Abstract
For screening a pool of potential substrates that load carrier domains found in non-ribosomal peptide synthetases, large molecule mass spectrometry is shown to be an ideal, unbiased assay. Combining the high resolving power of Fourier-Transform Mass Spectrometry with the ability of adenylation domains to select their own substrates, the mass change that takes place upon formation of a covalent intermediate thus identifies the substrate. This assay has an advantage over traditional radiochemical assays in that many substrates, the substrate pool, can be screened simultaneously. Using proteins on the nikkomycin, clorobiocin, coumermycin A1, yersiniabactin, pyochelin and enterobactin biosynthetic pathways as proof-of-principle, preferred substrates are readily identified from substrate pools. Furthermore this assay can be used to provide insight into the timing of tailoring events of biosynthetic pathways as demonstrated using the bromination reaction found on the jamaicamide biosynthetic pathway. Finally, this assay can provide insight into the role and function of orphan gene clusters for which the encoded natural product is unknown. This is demonstrated by identifying the substrates for two NRPS modules from the genes pksN and pksJ, that are found on an orphan NRPS/PKS hybrid cluster from Bacillus subtilis. This new assay format is especially timely for activity screening in an era when new types of thiotemplate assembly lines that defy classification are being discovered at an accelerating rate.
Keywords: Electrospray Fourier transform mass spectrometry, Non-ribosomal peptide synthetase, Polyketide synthase, Carrier domains, Post-translational modification
Almost weekly, new non-ribosomal peptide synthetase (NRPS) gene clusters encoding for proteins that biosynthesize bioactive compounds are discovered (1-4). Since many of the compounds produced by non-ribosomal peptide synthetase (NRPS) as well as polyketide synthase (PKS) paradigm have potent medicinal utility or are involved in virulence, and display unusual chemistry, they are of great academic and industrial interest. Therefore we wanted to develop an alternative method for substrate screening to complement the more traditional radioactive assays (5,6). In NRPS and PKS systems, the substrates and intermediates are loaded onto and processed while attached to the pantetheinyl functionality on a carrier domain (7,8). Examples of NRPS or hybrid NRPS/PKS natural products for which substrates load onto carrier domains are: the siderophore pyoverdine (9), a virulence factor excreted by pseudomonads, the antimicrobial agents penicillin (10), vancomycin (11) gramicidin (12) and the antitumor agent calicheamycin (13). With ∼300 genomes sequenced and available in the public domain, there are a large number of orphan PKS and NRPS gene clusters that have been identified (14,15). Even when the sequence of a gene cluster with a known natural product is available, progress to characterize the proteins in vitro has been slow. Among the limiting factors in our understanding of how these NRPS systems biosynthesize their respective natural products is the inefficiency in obtaining in vitro activity to verify and identify the substrates for these enzymes. One of the many reasons for this inefficient activity screening includes the unavailability of the proper radiolabeled substrates or intermediates. While excellent advancements have been made using bioinformatics to predict the substrate for adenylating domains (16,17,18), caution should be taken when relying solely on the predictions to assign a specific substrate (19). Furthermore new types of thiotemplate assembly lines that defy classification or thiotemplate assembly lines that do not follow the standard co-linearity rules are being discovered at an accelerating rate (1-4,20). As a general strategy, all bioinformatic predictions should be confirmed in vitro. With the recent push to develop NRPS-derived bioactive compounds in vitro (21,22) and to manipulate these systems in vivo to generate new pharmaceutically useful bioactive compounds (23-25), the in vitro characterization must be performed more efficiently. One of the ways one can improve the ability to screen for substrates is by being able to screen many substrates simultaneously. This assay would ideally also avoid using radiolabeled substrates which are used to load carrier domains usually used in autoradiographic analysis. Using nonradiolabeled substrates significantly broadens the accessibility of substrates that are commercially available or synthetically feasible. Here we describe a mass spectrometry based method to identify substrates based on mass changes that take place during acylation of a phosphopantetheinyl functionality on the carrier domain(s) of an NRPS protein from very complex substrate reaction mixtures. Observing such acylations by mass spectrometry on carrier domains is now becoming routine (5, 19, 26-36).
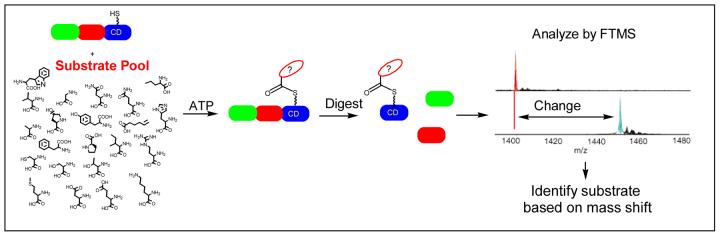
The general method to identify covalently loaded substrates or intermediates is shown in Figure 1. Overproduced protein that contains a carrier domain is purified and incubated with Sfp, a promiscuous phosphopantetheinyl transferase from B .subtilis, and CoA in order to generate the holo form of the carrier domain (37). A substrate pool and, when required, a separately purified activating domain is added to the holo carrier domain. Following an incubation period, the reaction is digested, quenched using formic acid and purified by HPLC prior to ESI-FTMS analysis.
Figure 1.
General approach of using FTMS to screen for acylation of a carrier domain using a substrate pool.
Results
NikP1, which is involved in the formation of the antibiotic nikkomycin (6), was incubated with Sfp and CoA to generate the holo form. The holo-NikP1 was incubated with ATP and all 19 proteinogenic L-amino-acids, glycine, L-selenocysteine, L-cystine, 4-trans-hydroxy-L-proline for 30 minutes before being quenched, digested using cyanogen bromide and analyzed by FTMS. The FTMS data indicated a mass shift of 137.2 Da, in agreement with loading of histidine onto the phosphopantetheinyl thiol (Figure 2C). When the same assay is repeated but L-histidine was omitted, NikP1 remained in its holo form, demonstrating that NikP1 is highly selective for loading of histidine relative to all other substrates present in the assay mixture (Figure 2B).
Figure 2.
