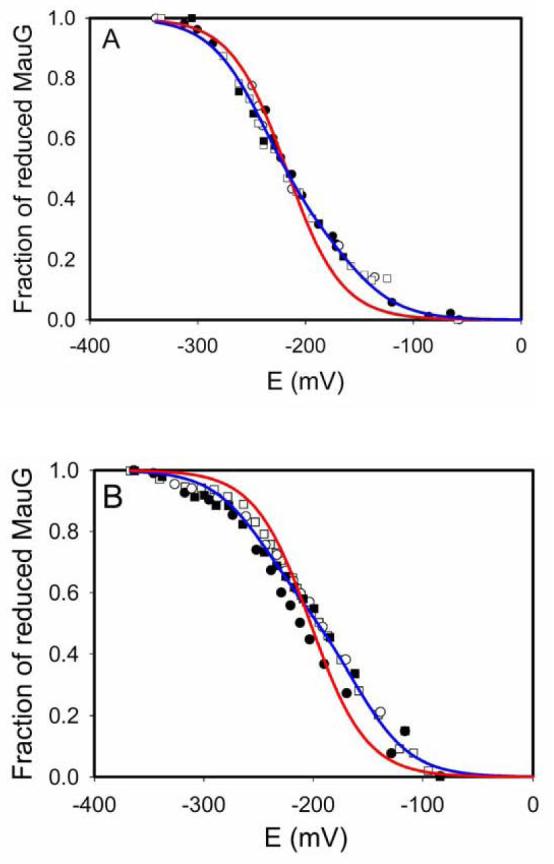
Determination of Em values of MauG. In each experiment the MauG sample was fully reduced by incremental addition of sodium dithionite (closed circles), then fully oxidized by incremental addition of ferricyanide (open circles), then re-reduced by addition incremental sodium dithionite (closed squares), and then re-oxidized by incremental addition of ferricyanide (open squares). The sets of data from each of these four reversible titrations are overlaid and were all included in the analysis. The fits of these data to equations 1 (red line, one-component fit) and 2 (blue line, two-component fit) are shown. A. This titration was performed in the presence of FMN as a mediator and Fraction reduced of MauG was calculated from the spectral changes at 550 nm. B. This titration was performed in the presence of FMN plus safranine T as mediators and Fraction reduced of MauG was calculated from the spectral changes at 406 nm.
Direct Electrochemistry of MauG
The redox properties of MauG were also analyzed by CV at a SWNT modified electrode (Figure 8A). The complexity of the system described in the spectrochemical studies is also evident in these data. The asymmetric cathodic (upper) peak is also suggestive of negative cooperativity. Computer simulations of the electrochemical data were performed to determine the basis for this asymmetry. The simulation which was most consistent with the experimental data describes a mechanism which occurs in three stages. The first step is an electrochemical process which involves the transfer of an electron to the protein at the electrode surface. This gives rise to a product which is converted into a second species which is itself electrochemically active within the potential window used in the experiment. A model consistent with this mechanism for MauG is shown in Fig. 9. It should be noted that it was only possible to obtain qualitative support for this model using these data. A more quantitative simulation required knowledge of parameters which could not be determined such as the surface concentration of the electroactive proteins. Another problem is that theoretical models of electron transfer kinetics that have been established for planar electrodes are not necessarily applicable to the surface of the SWNT modified electrode.
Figure 8.
