Abstract
Bis(3′-indolyl)methane (DIM) is a metabolite of the phytochemical indole-3-carbinol, and both compounds exhibit a broad spectrum of anticancer activities. We have developed a series of synthetic symmetrical ring-substituted DIM analogues, including 5,5′-dibromoDIM, which are more potent than DIM as inhibitors of cancer cell and tumor growth. In colon cancer cells, 5,5′-dibromoDIM decreased cell proliferation and inhibited G0-G1- to S-phase progression, and this was accompanied by induction of the cyclin-dependent kinase inhibitor p21 in HT-29 and RKO colon cancer cells. Mechanistic studies showed that induction of p21 in both RKO (p53 wild-type) and HT-29 (p53 mutant) cells by 5,5′-dibromoDIM was Krüppel-like factor 4 (KLF4) dependent, and induction of p53 in RKO cells was also KLF4 dependent. Analysis of the p21 promoter in p53-dependent RKO cells showed that 5,5′-dibromoDIM activated p21 gene expression through the proximal GC-rich sites 1 and 2, and chromatin immunoprecipitation assays showed that KLF4 and p53 bound to this region of the promoter, whereas in HT-29 cells unidentified upstream cis-elements were required for induction of p21. 5,5′-DibromoDIM (30 mg/kg/d) also inhibited tumor growth and induced p21 in athymic nude mice bearing RKO cells as xenografts, showing that ring-substituted DIM such as 5,5′-dibromoDIM represent a novel class of mechanism-based drugs for clinical treatment of colon cancer.
Introduction
Krüppel-like factor 4 (KLF4) or gut-enriched Krüppel-like factor is a member of the specificity protein/Krüppel-like factor family of transcription factors that are characterized by three COOH-terminal zinc fingers, which bind GC/GT-rich sequences (1, 2). KLF4 is developmentally regulated in multiple tissues and highly expressed in terminally differentiated intestinal epithelial cells (3–7). In mouse models where expression of KLF4 is abrogated, there is severe disruption of epidermal differentiation of skin development and offspring die shortly after birth (4, 8). KLF4 also plays a unique role in carcinogenesis and can act as a tumor suppressor gene in some tumors and an oncogene for others (9, 10). KLF4 is overexpressed in 70% of human breast cancers and also in squamous cell carcinomas (11, 12), and results of various genetic screens suggest that KLF4 transforms epithelial cells and can also be oncogenic in murine leukemias and lymphomas (11, 13, 14). At least one mechanism for the oncogenic action of KLF4 is associated with transcriptional repression of p53 in breast cancer (13).
In contrast, there is extensive evidence supporting the role of KLF4 as a tumor suppressor gene for multiple cancers, particularly those in the gastrointestinal tract (10, 11). KLF4 expression is decreased in various colon tumors, gastric cancers, esophageal squamous cell carcinomas, bladder and prostate cancer, lung cancer, and human glioma cells (12, 15–24). In addition, KLF4 expression is decreased in the Min mouse model, which spontaneously develop intestinal tumors due to inactivation of the APC gene (15). The mechanisms associated with the tumor suppressor activity of KLF4 are complex and involve interactions with multiple transcription factors and signaling pathways, particularly those associated with the tumor suppressor gene p53 and the cyclin-dependent kinase inhibitor p21 (9, 10).
The tumor suppressor gene p53 responds to different forms of cellular stress, including DNA damage to maintain genomic stability, and activation of p21 is a major p53-dependent response (25–27). Several studies show the relationship between KLF4, p53, and p21 where KLF4 appears to activate both genes (16, 26, 27). p53 induces transactivation in cells transfected with constructs containing both KLF4 and p21 promoter inserts, and both KLF4 and p53 synergistically activate p21 promoter constructs (16). Moreover, it was also shown that p53-dependent activation of KLF4 was important for inhibition of G1- to-S phase progress of colon cancer cells following DNA damage (27).
The importance of KLF4 as a tumor suppressor gene in colon and other cancer cell lines is also consistent with induction of this gene by cytotoxic agents, including 15-deoxy-Δ12,14-prostaglandin J2, sodium butyrate, and the peroxisome proliferator activator receptor γ agonist methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate (28–31). In this study, we show that cytotoxic ring-substituted bis(3′-indolyl)methane (DIM), including the 5,5′-dibromoDIM analogue, induced KLF4 expression in several colon cancer cell lines. Moreover, increased KLF4 was accompanied by induction of p21 and inhibition of G1- to S-phase progression in RKO and HT-29 colon cancer cells, which express wild-type and mutant p53, respectively. Subsequent analysis of the p21 promoter by transfection, chromatin immunoprecipitation, and RNA interference assays in RKO cells shows that KLF4 is directly recruited to the proximal GC-rich region of the p21 promoter and is required for activation of p21. Moreover, 5,5′-dibromoDIM also inhibits tumor growth and induces p21 expression in athymic nude mice bearing RKO cells as xenografts, showing that induction of KLF4 is associated with the antitumorigenic activity of this compound in both in vitro and in vivo models.
Materials and Methods
Cell Culture
RKO human colon carcinoma cell line was provided by M. D. Anderson Cancer Center, and HT-29 cells were purchased from the American Type Culture Collection. RKO and HT-29 cells were maintained in DMEM/Ham’s F-12 (Sigma-Aldrich) without phenol red supplemented with 0.22% sodium bicarbonate, 0.011% sodium pyruvate, 5% fetal bovine serum, and 10 mL/L of 100× antibiotics antimycotic solution (Sigma-Aldrich). Cells were maintained at 37°C in the presence of 5% CO2 and colon cancer cells used in these experiments were within passages 20 to 40.
Reagents
Antibodies for KLF4, p53, IgG, Sp1, Sp3, Sp4, and β-actin were purchased from Santa Cruz Biotechnology. The p21 antibody was purchased from BD PharMingen. Propidium iodide was obtained Calbiochem, and RNase A and corn oil was from Sigma-Aldrich. The kinase inhibitors SB203580, PD98059, and SP600125 were purchased from Calbiochem. Western Lightning chemiluminescence reagent was from Perkin-Elmer Life Sciences. p21 promoter reporter constructs pWWP, pWWP124, and pWWP101 were provided by Dr. Toshiyuki Sakai (Kyoto Prefectural University of Medicine). pWWP60 was generated by digesting pWWP with SmaI followed by religation of the purified vector (32). LipofectAMINE 2000 transfection reagent was purchased from Invitrogen. Reporter lysis buffer and luciferase reagent for luciferase assays were purchased from Promega. For RNA interference assays, we used a nonspecific scrambled (iScr) oligonucleotide and small inhibitory RNA for KLF4 (iKLF4) purchased from Dharmacon Research.
Cell Proliferation Assay
RKO and HT-29 cells were seeded in 12-well plates using DMEM/Ham’s F-12 plus 2.5% charcoal-stripped fetal bovine serum. After cells reached 50% to 60% confluence, which was usually observed 24 h after seeding, they were treated with DMSO and different concentrations of 5,5′-dibromoDIM for the indicated times. Both RKO and HT-29 cells were counted to evaluate the effects of 5,5′-dibromoDIM and DMSO(solvent control) on the number of viable cells using a Z1 Coulter particle counter (Beckman Coulter). Each experiment was carried out in triplicate and results are expressed as mean ± SD of each treatment group. In these and other experiments, the amount of DMSOwas 0.1%.
Fluorescence-Activated Cell Sorting Analysis for Cell Cycle Regulation
After treatment with 5,5′-dibromoDIM, detached cells (floaters) were collected by centrifugation and combined with adherent cells that were released by trypsinization. Cell cycle analysis was carried out by flow cytometry; cells were fixed in 70% ethanol overnight at -20°C. After centrifugation, ethanol was removed and the cells were subsequently stained with 0.02 mg/mL propidium iodide and subjected to DNA content analysis using a FACScan cytometer (Becton Dickinson).
Western Blot Analysis
Cell lysates were prepared using lysis buffer [20 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 0.1% SDS, 1 mmol/L sodium orthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride, 1 μmol/L leupeptin, and 1 μg/mL aprotinin]. After centrifugation of the lysate at 15,000 × g for 20 min, the supernatants were recovered, and protein was quantified by the Bradford protein assay using a reagent kit from Bio-Rad. Protein samples (20-60 μg) were size separated by electrophoresis on SDS-polyacrylamide gels under nonreducing conditions. Separated proteins were electroblotted onto nitrocellulose membranes. The blot was blocked by incubating in blocking buffer [5% skim milk, 10 mmol/L Tris (pH 7.5), 10 mmol/L NaCl, and 0.1% Tween 20] for 1 h at 20°C and then incubated with the primary antibody overnight at 4°C. Incubation with a horseradish peroxidase—conjugated anti-mouse or rabbit secondary antibody was then carried out at 20°C for 4 h. Antibody-bound proteins were detected by the Enhanced Chemiluminescence Western blotting analysis system (Perkin-Elmer Life and Analytical Sciences).