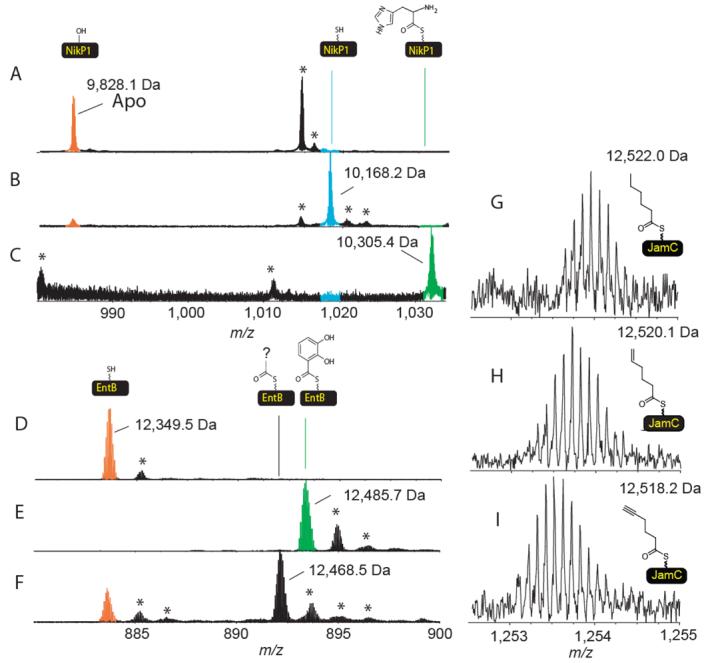
The identification of the preferred substrate by ESI- FTMS. A) The apo form of the carrier site of NikP1. B) Same as C but L-histidine omitted. C) The mass spectrum of the carrier site following incubation with CoA, Sfp and all 19 proteinogenic L-amino-acids, glycine, Lselenocysteine, L-cystine, 4-trans-hydroxy-L-proline. D) Holo-EntB(ArCP). E) Holo-EntB(ArCP) incubated with ATP and all 19 proteinogenic L-amino-acids, glycine, L-selenocysteine, L-cystine, 4-trans-hydroxy-L-proline and 2,3-dihydroxybenzoic acid F) Same as E but 2,3-dihydroxybenzoic acid omitted. G) Hexanoyl-S-JamC (only the N-term Met truncated version shown). H) Hexenoyl-S-JamC. I) Hexynoyl-S-JamC. * indicates salt adducts or non-active site peptides. The ions shown in A, B, C, G, H and I have a 10+ charge state. D,E and F are 14+
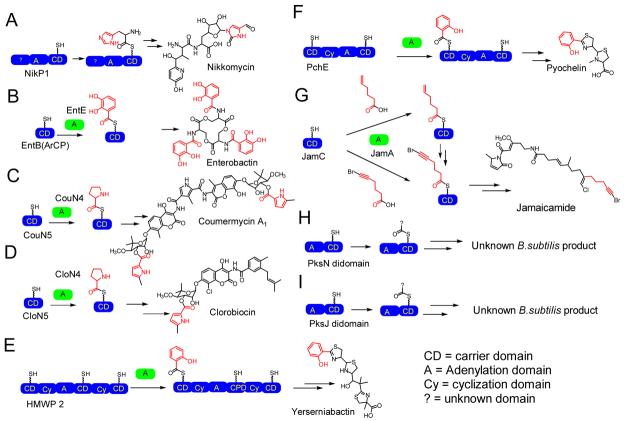
To illustrate that this method was not only valid for NikP1, the substrate loading for the following systems were examined: the CloN5/CloN4 carrier/adenylation domain pair involved in the formation of clorobiocin (Table 1) (5), CouN5/CouN4 involved in the formation of the antibiotic coumermycin (5), EntB(ArCP)/EntE involved in the formation of the siderophore enterobactin (24), HMWP2, involved in the formation of the siderophore yersiniabactin (27), PchE/PchD involved in the biosynthesis of the siderophore pyochelin (38), JamC/JamA involved in the formation of the neurotoxin jamaicamide (39), and two NRPS modules from the orphan NRPS/PKS gene cluster on Bacillus subtilis for which the product is unknown (40,41,42). The adenylation-carrier di-domains analyzed are from PksN (BG12652) and the second NRPS module from PksJ (BG10929, gene annotation from http://genolist.pasteur.fr/SubtiList/. All of the above systems were incubated with substrate pools and analyzed by ESI- FTMS.
Table 1.
Identification of substrate loading from defined substrate pools.
| System | Digested, method |
Mass holo (calc) |
Acylation substrate source |
Observed mass(es) after acylation (major species only) |
Mass change (Obs – Holoobs) |
Substrate ID based on mass change (* Natural substrate) |
|---|---|---|---|---|---|---|
| Nikkomycin NikP1 |
Y, CNBr |
10168.2 (10168.3) |
A | 10305.4 | 137.2 | L-histidine* |
|
B |
10168.2 |
0.0 |
N/A |
|||
| Clorobiocin CloN5/CloN4 |
N |
12520.1 (12520.1) |
A | 12616.9 | 96.8 | L-proline* |
|
C |
12633.0 |
112.9 |
4-trans-hydroxy- proline |
|||
| Coumermycin CouN5/CouN4 |
N |
12087.1 (12087.1) |
A | 12184.1 | 97.0 | L-proline* |
|
C |
12200.2 |
113.1 |
4-trans-hydroxy- proline |
|||
| Jamaicamide JamC/JamA |
N |
12544.9 (12544.9) |
D | 12650.9 | 96.0 | Hexenoic acid* |
| A | 12544.9 | 0.0 | N/A | |||
| Pyochelin PchE/PchD |
Y, CNBr |
17653.3 (17653.8) |
E |
17773.7 |
120.4 |
Salicylic acid* |
| Yersiniabactin HMWP2 |
Y, Trypsin |
3411.65 (3411.71) |
F | 3531.73 | 120.08 | Salicylic acid* |
| G | 3545.75 3515.74 |
134.10 104.09 |
4-Methylsalicylic acid Benzoic acid |
|||
| Enterobactin EntB(ArCP)/EntE |
N |
12349.5 (12349.3) |
A | 12468.5 | 119.0 | Unknown |
|
H |
12485.7 |
136.2 |
2,3-dihydroxybenzoic acid* |
A) All 19 proteinogenic L-amino-acids, glycine, L-selenocysteine, L-cystine, 4-trans-hydroxy-L-proline and ATP. B) Same as A but L-histidine omitted. C) Same as A but L-proline omitted. D) Same as A but with 5-hexenoic acid and 6-bromo-5-hexynoic acid added. E) Same as A with Salicylic acid added. F) Salicylic acid, 4-methylsalicylic acid, benzoic acid, methylsalicylic acid, benzoic acid, 4-hydroxybenzoic acid and p-toluic acid. G) Same as F but salicylic acid omitted. H) Same as A but with 2,3-dihydroxybenzoate added. Y = Yes digested, N = Not digested.