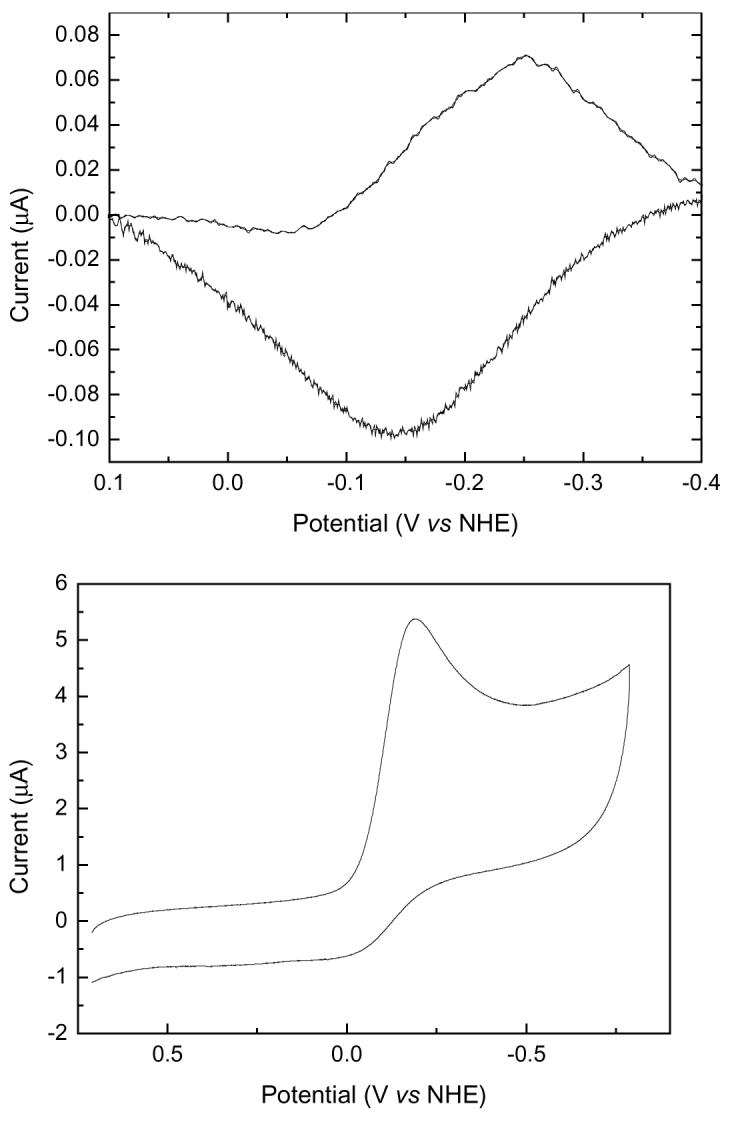
A. CV of MauG on a SWCN modified electrode in pH, 7.5 buffer under anaerobic conditions condition. B. CV of MauG on a SWCN modified electrode in pH, 7.5 buffer in the presence of oxygen.
Figure 9.
Model for cooperative redox behavior during the anaerobic reduction of MauG.
Although it was not possible to distinguish the two individual Em values by this technique, a single average Em value could be determined anaerobically at pH 7.5 of −198 mV versus NHE. This correlates very closely with the results of the spectrochemical titrations as this value is approximately equal to the average of the two Em values that were obtained in the spectrochemical study.
In the presence of oxygen, the CV of MauG (Figure 8B) shows a catalytic reduction peak of oxygen which is a characteristic of oxygen-binding heme proteins such as myoglobin and hemoglobin. This provides additional evidence for the oxygen-binding property of MauG.
DISCUSSION
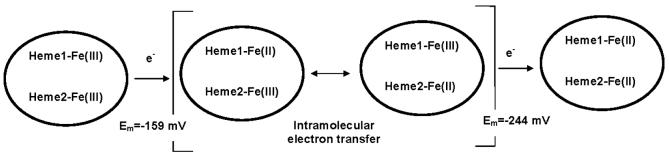
The data obtained during the spectrochemical titrations of MauG are clearly best fit to a two-component model with Em values of −159 and −244 mV. If these Em values described each of the two hemes of MauG in the absence of any cooperativity or interaction between hemes, then at the midpoint in the overall redox titration one heme should be predominantly reduced and the other predominantly oxidized. If the two hemes were reduced sequentially, then the difference spectra for the first and second halves of the titration should only be identical if the two hemes possess identical spectra. The high- and low-spin hemes of MauG have different spectral properties (Figs. 2 and 4), yet the data for MauG do show that these two difference spectra are essentially identical. Thus, the two hemes of MauG with distinct spectra appear to be reduced simultaneously, despite the observation of two Em values for the overall oxidation-reduction of MauG. This means that it is inappropriate to assign each of the Em values to one or the other heme. Instead the Em values should be viewed as describing the first and second one-electron reductions of an interacting di-heme system.