Reverse Transcriptase-PCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen), and 1 μg RNA was used to synthesize cDNA using Reverse Transcription System (Promega). The PCR conditions were as follows: initial denaturation at 94°C (2 min) followed by 26 cycles of denaturation for 30 s at 94°C, annealing for 1 min at 55°C, and extension at 72°C for 1 min and a final extension step at 72°C for 5 min. mRNA levels were normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal housekeeping gene. Primers obtained from IDT and used for amplification were KLF4 (sense 5′-CTATGGCAGGGAGTCCGCTCC-3′ and antisense 5′-ATGACCGACGGGCTGCCGTAC-3′) and GAPDH (sense 5′-ACGGATTTGGTCGTATTGGGCG-3′ and antisense 5′-CTCCTGGAAGATGGTGATGG-3′). PCR products were electrophoresed on 1% agarose gels containing ethidium bromide and visualized under UV transillumination.
Quantitative Real-time PCR
cDNA was prepared from RKO cell line using Reverse Transcription System (Promega). Each PCR was carried out in triplicate in a 30 μL volume using SYBR Green Master-Mix (Applied Biosystems) for 15 min at 95°C for initial denaturing followed by 40 cycles of 95°C for 30 s and 60°C for 1 min in the Applied Biosystems 7900HT Fast Real-time PCR System. The ABI Dissociation Curves software was used following a brief thermal protocol (95°C 15 s and 60°C 15 s followed by a slow ramp to 95°C) to control for multiple species in each PCR amplification. Values for each gene were normalized to expression levels of TATA-binding protein. The primers used for real-time PCR were obtained from Qiagen.
Transfection and Luciferase Assay
RKO and HT-29 cells were plated in 12-well plates at 1 × 105 per well in DMEM/Ham’s F12 supplemented with 2.5% charcoal-stripped fetal bovine serum. After growth for 16 h, various amounts of DNA [pWWP (0.25 μg), pWWP124 (0.25 μg), pWWP101 (0.25 μg), pWWP60 (0.25 μg), and β-galactosidase (0.05 μg)] were transfected by LipofectAMINE 2000 reagent (Invitrogen) according to the manufacturer’s protocol. After 5 h, cells were treated with complete medium containing either vehicle (DMSO) or the indicated ligands for 20 to 22 h. Cells were then lysed with 100 μL of 1× reported lysis buffer (Promega) and 30 μL cell extract was used for luciferase and β-galactosidase assays. Lumicount was used to quantitate luciferase and β-galactosidase activities, and the luciferase activities were normalized to β-galactosidase activity. Results are expressed as mean ± SD of at least three independent determinations for each treatment group.
Chromatin Immunoprecipitation Assay
RKO cells (2 × 107) were treated with DMSO (time 0) or 5,5-dibromoDIM (15 μmol/L) for 12 h. Cells were then fixed with 1.5% formaldehyde, and the cross-linking reaction was stopped by addition of 0.125 mol/L glycine. After washing twice with PBS, cells were scraped and pelleted. Nuclei were then sonicated to desired chromatin length (∼500 bp). The chromatin was precleared twice by addition of protein A—conjugated beads (Pierce Chemical) and then incubated at 4°C for 1 h with gentle agitation. The beads were pelleted, and the precleared chromatin supernatants were immunoprecipitated with antibodies specific to IgG, Sp1, Sp3, Sp4, p53, and KLF4 at 4°C overnight. Protein-antibody complexes were collected by addition of protein A—conjugated beads at room temperature for 1 h. Beads were extensively washed; the protein-DNA cross-links were eluted and reversed. DNA was purified by phenol extraction/ethanol precipitation followed by PCR amplification. The p21 primers are 5′-GCTGGCCTGCTGGAACTC-3′ (forward) and 5′-GGCAGCTGCTCACACCTC-3′ (reverse), and they amplify a 193-bp region of the human p21 promoter, which contains several GC-rich, Sp1 binding sites. The positive control primers are 5′-TACTAGCGGTTTTACGGGCG-3′ (forward) and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ (reverse), and they amplify a 167-bp region of human GAPDH gene. The negative control primers are 5′-atggttgccactggggatct-3′ (forward) and 5′-TGCCAAAGCCTAGGGGAAGA-3′ (reverse) and amplify a 174-bp region of genomic DNA between human GAPDH and CNAP1 genes. PCR products were resolved on a 2% agarose gel in the presence of SYBR gold (1:10,000).
RNA Interference Studies
Nonspecific oligonucleotide (iScr) and small inhibitory RNA for KLF4 (iKLF4) were purchased from Dharmacon Research, and equal amounts of each oligonucleotide were used in all experiments to maintain the same amount of nucleic acid in each treatment group. For transfection experiments, 100 nmol/L iScr or iKLF4 were transfected with LipofectAMINE 2000 reagent (Invitrogen); after 5 h, the medium was changed, cells were treated with DMSO or 5,5′-dibromoDIM for 20 to 22 h, and luciferase activity was determined as described above for the transfection and luciferase assays. RNA interference assays for protein knockdown used the same oligonucleotides, but treatment with DMSO or 5,5′-dibromoDIM was initiated 42 h after transfection.
Xenograft Studies in Athymic Mice
Male athymic nude mice (NCI-nu) were purchased from the Animal Production Area of the National Cancer Institute-Frederick Cancer Research and Development Center. The mice were housed and maintained under specific pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care and in accordance with current regulations and standards of the U.S. Department of Agriculture, U.S. Department of Health and Human Services. The mice were used in accordance with institutional guidelines when they were 8 to 12 weeks old. To produce tumors, RKO cells were harvested from subconfluent cultures by a brief exposure to 0.25% trypsin and 0.02% EDTA. Trypsinization was stopped with medium containing 10% fetal bovine serum, and the cells were washed once in serum-free medium and resuspended in HBSS. Only suspensions consisting of single cells with >90% viability were used for the injections. A xenograft was established by s.c. injection of the cells (7 × 106) into the flanks of individual mice. Tumors were allowed to grow for 6 days until they were palpable. Tumors in animals at the beginning of the experiment were randomized into three groups according to tumor size, and mice used in the control group and treated animals were randomly selected from these groups. Mice were randomized into two groups of five mice per group and dosed by oral gavage in corn oil or 30 mg/kg/d 5,5-dibromoDIM 5 days/wk for 21 days. The mice were weighed, and tumor size was measured every fourth day with calipers to permit calculation of tumor volumes: V = LW2 / 2, where L and W were length and width, respectively. Final body and tumor weights were determined at the end of the dosing regimen.
Terminal Deoxynucleotidyl Transferase — Mediated dUTP Nick End Labeling Assay, Histology, and p21 Immunostaining Studies
For the terminal deoxynucleotidyl transferase—mediated dUTP nick end labeling assay, tumor tissue was fixed in formalin and embedded in paraffin and used for terminal deoxynucleotidyl transferase—mediated dUTP nick end labeling staining, which was carried out using DeadEnd Colorimetric Terminal Deoxynucleotidyl Transferase—Mediated dUTP Nick End Labeling System (Promega,). Paraffinembedded sections (4-6 μm thick) were processed per manufacturer’s protocol. Briefly, sections were deparaffinized in xylene and then treated with a graded series of alcohol [100%, 95%, 85%, 70%, and 50% ethanol (v/v) in double-distilled H2O] and rehydrated in PBS (pH 7.5). Tissues were then treated proteinase K solution for permeabilization and then refixed with 4% paraformaldehyde solution. Slides were then treated with rTdT reaction mix and incubated at 37°C for 1 h. Reaction was terminated by immersing the slides in 2× SSC solutions for 15 min at room temperature. After blocking the endogenous peroxidase activity (by 0.3% H2O2), slides were washed with PBS and then incubated with streptavidin horseradish peroxidase solution for 30 min at room temperature. After washing, slides were incubated with 3,3′-diaminobenzidine (substrate) solution until a light-brown background appears (10 min) and then rinsed several times in deionized water. After mounting, slides were observed by light microscope. Immunostaining for p21 used deparaffinized tissue sections that were subjected to antigen retrieval with 0.1% pepsin in 0.01 N HCl at room temperature for 10 min followed by treatment with 0.1% H2O2 to block endogenous peroxidase activity. Sections were incubated with anti-p21 antibody (1:100) at 4°C overnight after blocking with normal goat serum at room temperature for 1 h. After washing in PBS, sections were incubated with biotinylated goat anti-rabbit IgG at room temperature for 30 min. Detection was made with Vectastain Elite ABC kit (Vector Laboratories) and 3,3′-diaminobenzidine (Biogenix Laboratories) as the chromagen following manufacturer’s protocol. The sections were counterstained with hematoxylin and dehydrated, and coverslips were mounted.