When PchE was presented with multiple substrates as well as the natural substrate salicylic acid and the activating enzyme PchD, the observed mass shift was 120 Da, in agreement with loading of salicylic acid (Table 1). When HMWP2 is incubated with six substrates known to independently load HMWP2 (27, 30), the mass shift corresponding to the authentic substrate salicylic acid was the major species observed. When salicylic acid was omitted, mass shifts corresponding to methyl salicylic acid and benzoic acid are the main species observed. When CloN5/CloN4 or CouN5/CouN4 carrier protein/activating protein pairs are incubated with substrates not including the natural substrate L-proline, a mass shift of 113 Da, corresponding to the 4-trans-hydroxy-proline is observed. However when proline is present, it is the only substrate loaded (+97 Da). In the case of EntB(ArCP)/EntE, a new species is formed that is 119 Da larger than the holo form of EntB(ArCP) when the authentic substrate is not present but when the natural substrate is present it is loaded exclusively (Table 1, Figure 2E and F). This mass shift was confirmed to be 119 Da by tandem mass spectrometry (data not shown). Next, the early steps in the biosynthesis of jamaicamide were investigated. The apo JamC protein showed two abundant forms of the protein, the WT and the post-translationally truncated form of the protein where the N-terminal methionine has been removed (loss of 131 Da). Simultaneous incubation of holo-JamC, JamA, a substrate pool that included 6-bromo-5-hexynoic acid, 5-hexenoic acid and ATP, produced a mass shift that was 96 Da larger than holo-JamC consistent with loading of hexenoic acid. Any attempts to load JamC with 6-bromo-5-hexynoic acid failed. 5-Hexanoic acid and 5-hexynoic acid are alternative substrates for this reaction (Figure 2G-I).
Even though as many as 24 substrates were screened simultaneously, the assay mixtures were still of defined nature. To further probe the limits of the assay, CloN5/CloN4 and NikP1 were incubated with ATP and a commercially available algal amino acid hydrolysate mixture as a representative undefined substrate pool. In both cases the correct mass shift corresponding to the natural substrate is observed (Table 2). This suggested that very complex mixtures/undefined assay conditions are possible. To further expand on this theme, a very complex substrate pool was tested, the E. coli metabolome, on NikP1, CloN5/CloN4, CouN5/CouN4, and EntB (ArCP)/EntE. In three of the four cases, even-though there is only partial acylation occupancy, the correct mass shift corresponding to the authentic substrate is observed. Loading onto holo-EntB(ArCP) was not observed, even when the metabolome is obtained from E. coli which was grown under iron limited conditions, suggesting that there is not enough free substrate available to observe the acylation reaction (Table 2).
Table 2.
Identification of substrate loading from undefined substrate pools.
|
System |
Digested, method |
Acylation substrate source |
New mass(es) after acylation |
Mass change (obs- holocalc) |
Substrate ID based on mass change (* Natural substrate) |
|---|---|---|---|---|---|
| NikP1 | Y, CNBr | Algal hydrolysate E.coli metabolome |
10306 10306 |
137.2 137.4 |
L-histidine* L-histidine* |
| CloN5/CloN4 | N | Algal hydrolysate E.coli metabolome |
12617 12617 |
97 97.1 |
L-proline* L-proline* |
| CouN5/CouN4 | N | E.coli metabolome | 12184 | 97 | L-proline* |
| EntB(ArCP)/EntE | N |
E.coli metabolome E.coli metabolomea |
NC NC |
NC NC |
Not loaded Not loaded |
NC = No mass Change observed, Y = Yes digested, N = Not digested.
Metabolome obtained from E. coli grown under iron limiting conditions.
To apply our method to orphan NRPS/PKS gene clusters, the adenylation and carrier di-domains from PksN and PksJ were cloned and overproduced. Because the di-domains are rather large (∼90 kDa) to observe by FTMS with sub-Dalton mass accuracy, the holo-forms of the di-domains were subjected to trypsin digestion and the active sites mapped by FTMS (Figure 4). The identity of the active sites were confirmed by tandem mass spectrometry as shown in Figure 4J for PksJ didomain and Figure 4K for the PksN didomain (43). Once mapped, the active sites were pantetheinylated using Sfp and CoA and compared to the apo form. In both cases, a mass shift of +340 Da is observed (Figure 4B and F). Subsequently, both of the proteins were incubated with ATP and an undefined substrate pool, the algal hydrolysate (Figure 4C and G). The PksJ domain increased by 57 Da, while the PksN domain increased by 71 Da. Incubation of PksJ with glycine and ATP resulted in the same mass shift (Figure 4D). Because the mass shift of PksN did not correspond to the predicted substrates cysteine orserine, alanine was incubated simultaneously with cysteine, serine and threonine. Again the observed mass shift was 71 Da (Figure 4H). When the substrate pool consisted of only threonine and serine, a mass shift of 87 was observed (Figure 4I), while no acylation of the carrier domain was observed when incubated with cysteine.
Figure 4.