A model that describes the redox behavior of MauG under anaerobic conditions is given in Figure 9. In this model the intrinsic Em value of each heme is very similar. The hemes are designated H1 and H2 since it is not possible to distinguish which serves as the entry point for the electron into MauG. After the first electron reduces the first heme of MauG, that electron rapidly equilibrates between the two hemes. The Em value associated with this first one-electron reduction of MauG is −159 mV. Once the first electron enters the system, this makes the further reduction of MauG more difficult, possibly due to electrostatic considerations. The Em value associated with the second one-electron reduction is −244 mV. Thus, the two redox potentials do not describe redox properties of distinct hemes, but the first and second one-electron reductions of a di-heme system with two hemes with similar intrinsic redox properties. Such behavior may be described as negative cooperativity.
The model in Figure 9 requires that the two hemes of MauG be well-coupled and suggests that they are positioned or oriented such that rapid electron transfer from one heme to the other may occur. This has mechanistic consequences. The mechanism by which MauG catalyzes the incorporation of oxygen into the biosynthetic precursor of TTQ has not yet been determined. A model for the activation of O2 by MauG that is based on the proposed mechanism of cytochrome P-450 enzymes and other heme-containing oxygenases (20) is shown in Figure 10. The high-spin and low-spin hemes are designated HemeH and HemeL, respectively. The reduced high-spin heme binds dioxygen and transfers an electron to it resulting in a covalent ferric-superoxide intermediate. This is followed by intraheme electron transfer followed by delivery of the second electron to this intermediate to form a ferric-peroxide intermediate. In the presence of substrate it is possible that this intermediate is directly involved in the oxygenation reaction required for TTQ biosynthesis (reaction A in Fig. 10). Alternatively this intermediate may convert to an oxo-ferryl intermediate which is used for the biosynthetic oxygenation reaction (reaction B in Fig. 10). Thus while oxygen is bound to the high-spin heme, the strong interaction with the low-spin heme allows the oxygen-binding heme to effectively function as a two-electron reductant. The ability to efficiently deliver the second electron to the ferricsuperoxide intermediate may be important mechanistically for two reasons. It prevents dioxygen from being released after reversible binding, thus effectively enhancing the affinity of reduced MauG for O2. It also prevents the accumulation of the potentially toxic superoxide intermediate which could potentially damage the host protein. In the absence of substrate, fully oxidized MauG may be regenerated by reaction C in Fig. 10. This accounts for the observation that reduced MauG is reoxidized by air (1).
Figure 10.
Possible reaction intermediates during the redox cycle of MauG in the presence of oxygen and substrate. HemeH and HemeL designate the high-spin and low-spin heme, respectively. A-C designate possible reactions involving intermediates which are described in the text.