Statistical Analysis
Statistical significance was assessed using Student’s t test. P < 0.05 compared with solvent control was considered statistically significant.
Results
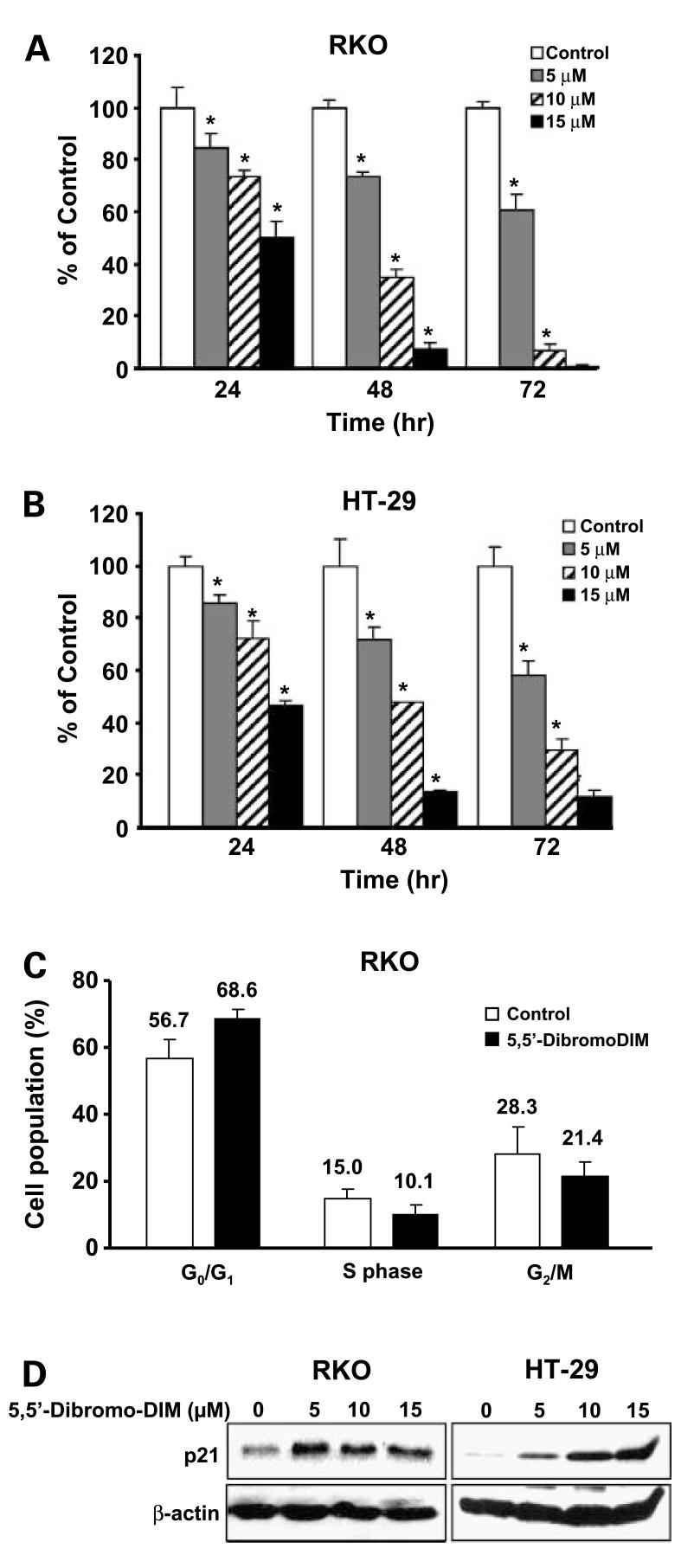
Ring-substituted DIM inhibit breast cancer cell proliferation and growth of carcinogen-induced mammary tumors in rats, and 5,5′-dibromoDIM was among the most active of these compounds in vivo and blocked tumor growth at doses ≤1.0 mg/kg every second day (33). Treatment of RKO and HT-29 cells with 5 to 15 μmol/L 5,5′-dibromoDIM for 72 h significantly inhibited cell proliferation (Fig. 1A and B). Similar results were observed in both cell lines, and treatment with 10 to 15 μmol/L concentrations also induced caspase-dependent apoptosis (data not shown). The effects of 15 μmol/L 5,5′-dibromoDIM on distribution of RKO cells in G0-G1, S, and G2-M phases of the cell cycle were determined by fluorescence-activated cell sorting analysis (Fig. 1C). The results showed that 5,5′-dibromoDIM increased cells in G0-G1 and decreased cells in S and G2-M phases, resulting in an overall G0-G1- to S-phase growth arrest. Data illustrated in the figure are means of three separate determinations and changes in G0-G1, G2-M, and S phases were significant (P < 0.05). Figure 1D shows that 5,5-dibromoDIM also decreased p21 expression in both RKO and HT-29 cells and these results are consistent with the inhibition of G0-G1- to S-phase progression in RKO cells treated with 5,5′-dibromoDIM.
Figure 1.
5,5′-DibromoDIM inhibits colon cancer cell proliferation and induces p21. Decreased proliferation of RKO (A) and HT-29 (B) cells. Cells were treated with DMSO or 5,5′-dibromoDIM (5, 10, or 15 μmol/L) for 24, 48, or 72 h, and cells were counted as described in Materials and Methods. The percent survival of treated cells was compared with DMSO (set at 100%). Mean ± SD of three determinations for each treatment. *, P < 0.05, significantly decreased survival. C, cell cycle progression. RKO cells were treated with DMSO or 15 μmol/L 5,5′-dibromoDIM for 24 h and analyzed by fluorescence-activated cell sorting as described in Materials and Methods. Mean ± SD of three determinations. *, significant 5,5′-dibromoDIM-induced changes. D, induction of p21 protein. RKO and HT-29 cells were treated with DMSO and different concentrations of 5,5′-dibromoDIM for 24 h, and p21 expression was determined by Western blot analysis as described in Materials and Methods.