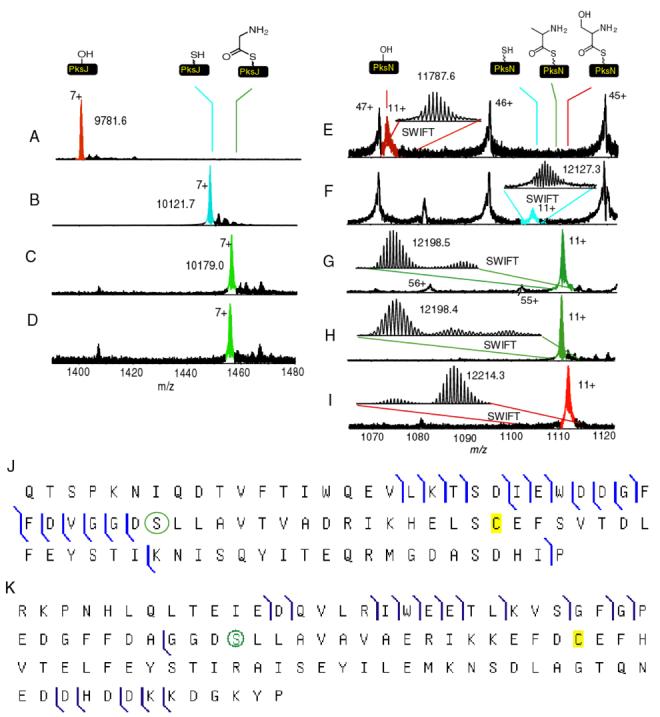
Active site mapping and substrate pool screening of the adenylation and carrier di-domains from PksN and PksJ. A) Apo-PksJ B) Holo-PksJ C) Algal hydrolysate substrate screen with holo-PksJ D) Glycyl-S-PksJ. E) Apo-PksN F) Holo-PksN G) Algal hydrolysate substrate screen with holo-PksN. H) Ala, Cyst, Ser, Thr screen with holo-PksN. I) Ser, Thr screen with holo-PksN. J) OCAD verification of the active site for holo-PksJ. K) IRMPD verification of the active site for holo-PksN. The green circle indicates the location of the phopshopantetheinyl functionality.
Discussion
This paper has presented yet another tool in the arsenal for the in vitro characterization of the NRPS and PKS substrate specificity. ESI-FTMS has been used in conjunction with substrate pools to screen for substrates that can be loaded onto carrier domains. Although the assay is performed on an FTMS instrument, substrate screening can likely be done on other mass spectrometers as well, but one will have to keep in mind that the ease in mapping the active sites and accuracy of the mass shifts observed increase with increasing resolution. Currently to look at protein domains, FT-MS, which requires considerable expertise, is the most accurate method for this. The custom build instrument used in this study has a mass accuracy of 5-25 ppm when it is externally calibrated, while some commercial FTMS instruments now give mass accuracies to within 2 ppm. To analyze substrates loaded onto carrier domains will be very difficult when the mass accuracy is lower than 100 ppm, but has been done before (34-36). But as there are increasingly more FT-MS instruments that are accessible to the scientific community, the implemention of our approach described here is likely to find wider application in the analysis of NRPS systems
The generality of this method has many applications and a few of these are demonstrated in this paper. The first application of the substrate pool is to identify the natural substrate. For the proof of concept experiments NikP1, EntB, HMWP2, CouN5, CloN5 and PchE were loaded with their natural substrate. In each case investigated, as long as the natural substrate was present, substrate loading by non-cognate substrates was negligible. In the absence of the natural substrate there was one unexplained result where EntB(ArCP) displayed a +119 Da species and none of the 98-99% pure amino acids used as a substrate matched this mass upon acylation. Therefore this is an unidentified non-cognate substrate, which must come from the remaining 1-2% of impurities found in commercially prepared amino acids. This is also in agreement with the observation that there was insufficient material for complete conversion of the holo form of the protein to the acylated species. When the authentic natural substrate, 2,3-dihydroxy-benzoic acid, was present with this same substrate pool, it was the major peak observed in the spectrum (Figure 2E). The identification of substrates is also possible from very complex substrate pools of undefined nature such as the algal hydrolysate or the E. coli metabolome.
Sometimes loading with a non-cognate substrate is desired, as would be the case in the development of new bioactive compounds. Replacing the pyrrole on clorobiocin or coumermycin may allow for the development of more soluble aminocoumarin antibiotics and therefore make them more applicable in a clinical setting (23). It was unknown if anything other than proline would load onto CloN5 or CouN5. When a substrate screen was done with 19 proteinogenic L-amino-acids, glycine, L-selenocysteine, L-cystine, 4-trans-hydroxy-L-proline but the native substrate, proline, was omitted, a mass shift of +113 Da was observed. This mass shift corresponds to the mass of 4-trans-hydroxy-L-proline being loaded. Subsequently, it was verified by the traditional radioactive pyrophosphate exchange assay that adenylating enzymes CloN4 and CouN4 could utilize 4-trans-hydroxy-L-proline as an alternative substrate (5). This makes 4-trans-hydroxy-L-proline a front-line candidate to generate more soluble clorobiocin and coumermycin antibiotics and was identified from a single assay.
The third utility of this FTMS-based assay is to establish the timing of tailoring events which take place on NRPS and PKS modules. In the jamaicamide biosynthetic pathway, it was unknown if the bromination took place before or after the loading of hexenoic acid onto JamC. Simultaneous incubation of holo-JamC, JamA, 6-bromo-5-hexynoic acid, 5-hexenoic acid and ATP only resulted in the loading of hexenoic acid. Any attempt to load 6-bromo-5-hexynoic acid by itself failed as well. This means that the bromination reaction either takes place while the substrate is attached to JamC or otherwise on one of the other NRPS or PKS proteins found on the jamaicamide biosynthetic pathway or after the jamaicamide is fully assembled. 5-Hexanoic acid and 5-hexynoic acid were also alternative substrates, in agreement with the classical pyrophosphate exchange assay (39).
The fourth application is to use the substrate pool method as an activity screen. This is particulary relevant when the substrate cannot be readily predicted by bioinformatic means. One example is the amine donor source for the aminotransferase domain of MycA, a protein responsible for the generation of the β-amino acid found on the mycosubtilin biosynthetic pathway. Because the substrate amine donor was unknown, all possible substrate candidates were screened simultaneously to see if the protein was active. Armed with its activity, it was ultimately established that Gln was the preferred amine donor in this reaction (33).