In general, deviations from simple Nernstian behavior in a system with multiple redox centers may be due to either intrinsic differences in Em values of the redox centers or cooperative interactions between the redox centers. Evidence has been presented for cooperative interactions in some proteins which possess multiple c-type hemes. Examples include the di-heme cytochrome c4 (21) and the tetra-heme cytochrome c554 from Nitrosomonas europa (22, 23). In contrast to MauG, these multi-heme c-type cytochromes function solely as electron transfer mediators. MauG exhibits some sequence similarity to di-heme cytochrome c peroxidases (1). These proteins also possess both high- and low-spin c-type hemes (24, 25). In contrast to MauG, the Em values of these two hemes are not equivalent but are separated by several hundred millivolts. Thus, among the multi-c-type heme proteins which have been thus far characterized, MauG remains unique in that it seems to take advantage of redox cooperativity between two c-type hemes to allow for activation of O2 by the high-spin heme. This description of MauG does bear strong similarity to the redox cooperativity that has been described for cytochrome c oxidase. Cooperative interactions between cytochrome a and cytochrome a3 of cytochrome c oxidase have long been considered to play an important role in the mechanism of that complex redox enzyme (26-28). In this case, rapid interheme electron transfer is important for the catalytic reduction of O2 which occurs at the cytochrome a3/CuB site. It has also been demonstrated that the relative timing of delivery of electrons and protons to this site of reaction is critical to achieve catalysis rather than suicide inactivation which likely results from damage from a reactive oxygen intermediate (29). Thus, the type of redox cooperativity described in this study may be of general importance to cofactor-cofactor interactions in enzymes which use a one-electron redox center to activate dioxygen.
The structure of MauG is not known but it is known from the sequence that each heme possesses at least one His axial ligand. The sequence contains two CXXCH motifs which are diagnostic of a c-type heme with covalent attachment via the two Cys residues and with the His following the second Cys serving as an axial ligand. The RR spectrum and Em value for the low spin six-coordinate heme suggests that a second His serves as the second axial ligand. Em values for most His-Met c-type hemes are greater than +200 mV, while those of His-His hemes are negative (30). The previously published EPR data for MauG (1) are also consistent with an imidazolate distal ligand. The Em value for the high-spin five-coordinate heme is somewhat negative compared to other five-coordinate hemes with an axial His ligand which usually have positive Emvalues (30). An exception is the 5-coordinate heme of di-heme cytochrome c peroxidase which exhibits an Em of −260 mV (24). This suggests that in MauG the protein environment of the proximal His ligand of the high-spin heme, and possibly hydrogen interactions, are responsible for the relatively negative Em value. Oxygen activation by heme proteins is typically catalyzed by cytochrome P450 enzymes. The heme cofactors of cytochrome P-450 enzymes possess a single axial ligand that is provided by a Cys residue. In MauG, the only Cys residues that are present in the sequence are those in the CXXCH motifs which are predicted to be involved in the covalent thioether linkage between the protein and the heme. Thus, it is not possible for Cys to provide an axial ligand for either heme of MauG. The results of this study suggest that O2 activation by MauG is catalyzed by a 5-coordinate high-spin c-type heme, with a single His axial ligand, which interacts closely with a second c-type heme. This suggests an atypical mechanism for a heme-catalyzed oxygenation reaction. Efforts to obtain the structure of MauG are in progress and additional studies are planned to further elucidate the physical properties and catalytic mechanism of MauG.
ACKNOWLEDGMENTS
The authors thank Yu Tang for technical assistance and Jonathan Hosler for helpful discussion.
†This work was supported by NIH grants GM-41574 (V.L.D.), S06G008047 (H.T.), and NIHRCMI 1G12RR12459-01 (H.T.)