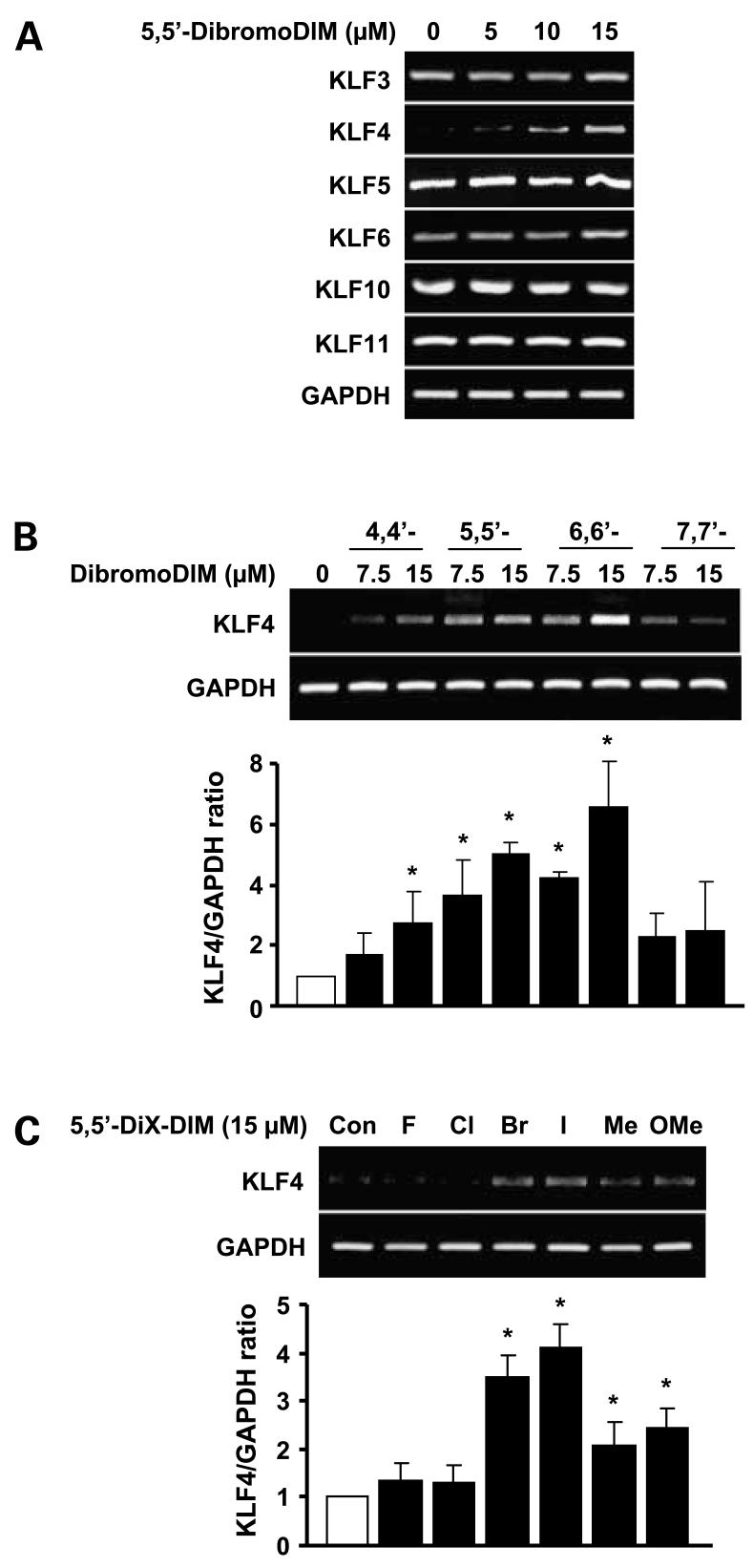
Induction of p21 by cytotoxic drugs including DIM is associated with modulation of several pathways, and in colon cancer cells, KLF4 and p53 play a role in the induction of this cyclin-dependent kinase inhibitor (16, 25–27). Using 5,5′-dibromoDIM as a model, this compound induced KLF4 mRNA in RKO cells but did not affect expression of KLF3, KLF5, KLF6, KLF10, or KLF11 mRNA levels in this cell line (Fig. 2A). Previous studies in pancreatic and breast cancer cells showed that 5,5′-dibromoDIM was a highly cytotoxic ring-substituted DIM (31, 34, 35). Structure activity studies among a series of symmetrical substituted dibromoDIM isomers as inducers of KLF4 mRNA showed that their relative order of potency was 6,6′-dibromo > 5,5′-dibromo ≥ 4,4′-dibromo > 7,7′-dibromoDIM as inducers in RKO cells (Fig. 2B). Moreover, for a series of 5,5′-difluoroDIM, 5,5′-dichloroDIM, 5,5′-dibromoDIM, 5,5′-diiodoDIM, 5,5′-dimethylDIM, and 5,5′-dimethoxyDIM analogues, the most active compound as inducers of KLF4 mRNA were 5,5′-dibromoDIM and 5,5′-diiodoDIM (Fig. 2C). The results show the importance of the substitutent group since even at the 15 μmol/L concentration, the fluoro and chloro analogues did not significantly induce KLF4 mRNA. We also investigated the induction of KLF4 mRNA by DIM, and significant induction was observed only at a high concentration (40 μmol/L; data not shown). Thus, 5,5′-dibromoDIM and 6,6′-dibromoDIM were among the most potent inducers of KLF4 mRNA in RKO cells. Because 5,5′-dibromoDIM is more readily synthesized from the commercially available 5-bromoindole-3-carboxaldehyde, this compound was selected as a prototype to further investigate the mechanisms of KLF4 induction by 5,5′-dibromoDIM and the role of this gene in mediating the growth-inhibitory effects of 5,5′-dibromoDIM.
Figure 2.
Induction of KLF by ring-substituted DIM. RKO cells were treated with DMSO or different concentrations of 5,5′-dibromoDIM (A), dibromoDIM isomers (B), or different 5,5′-disubstituted DIM analogues (C). KLF or KLF4 mRNAlevels were determined by reverse transcription-PCR as described in Materials and Methods. Mean ± SD of three determinations for each treatment group. *, P < 0.05, significantly increased activity.
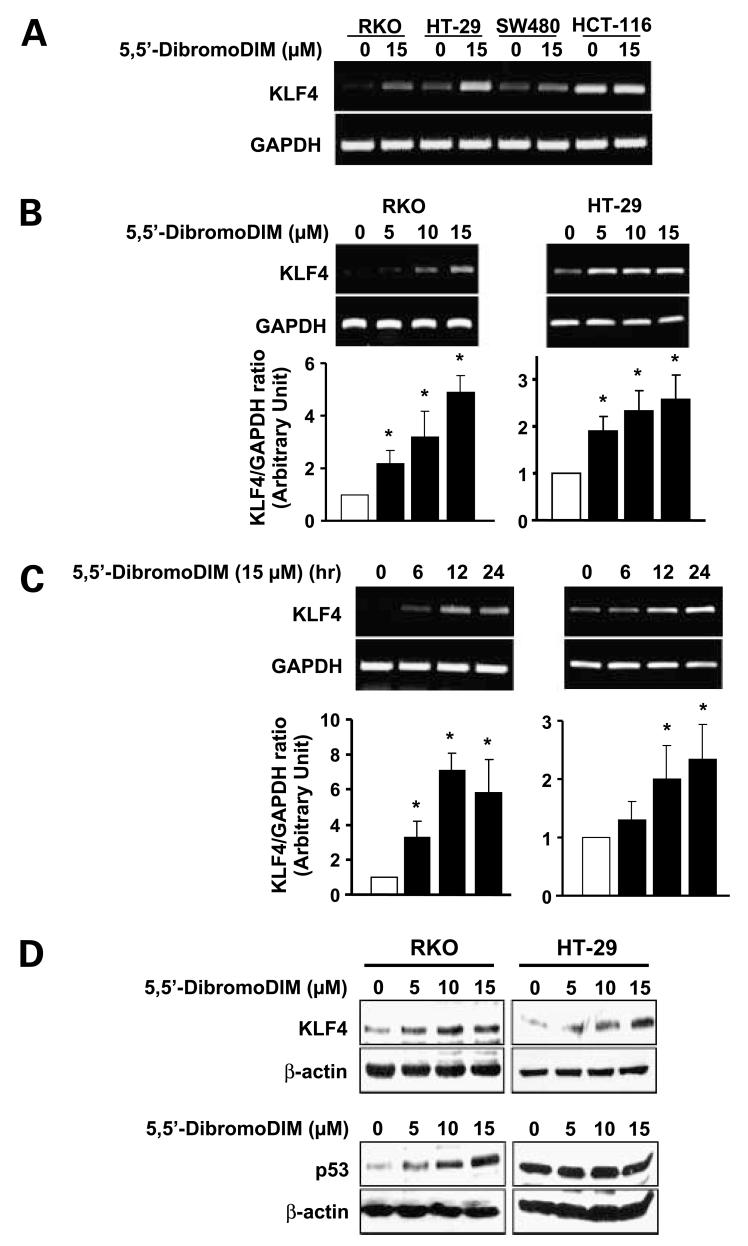
Among a series of four colon cancer cells (RKO, HT-29, SW480, and HCT-116), 5,5′-dibromoDIM significantly induced KLF4 mRNA in all of these cell lines. The highest induction was observed in RKO and HT-29 cells (Fig. 3A). Results in Fig. 3B and C show that 5,5′-dibromoDIM induced a concentration- and time-dependent increase in KLF4 mRNA levels. Significant induction of Krüppel-like factor mRNA was observed in RKO and HT-29 cells treated with 5 μmol/L 5,5′-dibromoDIM (Fig. 3B). In time course studies, 15 μmol/L 5,5′-dibromoDIM induced KLF4 after treatment of RKO or HT-29 cells for 6 or 12 h, respectively, and KLF4 mRNA levels remained elevated for 24 h in both cell lines (Fig. 3C). We also observed that 5,5′-dibromoDIM induced KLF4 protein in HT-29 and RKO cells (Fig. 3D); however, p53 protein was induced only in RKO cells, which expresses the wild-type form of this gene/protein. In contrast, the inactive mutant form of p53 expressed in HT-29 cells was not affected by treatment with 5,5′-dibromoDIM. Thus, induction of KLF4 and p21 was observed in colon cancer cells expressing wild-type or mutant (inactivated) p53, suggesting that wild-type p53 was not necessary for induction of KLF4 and p21 by 5,5′-dibromoDIM.