This method will be most useful in the characterization of gene clusters with unknown natural products (orphan gene clusters). As an example, Bacillus subtilis has one orphan NRPS/PKS gene cluster (40, 41). Even though this cluster has both NRPS and PKS modules, it has been suggested to be involved in the formation of difficidin (20, 41). Difficidin, however, does not have an amino acid in its structure and there is little experimental evidence to support that difficidin is produced by this gene cluster (41). Therefore, the activity screen was used to establish if the NRPS domains of this cluster are functional. When activity was observed, it was used to identify which substrates were loaded. Using bioinformatics, the substrate specificity for the second NRPS domain on PksJ was predicted to load glycine, while PksN was predicted to load cysteine (16,17) and BLAST analysis indicated that the adenylation domain may be similar also to serine loading domains. Once the active sites had been mapped, they were pantetheinylated using Sfp and CoA. Subsequently, both of the proteins were incubated with ATP and an unbiased substrate source, the algal hydrolysate. The PksJ domain increased by 57 Da, while the PksN domain increased by 71 Da (Figure 4D and G). These mass shifts correspond to glycine and alanine. Incubation of PksJ with glycine and ATP resulted in the same mass shift. Because the mass shift of PksN did not correspond to the predicted substrate cysteine or serine, L-alanine was incubated simultaneously with L-cysteine, L-serine and L-threonine (this substrate served as a negative control). Again, the only substrate that was loaded was alanine. When L-alanine was omitted, L-serine was a substrate for this protein. However, because alanine out-competes both cysteine and serine, the predicted substrates, we favor alanine to be the natural substrate. Having observed activity for the PksJ and PksN di-domains provides some insight into the role of this gene cluster. The first important observation is that the NRPS's found on the orphan gene cluster of Bacillus subtilis are active. Second, because we have observed activity, our data suggest that this gene cluster produces a product other than difficidin, since difficidin does not have a nitrogen in its structure that would have come from glycine or alanine. We cannot exclude the possibility that this gene cluster produces a modified form of difficidin or analogous natural products to difficidin such as hydroxymycotrienin A produced by Bacillus sp. BMJ958-62F4 (44), which has amino acid components in its structure. Lastly, this is the first demonstration of the utility of FTMS to provide insight into the functions of orphan gene cluster and is going to be applicable to many other orphan NRPS gene clusters that have been sequenced. This method is not only applicable to NRPS systems but also to systems of polyketide origin, fatty acid biosynthesis or any other system that can form covalent modifications to proteins.
Materials and methods
The sequencing grade trypsin was purchased from Promega, 1 U is defined as the amount of sequencing grade modified trypsin required to produce a ΔA253 of 0.001 per minute at 30°C with the substrate α-benzoyl-L-arginine ethyl ester. Cyanogen bromide (CNBr), amino-acids, HPLC solvents, CoA (trilithium salt), algal hydrolysate and all other substrates were purchased from SIGMA-ALDRICH. 6-Bromo-5-alkynoic acid was synthesized as described (45). Superflow nickel affinity resin was from Qiagen. PD10 gel filtration columns were obtained from Amersham Biosciences. The HPLC column used for all desalting steps and separations a Jupiter 5μ C4 300Å column from Phenomenex was used. The freeware PAWS was obtained from Proteometrics.
Construction of JamC, PksJ and PksN NRPS constructs
JamC gene was cloned from the pJam1 fosmid clone (39) using the following primers Forward primer 5′ CATGCCATGGAAAACTTAACCGTAG 3′ Reverse primer 5′ CCGCTCGAGTGCACCAAAGTGCTCTGC 3′ and PfuI polymerase. The resulting PCR product was cloned in frame with the carboxy-6xHis fusion at the NcoI and XhoI restriction sites of pET28a. The pksN and pksJ didomains were cloned in a similar fashion from the Bacillus subtilis strain NCIB 3610 (49). The pksJ-AT2 didomain was amplified using primers: Forward 5' AGCTAGCTTTGAACTGTGGGAAACAGA 3' and reverse 5' ACTCGAGTCATTTTTGCAATGTCCATAATCC 3'. The amplified fragment was digested with NheI and XhoI and cloned into pET28a to generate plasmid pPDS0372. The pksN-AT didomain was amplified using primers: forward 5' GAATTCACATATGGGCTTGCAAAAAGTGCTTG 3' and reverse 5' ACTCGAGTCACGGGTATTTTCCATCTTTTTTG 3'. The amplified fragment was digested with NdeI and XhoI and cloned into pET28a to generate plasmid pPDS0374.
Protein expression and protein purification
The proteins NikP1, HMWP2, PchE, PchD, JamA, CloN5, CloN4, CouN5, CouN4, EntB(ArCP), EntE were purified as described (5, 26, 27, 30, 39). An expression strain (E. coli BL21(DE3) star transformed with a plasmid encoding JamC) was incubated at 37°C until an OD 600 reached 0.7. At this point, IPTG was added (50 mg/L) and allowed to incubate for 4-6 hours at 28°C. The cells were then harvested by centrifugation, lysed by the addition of lysozyme and sonication. The insoluble materials were pelleted by centrifugation and the remaining supernatant loaded onto a column containing NTA superflow nickel affinity resin and the protein purified per instructions of the manufacturer (Qiagen). The overproduction and purification of Bacillus subtilis PksJ and PksN di-domains were done in a similar fashion. The purified proteins were buffer exchanged using PD-10 gel filtration columns that were equilibrated with 50 mM Tris buffer, pH = 7.5, with 1 mM TCEP. Stock solutions containing 10% glycerol were prepared and stored at −80°C.
Preparation of E. coli metabolomes
E. coli was grown in 150 mL of Luria Broth (LB) at 37°C to an OD600 = 0.6-0.8. The cells were harvested by centrifugation in a Sorvall RC-5C+ centrifuge (SLA-3000 rotor, 6000 rpm) at 4°C for 6 minutes. The pellet was resuspended in 2.5 mL of 25 mM Tris (pH 7.6 , 25 mM) and the cells were lysed by sonication in the presence of lysozyme. The lysate was clarified by centrifugation in a Sorvall RC-C5+ centrifuge (SS-34, 16000 rpm, 4°C for 25 minutes). About 1 mL of lysed crude extract (CE) was gel filtered by using a PD10 column equilibrated with 25 mM Tris (pH 7.6 , 25 mM). This was achieved by loading the column with 1.0 mL of CE and allowed to flow through. Then 5.0 ml of Tris-Cl (pH 7.6 , 25 mM) was added and again allowed to flow through to remove all the proteins. After this volume, the small molecules of the lysed extract were collected in the following 3.0 mL. These 3.0 mL were collected as 1 mL fractions, frozen at −80°C and lyophilized. The dried sample was resuspended in 50-100 μL of 50 mM Tris, pH 7.6 so it could be used in the substrate identifications studies.