Abbreviations
- MADH
methylamine dehydrogenase
- TTQ
tryptophan tryptophylquinone
- Em
oxidation-reduction midpoint potential
- NHE
normal hydrogen electrode
- RR
resonance Raman
- CV
cyclic voltammogram
- SWNT
single-walled carbon nanotube
REFERENCES
- 1.Wang Y, Graichen ME, Liu A, Pearson AR, Wilmot CM, Davidson VL. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42:7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 2.McIntire WS, Wemmer DE, Chistoserdov A, Lidstrom ME. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991;252:817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- 3.Davidson VL. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv. Protein Chem. 2001;58:95–140. doi: 10.1016/s0065-3233(01)58003-1. [DOI] [PubMed] [Google Scholar]
- 4.Pearson AR, de la Mora-Rey T, Graichen ME, Wang Y, Jones LH, Marimanikkupam S, Aggar SA, Grimsrud PA, Davidson VL, Wilmot CM. Further insights into quinone cofactor biogenesis: Probing the role of mauG in methylamine dehydrogenase TTQ formation. Biochemistry. 2004;43:5494–5502. doi: 10.1021/bi049863l. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Li X, Jones LH, Pearson AR, Wilmot CM, Davidson VL. MauG-dependent in vitro biosynthesis of tryptophan tryptophylquinone in methylamine dehydrogenase. J. Am. Chem. Soc. 2005;127:8258–8259. doi: 10.1021/ja051734k. [DOI] [PubMed] [Google Scholar]
- 6.van der Palen CJ, Reijnders WN, de Vries S, Duine JA, van Spanning RJ. MauE and MauD proteins are essential in methylamine metabolism of Paracoccus denitrificans. Antonie Van Leeuwenhoek. 1997;72:219–228. doi: 10.1023/a:1000441925796. [DOI] [PubMed] [Google Scholar]
- 7.van der Palen CJ, Slotboom DJ, Jongejan L, Reijnders WN, Harms N, Duine JA, van Spanning RJ. Mutational analysis of mau genes involved in methylamine metabolism in Paracoccus denitrificans. Eur. J. Biochem. 1995;230:860–871. doi: 10.1111/j.1432-1033.1995.tb20629.x. [DOI] [PubMed] [Google Scholar]
- 8.Husain M, Davidson VL, Smith AJ. Properties of Paracoccus denitrificans amicyanin. Biochemistry. 1986;25:2431–2436. doi: 10.1021/bi00357a020. [DOI] [PubMed] [Google Scholar]
- 9.Cammack R. In: Bioenergetics: A Practical Approach. Brown GC, Cooper CE, editors. IRL Press; New York: 1995. pp. 85–109. [Google Scholar]
- 10.Liu Z, Shen Z, Zhu T, Hou S, Ying L, Shi Z, Gu Z. Organizing single-walled carbon nanotubes on gold using a wet chemical self-assembling technique. Langmuir. 2000;16:3569–3573. [Google Scholar]
- 11.Feng M, Tachikawa H. Raman spectroscopic and electrochemical characterization of myoglobin thin film: implication of the role of histidine 64 for fast heterogeneous electron transfer. J. Am. Chem. Soc. 2001;123:3013–3020. doi: 10.1021/ja003088p. [DOI] [PubMed] [Google Scholar]
- 12.Chottard G, Kazanskaya I, Bruschi M. Resonance Raman study of multihemic c-type cytochromes from Desulfuromonas acetoxidans. Eur. J. Biochem. 2000;267:1050–1058. doi: 10.1046/j.1432-1327.2000.01096.x. [DOI] [PubMed] [Google Scholar]
- 13.Andersson KK, Babcock GT, Hooper AB. Diheme cytochrome c-554 from Nitrosomonas. Soret resonance Raman indication of an unusual ferric 5-coordinate structure. FEBS Lett. 1984;170:331–334. doi: 10.1016/0014-5793(84)81338-1. [DOI] [PubMed] [Google Scholar]
- 14.Oellerich S, Wackerbarth H, Hildebrandt P. Spectroscopic characterization of nonnative conformational states of cytochrome c. J. Phys. Chem. B. 2002;106:6566–6580. [Google Scholar]
- 15.Eng LH, Schlegel V, Wang D, Neujahr H, Stankovich MT, Cotton T. Resonance Raman scattering and surface enhanced resonance Raman scattering studies of oxidoreduction of cytochrome c3. Langmuir. 1996;12:3055–3059. [Google Scholar]