Figure 3.
Effects of 5,5′-dibromoDIM on KLR4 and p53 expression in RKO and HT-29 cells. Induction of KLF4 mRNAin multiple (A) and RKO/HT-29 (B or C) cells. Cells were treated with DMSO or the indicated concentrations of 5,5′-dibromoDIM for 24 h (A and B) or different time points (C) and mRNA levels were determined by reverse transcription-PCR as described in Materials and Methods. Mean ± SD of three replicate experiments for each treatment group. *, P < 0.05, significant induction. D, induction of KLF4 and p53 proteins. Cells were treated with DMSO or 5, 10, or 15 μmol/L 5,5′-dibromoDIM for 24 h, and whole-cell lysates were examined by Western blot analysis as described in Materials and Methods. Blots are representative of duplicate experiments.
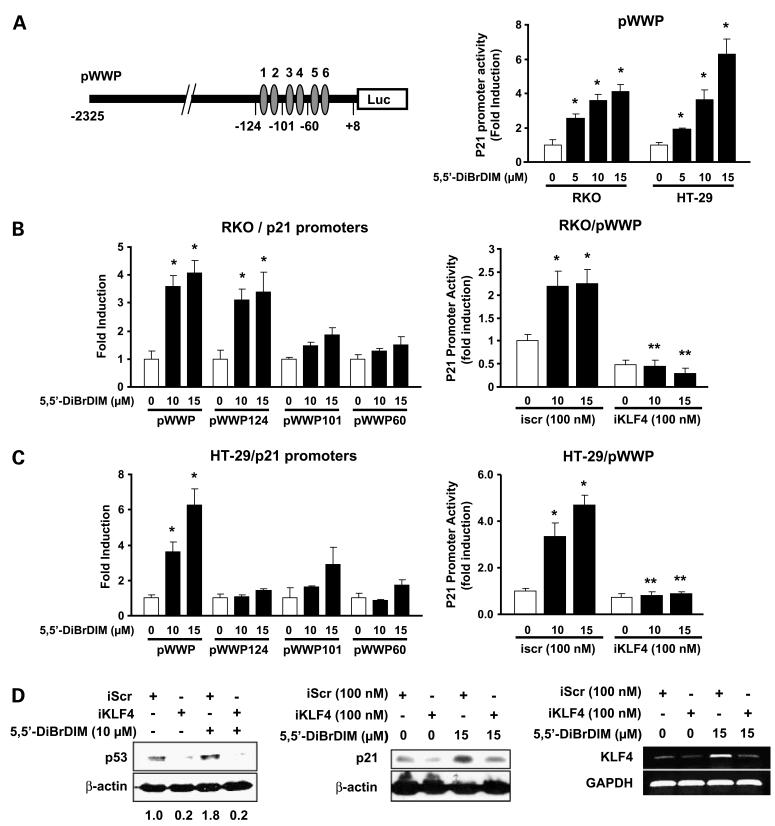
Interactions between KLF4 and p21 in the induction of p21 by 5,5′-dibromoDIM was further investigated in colon cancer cells. Results in Fig. 4A show that 5,5′-dibromoDIM also induces transactivation in both RKO and HT-29 cells transfected with pWWP, a construct containing a -2,325 to +8 p21 promoter insert. 5,5′-DibromoDIM also induces luciferase activity in RKO cells transfected with pWWP124, but not pWWP101 or pWWP60 constructs, which contain -124 to +8, -101 to +8, and -60 to +8 p21 promoter inserts, respectively (Fig. 4B). These constructs contain the six proximal GC-rich sites in the p21 promoter, and results of the promoter deletion studies suggest that loss of distal (5′) sites 1 and 2 to give pWWP101 resulted in loss of inducibility by 5,5′-dibromoDIM. Previous studies indicate that this region of the promoter was also required for activation of p21 promoter constructs in HEK293 cells transfected with KLF4 expression plasmid (16). RNA interference using a small-inhibitory RNA for KLF4 (iKLF4) decreased luciferase activity in RKO cells transfected with pWWP. 5,5′-DibromoDIM also activates luciferase activity in HT-29 cells transfected with pWWP but not pWWP124, pWWP101, or pWWP60 (Fig. 4C), showing that activation of p21 in this p53-independent cell line requires upstream (5′) promoter sequences and not the more proximal GC-rich motifs. However, 5,5′-dibromo-DIM-dependent induction of luciferase activity in HT-29 cells transfected with pWWP was also inhibited by cotransfection with iKLF4. We also determined the effects of KLF4 knockdown on p21 and p53 expression in RKO cells (Fig. 4D) and show that iKLF4 decreased constitutive and 5,5′-dibromoDIM-induced p21 and p53 proteins, showing a role for KLF4 in regulating p21 and p53 expression in this cell line. In addition, iKLF4 decreases expression of endogenous and 5,5′-dibromoDIM-induced KLF4 mRNA levels (Fig. 4D).
Figure 4.
5,5′-DibromoDIM activation of KLF4, p21, and p53. 5,5′-DibromoDIM activates pWWP (A) and p21 promoter deletion constructs in RKO (B) and HT-29 (C) cells. Cells were treated with DMSO or different concentrations of 5,5′-dibromoDIM transfected with pWWP (A) and other deletion constructs (B and C) in the presence or absence of nonspecific iScr or iKLF4 as indicated, and luciferase activity was determined as described in Materials and Methods. Mean ± SD of three replicate determinations for each treatment group. *, P < 0.05, significant induction of induced activity by iKLF4; **, P < 0.05, significant inhibition of induced activity by iKLF4. D, effects of iKLF4 on p21 and p53 expression. RKO cells were treated with DMSO or 10 μmol/L 5,5′-dibromoDIM and transfected with nonspecific iScr or 100 nmol/L iKLF4, and 24 h after treatment, whole-cell lysates were analyzed by Western blotting. The pattern of protein expression was similar in duplicate experiments. The effects of iKLF4/iScr on endogenous and 5,5′-dibromoDIM-induced KLF4 mRNAlevels are also shown. Similar results were observed in duplicate experiments.
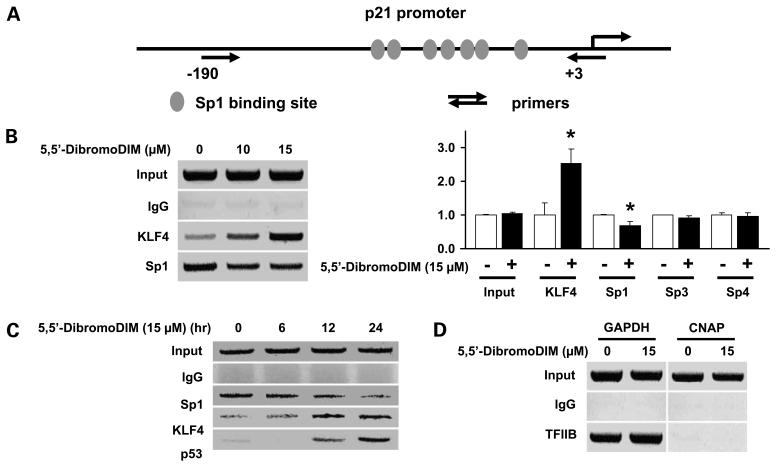
The potential interactions of KLF4 with the proximal GC-rich region of the p21 promoter (Fig. 5A) were determined in a chromatin immunoprecipitation assay. Treatment of RKO cells with 5,5′-dibromoDIM for 12 h resulted in recruitment of KLF4 to the p21 promoter, and a slight decrease in Sp1 protein interacting with the p21 promoter was also observed (Fig. 5B). This experiment was replicated (3×) and interactions of KLF4, Sp1, Sp3, and Sp4 to the p21 promoter were quantitated. 5,5′-DibromoDIM significantly enhanced (>2.5-fold) KLF4 and decreased (<25%) Sp1 interactions with the p21 promoter, whereas Sp3 and Sp4 interactions were unchanged (Fig. 5B). RKO cells were treated with 15 μmol/L 5,5′-dibromoDIM for 6, 12, and 24 h, and the recruitment of KLF4 and slight loss of Sp1 interactions with the p21 promoter were also observed (Fig. 5C). Moreover, p53 was also recruited to this region of the p21 promoter, confirming an association of both p53 and KLF4 with the p21 promoter, although cis-elements that bind p53 are located in a distal region of this promoter at -1,394. As a control for this experiment, we also showed that the transcription factor TFIIB bound to the proximal promoter region of the GAPDH gene but not to exon 1 of the CNAP gene (negative control; Fig. 5D). The results show that 5,5′-dibromoDIM-mediated induction of p21 involves induction of KLF4, which is recruited to the p21 promoter to activate transcription.