Acylation of CloN5 using undefined metabolite pools
As a representative acylation we describe the acylation of CloN5, all other assays were carried out in a similar fashion except that the substrate pool was varied. The acylation of CloN5 was carried out in two steps. First, the apo form of CloN5 in a concentration of 25 μM was reacted for one hour at room temperature in the presence of 3.6 μM Sfp, 8 mM MgCl2, and 250 μM CoA in a total reaction volume of 100 μL, which resulted in the generation of the holo form of the enzyme. In the second step, 4 μL of 0.1 M ATP, 2 μL of 1.3 mg/mL of CloN4 and 10-50 μL of an E. coli metabolome were added to the holo enzyme generated in the first step. This was incubated at room temperature for 30 minutes before quenching with 1:1 (v/v) with 10% formic acid. This was repeated for each (5 total) E. coli metabolome. Other substrate pools included the defined mixtures of substrates which were present at 0.5 to 1 mM (often some precipitation would be observed), the algal hydrolysate mixture was present at 0.2-0.5 mg/mL.
Acylation and CNBr digestion of NikP1
The acylation of NikP1 was also carried out in two steps. The generation of the holo form of the enzyme was performed as described for CloN5. In the second step, 4 μL of 0.1 M ATP and 5 μL of an E. coli metabolome were added to the solution. This was incubated at room temperature for 30 minutes before stopping the reaction by quenching 1:1 (v/v) with 10% formic acid. The acylated forms of NikP1 were purified by HPLC on an HP1090M HPLC using a 30 minute gradient of 90% water with 0.1% TFA and 10% ACN (0.1% TFA) to 5% water (0.1% TFA) and 95% ACN (0.1% TFA), collecting peaks between 21.5 and 23.0 minutes (This additional purification step of NikP1, before CNBr digestion, was done because the metabolome introduced contaminations that overlapped with the active site after digestion, this step was not necessary when defined substrate sources were used in the substrate screen). The resulting samples were frozen at −80°C and lyophilized overnight. The samples were redissolved in 100 mM NH4Oac (pH 4), 6M urea, 10% CH3CN, 10 mM TCEP and digested with 1.0 M CNBr in ACN (400 μL reaction volume) for 12-18 hours in the dark. Each sample was then frozen at −80°C and lyophilized overnight in the dark, rechromatographed using the HPLC conditions describe above and the fractions eluting at 18 to 20 min were frozen, lyophilized and analyzed by FTMS. As with CloN5, each E. coli metabolome was tested as well as a control containing 22 amino acids. PchE was digested using CNBr using an identical protocol as described for HMWP2 (30), PksJ and PksN didomains were digested using trypsin in an identical fashion as described for MycA on the mycosubtilin biosynthetic pathway (33). All other proteins did not need digestion in order to be seen them by ESI-FTMS.
Peptide mapping of the PksJ and PksN constructs
Each didomain of PksJ and PksN 1 mg/mL was incubated with 3.6 μM Sfp, 8 mM MgCl2, and 320 μM coenzyme A for 1 hour (reaction volume 300 μL). The final product was digested with trypsin at pH 8.0, 85 U, for 10 minutes, and quenced by acidifying with 50 μL of 10 % formic acid. The reaction was purified by HPLC on an HP1100 HPLC using a 60 minute gradient (Table 3) collecting fractions at 1 min intervals. A peak-list of all the observed ions was generated using the software THRASH and/or manual deconvolution of the charge states. The generated peak-list was imported into the freeware PAWS to identify the active sites. Once a match to the active site was obtained, the active sites were subjected to OCAD or IRMPD (46). The resulting fragment ions from OCAD and IRMPD were then analyzed using ProSight PTM to verify that they were indeed the active sites (47).
Table 3.
HPLC Gradient for active site purification, Solvent A is water (0.1% TFA) and Solvent B is acetonitrile (ACN, 0.1% TFA). The reason for the unusual gradient above 60 min is to wash the column prior to the next injection to avoid contamination from a previous run.
| Time (min) |
0.00 | 10.0 | 15.0 | 55.0 | 60.0 | 60.1 | 60.2 | 62.6 | 63.0 | 65.0 | 66.0 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| %A | 90 | 90 | 70 | 30 | 10 | 10 | 95 | 95 | 5 | 5 | 90 |
| %B | 10 | 10 | 30 | 70 | 90 | 90 | 5 | 5 | 95 | 95 | 10 |
MS analysis
HPLC fractions containing the active sites prepared as described above were redissolved in 100 μL of 78 % ACN, 0.1% acetic acid or 49% methanol, 1% formic acid and analyzed by ESI-FTMS. For mass spectrometric analysis, a custom 8.5 Tesla ESI-FTMS mass spectrometer was used which was equipped with a front-end quadrupole (46). The samples were introduced into the FTMS using a NanoMate 100 for automated nanospray (Advion Biosciences, Ithaca, NY). Typically 500 ms ion accumulation per scan was used and 50-200 scans were acquired per spectrum. The instrument was externally calibrated using ubiquitin, 8560.65 Da monoisotopic Mr value (Sigma). For the calculation of the masses of the proteins, the MIDAS analysis data-station was used (48). A mass peak-list of all the observed ions was generated using the software THRASH (embedded in the MIDAS data-station) and/or manual deconvolution. All masses reported in this manuscript are reported as the neutral monoisotopic masses.
Figure 3.
Systems interrogated in this study.