- 16.Verma AL, Kimura K, Nakamura A, Yagi T, Inokuchi H, Kitagawa T. Resonance Raman studies of hydrogenase catalyzed Reduction of cytochrome c3 by hydrogen. Evidence for heme heme interactions. J. Am. Chem. Soc. 1988;110:6617–6623. [Google Scholar]
- 17.Othman S, Richaud P, Vermeglio A, Desbois A. Evidence for a proximal histidine interaction in the structure of cytochromes c in solution: a resonance Raman study. Biochemistry. 1996;35:9224–9234. doi: 10.1021/bi952818g. [DOI] [PubMed] [Google Scholar]
- 18.Smulevich G, Mauro JM, Fishel LA, English AM, Kraut J, Spiro TG. Heme pocket interactions in cytochrome c peroxidase studied by site-directed mutagenesis and resonance Raman spectroscopy. Biochemistry. 1988;27:5477–5485. doi: 10.1021/bi00415a014. [DOI] [PubMed] [Google Scholar]
- 19.Feng M, Tachikawa H, Wang X, Pfister TD, Gengenbach A,J, Lu Y. Resonance Raman spectroscopy of cytochrome c peroxidase variants that mimic manganese peroxidase. J. Biol. Inorg. Chem. 2003;8:699–706. doi: 10.1007/s00775-003-0460-9. [DOI] [PubMed] [Google Scholar]
- 20.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-containing oxygenases. Chem. Rev. 1996;96:28441–22887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 21.Leitch FA, Brown KR, Pettigrew GW. Complexity in the redox titration of the dihaem cytochrome c4. Biochim. Biophys. Acta. 1985;808:213–218. doi: 10.1016/0005-2728(85)90001-5. [DOI] [PubMed] [Google Scholar]
- 22.Arciero DM, Collins MJ, Haladjian J, Bianco P, Hooper AB. Resolution of the four hemes of cytochrome c554 from Nitrosomonas europaea by redox potentiometry and optical spectroscopy. Biochemistry. 1991;30:11459–11465. doi: 10.1021/bi00112a013. [DOI] [PubMed] [Google Scholar]
- 23.Upadhyay AK, Petasis DT, Arciero DM, Hooper AB, Hendrich MP. Spectroscopic characterization and assignment of reduction potentials in the tetraheme cytochrome c554 from Nitrosomonas europaea. J. Am. Chem. Soc. 2003;125:1738–1747. doi: 10.1021/ja020922x. [DOI] [PubMed] [Google Scholar]
- 24.Arciero DM, Hooper AB. A di-heme cytochrome c peroxidase from Nitrosomonas europaea catalytically active in both the oxidized and half-reduced states. J. Biol. Chem. 1994;269:11878–11886. [PubMed] [Google Scholar]
- 25.Gilmour R, Goodhew CF, Pettigrew GW, Prazeres S, Moura I, Moura JJ. Spectroscopic characterization of cytochrome c peroxidase from Paracoccus denitrificans. Biochem. J. 1993;294:745–752. doi: 10.1042/bj2940745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendler RW, Westerhoff HV. Redox interactions in cytochrome c oxidase: from the “neoclassical” toward “modern” models. Biophys. J. 1992;63:1586–1604. doi: 10.1016/S0006-3495(92)81748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wikstrom KF, Harmon HJ, Ingledew WJ, Chance B. A re-evaluation of the spectral, potentiometric and energy-linked properties of cytochrome c oxidase in mitochondria. FEBS Lett. 1976;65:259–277. doi: 10.1016/0014-5793(76)80127-5. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls P, Petersen LC. Haem-haem interactions in cytochrome aa3 during the anaerobic-aerobic transition. Biochim. Biophys. Acta. 1974;357:462–467. doi: 10.1016/0005-2728(74)90038-3. [DOI] [PubMed] [Google Scholar]
- 29.Mills DA, Hosler JP. Slow proton transfer through the pathways for pumped protons in cytochrome c oxidase induces suicide inactivation of the enzyme. Biochemistry. 2005;44:4656–4666. doi: 10.1021/bi0475774. [DOI] [PubMed] [Google Scholar]
- 30.Reedy CJ, Gibney BR. Heme protein assemblies. Chem. Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]