Figure 5.
Interaction of KLF4 with the p21 promoter. A, KLF4 proximal promoter GC-rich sites and PCR primer locations. B, chromatin immunoprecipitation analysis. RKO cells were treated with 15 μmol/L 5,5′-dibromoDIM for 12 h, and interactions of KLF4 and specificity protein proteins with the p21 promoter were determined as described in Materials and Methods. The chromatin immunoprecipitation experiments were replicated (3×), and relative intensities in the treated cells compared with DMSO (set at 1.0) are given as mean ± SD. *, P < 0.05, significantly different activities. C, time-course chromatin immunoprecipitation assay. RKO cells were treated with 15 Amol/L 5,5′-dibromoDIM for 6, 12, and 24 h, and interactions of KLF4, p53, and Sp1 with the p21 promoter were determined as described in Materials and Methods. D, chromatin immunoprecipitation control experiment. The binding of the transcription factor TFIIB to the GAPDH promoter (positive control) and exon 1 of the CNAP promoter (negative control) were determined in a chromatin immunoprecipitation assay as described in Materials and Methods.
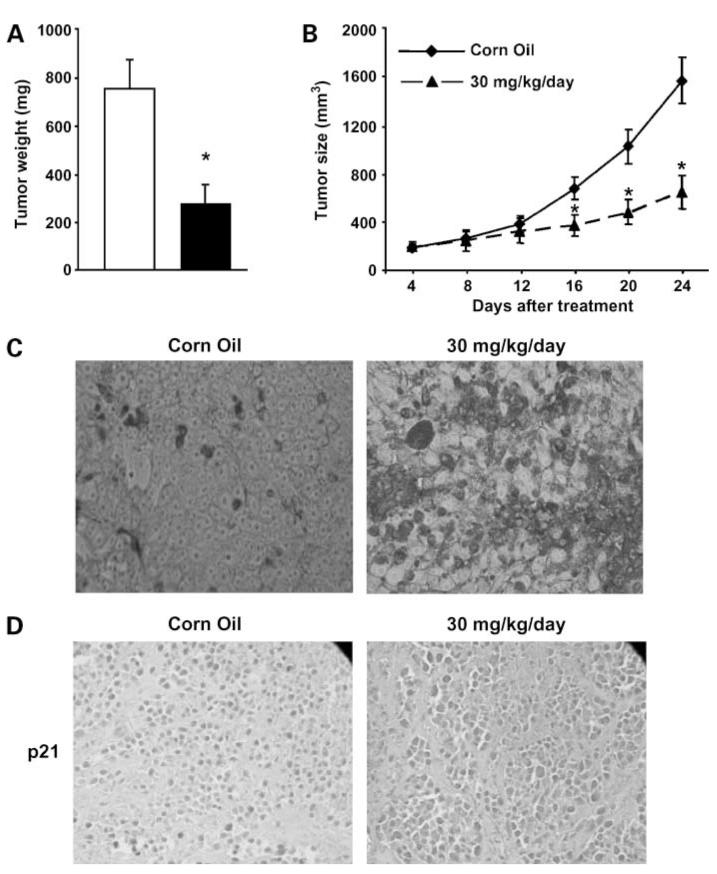
The in vivo anticarcinogenic activity of 5,5′-dibromoDIM was further investigated in athymic nude mice bearing RKO cells as xenografts, and treatment with 30 mg/kg/d over a period of 24 days significantly inhibited tumor weight (Fig. 6A) and volume (Fig. 6B). After the experiment was terminated, the results showed no changes in body or organ weights in treated versus untreated (corn oil vehicle control) mice; however, there was a significant decrease in tumor weight in animals receiving 5,5′-dibromoDIM. We also observed the 5,5′-dibromoDIM-induced apoptosis (Fig. 6C) and p21 expression (Fig. 6D) compared with animals treated with corn oil (vehicle control) and this correlated with results of the in vitro studies (Fig. 1).
Figure 6.
In vivo inhibition of tumor growth by 5,5′-dibromoDIM. A, tumor weights; B, tumor volume. Athymic nude mice bearing RKO cells as xenografts were treated with 5,5′-dibromoDIM (30 mg/kg/d) and tumor weights and volumes were determined as described in Materials and Methods. C, immunostaining for apoptosis. Tumor tissue from animals treated with corn oil (control) or 5,5′-dibromoDIM were stained using the terminal deoxynucleotidyl transferase — mediated dUTP nick end labeling assay to detect apoptosis. D, immunostaining for p21 protein expression. Paraffin-embedded sections obtained from treated and untreated tumors were immunostained for p21 as described in Materials and Methods.
Discussion
DIM is a dimerization product of the chemoprotective phytochemical indole-3-carbinol, which is found as a conjugate in cruciferous vegetables (36). DIM activates diverse growth-inhibitory and proapoptotic pathways in various cancer cell lines (36–40). Research in this laboratory has modified the DIM structure to give a series of 1,1-bis(3′-indolyl)-1-(p-substituted phenyl)methanes, which also inhibit cancer cell and tumor growth through activation of peroxisome proliferator-activated receptor γ and the orphan receptor Nur77 and through receptor-independent pathways (32, 41–45). In addition, several symmetrical ring-substituted DIM, including 5,5′-dibromoDIM, are potent inhibitors of carcinogen-induced rat mammary tumor growth and, like DIM, exhibit antiandrogenic activity (33, 35, 40, 46). However, our studies on a series of 4,4′-, 5,5′-, 6,6′-, and 7,7′-dibromoDIM and 4,4′-, 5,5′-, 6,6′-, and 7,7′-dichloroDIM as antiandrogens indicated that there were structure-dependent differences in their activities; 7,7′-dibromoDIM decreased AR expression, whereas 4,4′-dibromoDIM did not affect expression of this receptor and similar structure-dependent effects were observed for the dichloroDIM (46). Thus, DIM and ring-substituted DIM exhibit overlapping and different activities (33–35); however, most ring-substituted DIM, such as 5,5′-dibromoDIM, are more active than DIM in bothin vivo and in vitro studies.
1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methane compounds decreased colon cancer cell proliferation (41, 42, 45) and inhibited G0-G1- to S-phase progression in HT-29 cells, and these results were comparable with those observed for 5,5′-dibromoDIM (Fig. 1). 5,5′-DibromoDIM induced p21 expression in both RKO and HT-29 cells (Fig. 1D), and this was consistent with the inhibitory effects of this compound on the cell cycle where there was significant inhibition of G0-G1- to S-phase progression (Fig. 1C). In contrast, 1,1-bis(3′-indolyl)-1-(p-substituted phenyl)methane did not induce p21 in colon cancer cells and this represents an important difference between these two classes of synthetic DIM analogues (41, 42, 45). This suggests that induction of p21 in colon cancer cells by 5,5′-dibromoDIM may be important for mediating the antiproliferative activity of this compound in colon cancer cells; therefore, the mechanism of p21 induction by 5,5′-dibromoDIM was further investigated.
Several reports show that p21 is activated by both p53 and KLF4 in colon cancer cell lines (9, 10, 16), and we initially investigated induction of KLF4 gene expression by a series of ring-substituted DIM analogues in HT-29 and RKO cells. There was a structure-dependent activation of KLF4 by several isomeric dibromoDIM (Fig. 2B) and 5,5′-substituted DIM (Fig. 2D), and the most active compounds were 5,5′- and 6,6′-dibromoDIM and 5,5′-diiodoDIM. 5,5′-DibromoDIM was selected as the prototypical compound for this study in colon cancer cells due to its relative ease of synthesis and well-characterized in vivo and in vitro activity as an inhibitor of pancreatic and breast cancer cell growth and carcinogen-induced rat mammary tumor growth (33, 35).