Acknowledgments
This work was supported in part by grants from NIH GM 49338 (CTW), NIH 067725 (NLK), NIH GM 58213 (RK), NIH CA 83155 (WCG), NIH Kirschstein NRSA postdoctoral fellowship F32-GM 073323-01 (PCD), National Science Foundation Postdoctoral Fellowship in Microbial Biology DBI-0200307 (PDS) and the Hertz Foundation Graduate Fellowship (MAF).
ABBREVIATIONS
- HPLC
high pressure liquid chromatography
- ESI
electrospray
- FTMS
Fourier transform mass spectrometry
- ATP
adenosine triphosphate
- CoA
CoenzymeA
- NRPS
non-ribosomal peptide synthetase
- PKS
polyketide synthase
- WT
wild-type
- CNBr
cyanogen bromide
- OCAD
octopole collisional activated dissociation
- IRMPD
infrared multiphoton dissociation
- SWIFT
stored waveform inverse Fourier transform
- PPi
pyrophosphate
REFERENCES
- 1.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q.u, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, Thomashow LS, Loper JE. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nature Biotechnology. 2005;23(7):873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nature Chemical Biology. 2005;1(5):265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 3.Liu W, Nonaka K, Nie L, Zhang J, Christenson SD, Bae J, Van Lanen SG, Zazopoulos E, Farnet CM, Yang CF, Shen B. The neocarzinostatin biosynthetic gene cluster from Streptomyces carzinostaticus ATCC 15944 involving two iterative type I polyketide synthases. Chemistry & Biology. 2005;12(3):293–302. doi: 10.1016/j.chembiol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Rondon MR, Ballering KS, Thomas MG. Identification and analysis of a siderophore biosynthetic gene cluster from Agrobacterium tumefaciens C58. Microbiology. 2004;150(11):3857–3866. doi: 10.1099/mic.0.27319-0. [DOI] [PubMed] [Google Scholar]
- 5.Garneau S, Dorrestein PC, Kelleher NL, Walsh CT. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry. 2005;44(8):2770–2780. doi: 10.1021/bi0476329. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Hubbard BK, O'Connor SE, Walsh CT. Formation of β-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chemistry & Biology. 2002;9(1):103–112. doi: 10.1016/s1074-5521(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science. 2004;303(5665):1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 8.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annual Review of Microbiology. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 9.Smith EE, Sims EH, Spencer DH, Kaul R, Olson MV. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. Journal of Bacteriology. 2005;187(6):2138–2147. doi: 10.1128/JB.187.6.2138-2147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keszenman-Pereyra D, Lawrence S, Twfieg M-E, Price J, Turner G. The npgA/ cfwA gene encodes a putative 4'-phosphopantetheinyl transferase which is essential for penicillin biosynthesis in Aspergillus nidulans. Current Genetics. 2003;43(3):186–190. doi: 10.1007/s00294-003-0382-7. [DOI] [PubMed] [Google Scholar]
- 11.Zerbe K, Woithe K, Li DB, Vitali F, Bigler L, Robinson JA. An oxidative phenol coupling reaction catalyzed by OxyB, a cytochrome P450 from the vancomycin-producing microorganism. Angewandte Chemie, International Edition. 2004;43(48):6709–6713. doi: 10.1002/anie.200461278. [DOI] [PubMed] [Google Scholar]
- 12.Vater J, Stein TH. Structure, function, and biosynthesis of gramicidin S synthetase. Comprehensive Natural Products Chemistry. 1999;4:319–352. [Google Scholar]
- 13.Ahlert J, Shepard E, Lomovskaya N, Zazopoulos E, Staffa A, Bachmann BO, Huang K, Fonstein L, Czisny A, Whitwam RE, Farnet CM, Thorson JS. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science. 2002;297(5584):1173–1176. doi: 10.1126/science.1072105. [DOI] [PubMed] [Google Scholar]
- 14.Walsh CT. Natural insights for chemical biologists. Nature Chemical Biology. 2005;1(3):122–124. doi: 10.1038/nchembio0805-122. [DOI] [PubMed] [Google Scholar]
- 15.Fortman JL, Sherman DH. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. ChemBioChem. 2005;6(6):960–978. doi: 10.1002/cbic.200400428. [DOI] [PubMed] [Google Scholar]
- 16.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chemistry & Biology. 1999;6(8):493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 17.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chemistry & Biology. 2000;7(3):211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 18.Di Vincenzo L, Grgurina I, Pascarella S. In silico analysis of the adenylation domains of the freestanding enzymes belonging to the eucaryotic nonribosomal peptide synthetase-like family. FEBS Journal. 2005;272(4):929–941. doi: 10.1111/j.1742-4658.2004.04522.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Lanen SG, Dorrestein PC, Christenson SD, Liu W, Ju J, Kelleher NL, Shen B. Biosynthesis of the β-amino acid moiety of the enediyne antitumor antibiotic C-1027 featuring β-amino acyl-S-carrier protein intermediates. Journal of the American Chemical Society. 2005;127(33):11594–11595. doi: 10.1021/ja052871k. [DOI] [PubMed] [Google Scholar]
- 20.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of Curacin A, an antitubulin natural product from the tropical marine Cyanobacterium Lyngbya majuscula. Journal of Natural Products. 2004;67(8):1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 21.Kohli RM, Walsh CT, Burkart MD. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. 2002;418(6898):658–61. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 22.Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Peptide cyclization catalysed by the thioesterase domain of tyrocidine synthetase. Nature. 2000;407(6801):215–218. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- 23.Galm U, Dessoy MA, Schmidt J, Wessjohann LA, Heide L. In Vitro and In Vivo Production of new aminocoumarins by a combined biochemical, genetic, and synthetic approach. Chemistry & Biology. 2004;11(2):173–183. doi: 10.1016/j.chembiol.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Eustaquio AS, Gust B, Galm U, Li S-M, Chater KF, Heide L. Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters. Applied and Environmental Microbiology. 2005;71(5):2452–2459. doi: 10.1128/AEM.71.5.2452-2459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freitag A, Galm U, Li S-M, Heide L. New aminocoumarin antibiotics from a cloQ-defective mutant of the clorobiocin producer Streptomyces roseochromogenes DS12.976. Journal of Antibiotics. 2004;57(3):205–209. doi: 10.7164/antibiotics.57.205. [DOI] [PubMed] [Google Scholar]