5,5′-DibromoDIM specifically induced KLF4 and not other KLF in RKO cells (Fig. 2A) and KLF4 mRNA and protein was induced in p53 wild-type RKO and p53 mutant HT-29 cells (Fig. 3A-C). The relationship between KLF4, p53, and p21 is complex and cell context dependent (16, 17, 27). In breast cancer cells, KLF4-dependent suppression of p53 and induction of p21 may be associated with cellular transformation under certain conditions (9). In contrast, DNA damage-dependent activation of p53 in colon cancer cells resulted in the stepwise induction of KLF4 and then p21 (16). In HCT116 colon cancer cells expressing wild-type p53 (HCT116p53+/+), irradiation induced p53, KLF4, and p21, whereas in HCT116p53-/- cells irradiation did not induce either KLF4 or p53 (27). Moreover, knockdown of KLF4 by RNA interference decreased irradiation-induced p21, increased the percentage of cells in G1, but did not affect p53 induction in HCT116p53+/+ cells. These results show that KLF4 is required for p53-dependent activation of p21 in some colon cancer cell lines. In contrast, induction of KLF4 by 5,5′-dibromoDIM is observed in RKO and HT-29 cells expressing wild-type and inactive mutant p53, respectively (Figs. 2 and 3), suggesting that, at least in the latter cell line, induction of KLF4 and p21 are not p53 dependent.
Because 5,5′-dibromoDIM induces p21 in RKO and HT-29 cells (Fig. 1D), we investigated the role of KLF4 in mediating the induction response. Results of transient transfection studies in RKO and HT-29 cells show that 5,5′-dibromoDIM induced luciferase activity in both cell lines transfected with the pWWP construct containing the -2,325 to +8 region of the p21 promoter. RNA interference studies using a small-inhibitory RNA for KLF4 (iKLF4) show that activation of the p21 promoter by 5,5′-dibromoDIM was significantly inhibited in cells cotransfected with iKLF4, showing that KLF4 was essential for induction of p21 in cells expressing either wild-type (RKO) or mutant (HT-29) p53. Thus, at least in HT-29 cells, p53 was not required for induction of p21 or KLF4 by 5,5′-dibromoDIM. However, in RKO cells, KLF4 knockdown by RNA interference decreased both constitutive and 5,5′-dibromoDIM-induced p21 and p53 expression (Fig. 4D), showing the important role of KLF4 in regulating expression of both genes in this cell line. These observations in RKO and HT-29 cells are in contrast to results in HCT116p53+/+ cells where KLF4 knockdown did not affect constitutive or radiation-induced p53 expression (27), suggesting that the interdependent regulation of p53 and KLF4 in colon cancer cells is cell context dependent.
The p21 promoter contains six proximal GC-rich motifs (47–49) that bind specificity protein proteins, and previous studies show that the upstream GC-rich sites 1 and 2 are required for KLF4-dependent activation of p21 promoter constructs (16). Deletion analysis of the p21 promoter in RKO cells shows that 5,5′-dibromoDIM induces transactivation in cells transfected with pWWP (-2,325 to +8) and pWWP124, which contains GC-rich sites 1 to 6. However, deletion of sites 1 and 2 resulted in loss of transactivation (Fig. 4B). These data are consistent with previous studies, which suggest a critical role for these specific GC-rich sites (16). We have also confirmed in a chromatin immunoprecipitation assay that KLF4 and p53 are recruited to the GC-rich proximal region of the p21 promoter in RKO cells (Fig. 5). The mechanism of KLF4-dependent activation of p21 by 5,5′-dibromoDIM in p53-mutant HT-29 cells is clearly different from that observed in RKO cells. Deletion analysis of the p21 promoter in HT-29 cells shows that transactivation is induced only in cells transfected with pWWP and not with constructs containing only the proximal GC-rich sites 1 to 6 (Fig. 4C). Thus, p53 status of colon cancer cells is at least one determinant for the mechanism of KLF4-mediated activation of p21 in cells treated with 5,5′-dibromoDIM. Current studies are further investigating KLF4-p21-p53 interactions in colon cancer cells expressing wild-type p53 and the identification of critical cis-elements required for induction of p21 in p53 mutant colon cancer cells.
5,5′-DibromoDIM also inhibited tumor growth and tumor weights in athymic nude mice bearing RKO cells as xenografts, and this was not accompanied by any changes in organ or body weights or evidence for compound-induced toxicity (Fig. 6). This shows the antitumorigenic activity of 5,5′-dibromoDIM in both a mouse xenograft and carcinogen-induced rat mammary tumor model (33). We also observed an increase in p21 and apoptosis and p21 protein expression in tumors from 5,5′-dibromoDIM-treated mice (Fig. 6C and D), and the proapoptotic activity of this compound was also observed in both RKO and HT-29 cells in culture (data not shown). Immunohistochemical analysis for KLF4 gave inconsistent results due to problems with antibodies, and improvements in the detection of KLF4 protein and mRNA in tumor samples are under way. Induction of p53/p21 directly leads to apoptosis; however, in pancreatic cancer cells, 5,5′-dibromoDIM induces endoplasmic reticulum stress and activation of death receptor 5 (34), and preliminary studies (data not shown) indicate that this pathway may also be activated in colon cancer cells and could contribute to the observed apoptosis and growth inhibition. 5,5′-DibromoDIM also decreases cyclin D1 expression in colon cancer cells (data not shown) as reported previously in breast cancer cells (35), suggesting that p21 induction is one of several growth-inhibitory/ proapoptotic responses induced by this compound.
In summary, this study shows that the anticarcinogenic activity of 5,5′-dibromoDIM in colon cancer cells is associated with the induction of KLF4. Moreover, we show that induction of p21 is KLF4 dependent in p53-mutant HT-29 and p53 wild-type RKO cells, and induction of p53 in the latter cell line is also KLF4 dependent. These results, coupled with the in vivo anticarcinogenic activity of 5,5′-dibromoDIM in the mouse xenograft model, shows that specific ring-substituted DIM represent a novel class of drugs that activate KLF4, and these compounds are currently being developed for clinical applications.
Acknowledgments
Grant support: NIH grants ES09106 and CA112337, Chonbuk National University, and Texas Agricultural Experiment Station.
Footnotes
Requests for reprints: Stephen Safe, Department of Veterinary Physiology and Pharmacology, Texas A&M University, 4466 TAMU, Veterinary Research Building 410, College Station, TX 77843-4466. Phone: 979-845-5988; Fax: 979-862-4929. E-mail: ssafe@cvm.tamu.edu
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–6. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–8. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 3.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–17. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–60. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 5.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de CB. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–90. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein-Barr virus ED-L2 promoters by gut-enriched Kruppel-like factor. J Biol Chem. 1998;273:10747–54. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 7.Ehlermann J, Pfisterer P, Schorle H. Dynamic expression of Kruppel-like factor 4 (Klf4), a target of transcription factor AP-2a during murine mid-embryogenesis. Anat Rec A Discov Mol Cell Evol Biol. 2003;273:677–80. doi: 10.1002/ar.a.10089. [DOI] [PubMed] [Google Scholar]
- 8.Jaubert J, Cheng J, Segre JA. Ectopic expression of Kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–77. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 9.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nat Rev Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 10.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 11.Foster KW, Ren S, Louro ID, et al. Oncogene expression cloning by retroviral transduction of adenovirus E1A-immortalized rat kidney RK3E cells: transformation of a host with epithelial features by c-MYC and the zinc finger protein GKLF. Cell Growth Differ. 1999;10:423–34. [PubMed] [Google Scholar]
- 12.Foster KW, Frost AR, Kie-Bell P, et al. Increase of GKLF messenger RNAand protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–95. [PubMed] [Google Scholar]