- 26.Shaw-Reid CA, Kelleher NL, Losey HC, Gehring AM, Berg C, Walsh CT. Assembly line enzymology by multimodular nonribosomal peptide synthetases: the thioesterase domain of E. coli EntF catalyzes both elongation and cyclolactonization. Chemistry & Biology. 1999;6(6):385–400. doi: 10.1016/S1074-5521(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 27.Mazur MT, Walsh CT, Kelleher NL. Site-specific observation of acyl intermediate processing in thiotemplate biosynthesis by Fourier transform mass spectrometry: The Polyketide Module of Yersiniabactin Synthetase. Biochemistry. 2003;42(46):13393–13400. doi: 10.1021/bi035585z. [DOI] [PubMed] [Google Scholar]
- 28.Hicks L, Weinreb P, Konz D, Marahiel MA, Walsh CT, Kelleher NL. Fourier-transform mass spectrometry for detection of thioester-bound intermediates in unfractionated proteolytic mixtures of 80 and 191 kDa portions of Bacitracin A synthetase. Analytica Chimica Acta. 2003;496(12):217–224. [Google Scholar]
- 29.Hicks LM, O'Connor SE, Mazur MT, Walsh CT, Kelleher NL. Mass Spectrometric interrogation of thioester-bound intermediates in the initial stages of epothilone biosynthesis. Chemistry & Biology. 2004;11(3):327–335. doi: 10.1016/j.chembiol.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 30.McLoughlin SM, Kelleher NL. Kinetic and regiospecific interrogation of covalent intermediates in the nonribosomal peptide synthesis of Yersiniabactin. Journal of the American Chemical Society. 2004;126(41):13265–13275. doi: 10.1021/ja0470867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatto GJ, Jr., McLoughlin SM, Kelleher NL, Walsh CT. Elucidating the substrate specificity and condensation domain activity of FkbP, the FK520 pipecolate-incorporating enzyme. Biochemistry. 2005;44(16):5993–6002. doi: 10.1021/bi050230w. [DOI] [PubMed] [Google Scholar]
- 32.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis; Proceedings of the National Academy of Sciences of the United States of America; 2005. pp. 13843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aron ZD, Dorrestein PC, Blackhall JR, Kelleher NL, Walsh CT. Characterization of a new tailoring domain in polyketide biogenesis: the amine transferase domain of MycA in the Mycosubtilin gene cluster. Journal of the American Chemical Society. 2005;127(43):14986–14987. doi: 10.1021/ja055247g. [DOI] [PubMed] [Google Scholar]
- 34.Hong H, Appleyard AN, Siskos AP, Garcia-Bernardo J, Staunton J, Leadlay PF. Chain initiation on type I modular polyketide synthases revealed by limited proteolysis and ion-trap mass spectrometry. FEBS Journal. 2005;272(10):2373–2387. doi: 10.1111/j.1742-4658.2005.04615.x. [DOI] [PubMed] [Google Scholar]
- 35.Schnarr NA, Chen AY, Cane DE, Khosla C. Analysis of covalently bound polyketide intermediates on 6-Deoxyerythronolide B synthase by tandem proteolysis-mass spectrometry. Biochemistry. 2005;44(35):11836–11842. doi: 10.1021/bi0510781. [DOI] [PubMed] [Google Scholar]
- 36.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris HR. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. Journal of Biological chemistry. 1996;271(26):15428–35. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 37.Reuter K, Mofid MR, Marahiel MA, Ficner R. Crystal structure of the surfactin synthetase-activating enzyme Sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO Journal. 1999;18(23):6823–6831. doi: 10.1093/emboj/18.23.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadri LEN, Keating TA, Patel HM, Walsh CT. Assembly of the pseudomonas aeruginosa nonribosomal peptide siderophore Pyochelin: in vitro reconstitution of Aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry. 1999;38(45):14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- 39.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. structure and biosynthesis of the Jamaicamides, new mixed polyketide-peptide neurotoxins from the marine Cyanobacterium Lyngbya majuscula. Chemistry & Biology. 2004;11(6):817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 40.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the gram positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 41.Hofemeister J, Conrad B, Adler B, Hofemeister B, Feesche J, Kucheryava N, Steinborn G, Franke P, Grammel N, Zwintscher A, Leenders F, Hitzeroth G, Vater J. Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Molecular Genetics and Genomics. 2004;272(4):363–378. doi: 10.1007/s00438-004-1056-y. [DOI] [PubMed] [Google Scholar]
- 42.Stein T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Molecular Microbiology. 2005;56(4):845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 43.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrometry Reviews. 1998;17(1):1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 44.Hosokowa N, Naganawa H, Hamada M, Takeuchi T, Ikeno S, Hori M. Hydroxymycotrienins A and B, new ansamycin group antibiotics. Journal of Antibiotics. 1996;49(5):425–431. doi: 10.7164/antibiotics.49.425. [DOI] [PubMed] [Google Scholar]
- 45.Gung BW, Dickson H. Total synthesis of (−)-Minquartynoic Acid: an anti-cancer, anti-HIV natural product. Organic Letters. 2002;4(15):2517–2519. doi: 10.1021/ol026145n. [DOI] [PubMed] [Google Scholar]
- 46.Patrie SM, Charlebois JP, Whipple D, Kelleher NL, Hendrickson CL, Quinn JP, Marshall AG, Mukhopadhyay B. Construction of a hybrid quadrupole/fourier transform ion cyclotron resonance mass spectrometer for versatile MS/MS above 10 kDa. Journal of the American Society for Mass Spectrometry. 2004;15:1099–1108. doi: 10.1016/j.jasms.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 47.LeDuc RD, Taylor GK, Kim Y-B, Januszyk TE, Bynum LH, Sola JV, Garavelli JS, Kelleher NL. ProSight PTM: An integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Research. 2004;32:W340–W345. doi: 10.1093/nar/gkh447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senko MW, Canterbury JD, Guan S, Marshall AG. A high-performance modular data system for Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 1996;10:1839–1844. doi: 10.1002/(SICI)1097-0231(199611)10:14<1839::AID-RCM718>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 49.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]