- 13.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–82. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Shen H, Akagi K, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–74. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 15.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang VW. Expression of the gut-enriched Kruppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997;419:239–43. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Geiman DE, Shields JM, et al. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–8. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei D, Gong W, Kanai M, et al. Drastic down-regulation of Krüppel-like factor 4 expression is critical in human gastric cancer development and progression. Cancer Res. 2005;65:2746–54. doi: 10.1158/0008-5472.CAN-04-3619. [DOI] [PubMed] [Google Scholar]
- 18.Wang N, Liu ZH, Ding F, et al. Down-regulation of gut-enriched Kruppel-like factor expression in esophageal cancer. World J Gastroenterol. 2002;8:966–70. doi: 10.3748/wjg.v8.i6.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo A, Kong J, Hu G, et al. Discovery of Ca2+-relevant and differentiation-associated genes downregulated in esophageal squamous cell carcinoma using cDNAmicroarray. Oncogene. 2004;23:1291–9. doi: 10.1038/sj.onc.1207218. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi S, Ohnami S, Laub F, et al. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem Biophys Res Commun. 2003;308:251–6. doi: 10.1016/s0006-291x(03)01356-1. [DOI] [PubMed] [Google Scholar]
- 21.Luo J, Dunn T, Ewing C, et al. Gene expression signature of benign prostatic hyperplasia revealed by cDNA microarray analysis. Prostate. 2002;51:189–200. doi: 10.1002/pros.10087. [DOI] [PubMed] [Google Scholar]
- 22.Bianchi F, Hu J, Pelosi G, et al. Lung cancers detected by screening with spiral computed tomography have a malignant phenotype when analyzed by cDNAmicroarray. Clin Cancer Res. 2004;10:6023–8. doi: 10.1158/1078-0432.CCR-04-0619. [DOI] [PubMed] [Google Scholar]
- 23.Yasunaga J, Taniguchi Y, Nosaka K, et al. Identification of aberrantly methylated genes in association with adult T-cell leukemia. Cancer Res. 2004;64:6002–9. doi: 10.1158/0008-5472.CAN-04-1422. [DOI] [PubMed] [Google Scholar]
- 24.Madden SL, Cook BP, Nacht M, et al. Vascular gene expression in nonneoplastic and malignant brain. Am J Pathol. 2004;165:601–8. doi: 10.1016/s0002-9440(10)63324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Johns DC, Geiman DE, et al. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–8. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNAdamage. J Biol Chem. 2003;278:2101–5. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shie JL, Chen ZY, O’Brien MJ, et al. Role of gut-enriched Krüppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 29.Chen ZY, Rex S, Tseng CC. Kruppel-like factor 4 is transactivated by butyrate in colon cancer cells. J Nutr. 2004;134:792–8. doi: 10.1093/jn/134.4.792. [DOI] [PubMed] [Google Scholar]
- 30.Chen ZY, Tseng CC. 15-Deoxy-D12,14 prostaglandin J2 up-regulates Krüppel-like factor 4 expression independently of peroxisome proliferator-activated receptor g by activating the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signal transduction pathway in HT-29 colon cancer cells. Mol Pharmacol. 2005;68:1203–13. doi: 10.1124/mol.105.014944. [DOI] [PubMed] [Google Scholar]
- 31.Chintharlapalli S, Papineni S, Jutooru I, McAlees A, Safe S. Structure-dependent activity of glycyrrhetinic acid derivatives as peroxisome proliferator-activated receptor g (PPARg) agonists in colon cancer cells. Mol Cancer Ther. 2007;6:1588–98. doi: 10.1158/1535-7163.MCT-07-0022. [DOI] [PubMed] [Google Scholar]
- 32.Hong J, Samudio I, Liu S, Abdelrahim M, Safe S. Peroxisome proliferator-activated receptor γ-dependent activation of p21 in Panc-28 pancreatic cancer cells involves Sp1 and Sp4 proteins. Endocrinology. 2004;145:5774–85. doi: 10.1210/en.2004-0686. [DOI] [PubMed] [Google Scholar]
- 33.McDougal A, Sethi-Gupta M, Ramamoorthy K, Sun G, Safe S. Inhibition of carcinogen-induced rat mammary tumor growth and other estrogen-dependent responses by symmetrical dihalo-substituted analogs of diindolylmethane. Cancer Lett. 2000;151:169–79. doi: 10.1016/s0304-3835(99)00406-1. [DOI] [PubMed] [Google Scholar]
- 34.Abdelrahim M, Newman K, Vanderlaag K, Samudio I, Safe S. 3,3′Diindolylmethane (DIM) and derivatives induce apoptosis in pancreatic cancer cells through endoplasmic reticulum stress-dependent upregulation of DR5. Carcinogenesis. 2006;27:717–28. doi: 10.1093/carcin/bgi270. [DOI] [PubMed] [Google Scholar]
- 35.Vanderlaag K, Samudio I, Burghardt R, Barhoumi R, Safe S. Inhibition of breast cancer cell growth and induction of cell death by 1,1-bis(3′indolyl)methane (DIM) and 5,5′-dibromoDIM. Cancer Lett. 2005;236:198–212. doi: 10.1016/j.canlet.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 36.Shertzer HG, Senft AP. The micronutrient indole-3-carbinol: implications for disease and chemoprevention. Drug Metabol Drug Interact. 2000;17:159–88. doi: 10.1515/dmdi.2000.17.1-4.159. [DOI] [PubMed] [Google Scholar]
- 37.Hong C, Kim HA, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane (DIM) induces a G1 cell cycle arrest in human breast cancer cells that is accompanied by Sp1-mediated activation of p21WAF1/CIP1 expression. Carcinogenesis. 2002;23:1297–305. doi: 10.1093/carcin/23.8.1297. [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Sohn H, Xue L, Firestone GL, Bjeldanes LF. 3,3′-Diindolylmethane is a novel mitochondrial H+-ATP synthase inhibitor that can induce p21Cip1/Waf1 expression by induction of oxidative stress in human breast cancer cells. Cancer Res. 2006;66:4880–7. doi: 10.1158/0008-5472.CAN-05-4162. [DOI] [PubMed] [Google Scholar]
- 39.Rahman KW, Li Y, Wang Z, Sarkar SH, Sarkar FH. Gene expression profiling revealed survivin as a target of 3,3′-diindolylmethane-induced cell growth inhibition and apoptosis in breast cancer cells. Cancer Res. 2006;66:4952–60. doi: 10.1158/0008-5472.CAN-05-3918. [DOI] [PubMed] [Google Scholar]
- 40.Le HT, Schaldach CM, Firestone GL, Bjeldanes LF. Plant-derived 3,3′diindolylmethane is a strong androgen antagonist in human prostate cancer cells. J Biol Chem. 2003;278:21136–45. doi: 10.1074/jbc.M300588200. [DOI] [PubMed] [Google Scholar]
- 41.Chintharlapalli S, Papineni S, Baek SJ, Liu S, Safe S. 1,1-Bis(3′indolyl)-1-(p-substituted phenyl)methanes are peroxisome proliferator-activated receptor gamma agonists but decrease HCT-116 colon cancer cell survival through receptor-independent activation of early growth response-1 and NAG-1. Mol Pharmacol. 2005;68:1782–92. doi: 10.1124/mol.105.017046. [DOI] [PubMed] [Google Scholar]
- 42.Chintharlapalli S, Papineni S, Safe S. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes inhibit colon cancer cell and tumor growth through PPARg-dependent and PPARg-independent pathways. Mol Cancer Ther. 2006;5:1362–70. doi: 10.1158/1535-7163.MCT-06-0002. [DOI] [PubMed] [Google Scholar]
- 43.Qin C, Morrow D, Stewart J, et al. A new class of peroxisome proliferator-activated receptor γ (PPARγ) agonists that inhibit growth of breast cancer cells: 1,1-bis(3′-indolyl)-1-(p-substituted phenyl)methanes. Mol Cancer Ther. 2004;3:247–59. [PubMed] [Google Scholar]
- 44.Chintharlapalli S, Papineni S, Safe SH. 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes inhibit growth, induce apoptosis, and decrease the androgen receptor in LNCaP prostate cancer cells through PPARγ-independent pathways. Mol Pharmacol. 2007;71:558–69. doi: 10.1124/mol.106.028696. [DOI] [PubMed] [Google Scholar]
- 45.Chintharlapalli S, Smith R, III, Samudio I, Zhang W, Safe S. 1,1-Bis(3′indolyl)-1-(p-substituted phenyl)methanes induce peroxisome proliferator-activated receptor γ-mediated growth inhibition, transactivation and differentiation markers in colon cancer cells. Cancer Res. 2004;64:5994–6001. doi: 10.1158/0008-5472.CAN-04-0399. [DOI] [PubMed] [Google Scholar]
- 46.Kotha L. Inhibition of breast and prostate cancer cell growth by 3,3′diindolylmethane and related compounds. Texas A&M University. 2007.
- 47.Gartel AL, Tyner AL. Transcriptional regulation of the p21WAF1/CIP1 gene. Exp Cell Res. 1999;246:280–9. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 48.Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–84. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 49.Koutsodontis G, Tentes I, Papakosta P, Moustakas A, Kardassis D. Sp1 plays a critical role in the transcriptional activation of the human cyclin-dependent kinase inhibitor p21(WAF1/Cip1) gene by the p53 tumor suppressor protein. J Biol Chem. 2001;276:29116–25. doi: 10.1074/jbc.M104130200. [DOI] [PubMed] [Google Scholar]