Abstract
Objective:
The RCAα block (Regulators of Complement Activation, 1q32) contains critical complement regulatory genes such as CR1 and MCP. This study examined RCAα block haplotype associations with both disease susceptibility and diversification of the anti-Ro/La autoantibody response in primary Sjögren syndrome (pSS).
Methods:
115 patients with pSS and 98 controls were included in the study. 93 of 109 (85%) of the patients with pSS were seropositive for Ro/La autoantibodies. The Genomic Matching Technique (GMT) was used to define RCAα block ancestral haplotypes (AH).
Results:
RCAα block haplotypes, AH1 and AH3, were both associated with autoantibody-positive pSS (p = 0.0003). Autoantibody associations with both HLA DR3 and DR15 have been previously defined. There was an epistatic interaction (p = 0.023) between RCAα AH1 and HLA DR3, and this genotypic combination was present in 48% of autoantibody-positive patients with pSS compared with 8% of controls. This epistasis is most simply attributable to an interaction between C4 and its receptor, CR1, encoded within the RCAα block. Both DR3 and a relative C4 deficiency are carried on the major histocompatibility complex 8.1 ancestral haplotype. Only four of 92 (4%) autoantibody-positive patients with pSS did not carry any risk RCAα or HLA haplotype, compared with 36 of 96 (38%) controls, and there were differences in haplotype frequencies within autoantibody subsets of pSS.
Conclusions:
Normal population variation in the RCAα block, in addition to the major histocompatibility complex, contributes genetic susceptibility to systemic autoimmune disease and the autoantibody response. This finding provides evidence for the role of regulation of complement activation in disease pathogenesis.
Primary Sjögren syndrome (pSS) is a systemic autoimmune disease, which shares a number of clinical, serological and genetic features with systemic lupus erythematosus (SLE).1 One of the features of this disease is the high prevalence of autoantibodies to the Ro and La components of a ribonuclear protein (RNP) complex. Ro/La autoantibodies occur in individual patients as targeting either Ro alone, or with specificity to both Ro and La, and autoantibody subgroups are stable over time. Major histocompatibility complex (MHC) disease associations have been known for many years,2 3 and more recently it has been recognised that these MHC haplotypes regulate the diversification of the autoantibody response.4 5 Ro/La autoantibodies occur in other rheumatological diseases such as SLE; however, they are highly specific for pSS and constitute one of the classification criteria for this disease.6 Serum Ro/La autoantibodies have been shown to pre-date disease onset, often for many years, and seropositivity predicts disease severity and extraglandular features in patients with pSS.7 More recently, a pathogenic role has been proposed as Ro/La autoantibodies form nucleic-acid-containing immune complexes that can trigger prolonged type I interferon production, leading to a self-perpetuating autoimmune reaction.8
Deficiencies in the classical pathway of the complement system have been implicated in the aetiology and pathogenesis of autoimmune diseases such as SLE.9 Human complement receptor 1 (CR1, CD35) is an integral membrane complement control protein (CCP) whose primary role on erythrocytes is the non-inflammatory clearance of immune complexes opsonised with C3a and C4b. There is longstanding evidence, over 25 years in our own papers,10 for an acquired CR1 deficiency in SLE. A recent meta-analysis has demonstrated an association between a molecular weight variant of CR1 and SLE.11 Further, in the mouse, CR1/CR2 (encoded by alternatively spliced forms of the same gene) is a lupus susceptibility gene12 13 important in the modulation of the antinuclear autoantibody response.14 15
Human CR1 is located at 1q32 in the regulators of complement activation (RCA) complex, which also includes other complement regulatory encoding genes, namely factor H (CFH), complement 4 α and β binding protein’s (C4ABP and C4BBP), decay accelerating factor (DAF, CD56), complement receptor 2 (CR2, CD21) and membrane cofactor protein (MCP, CD46).
Encompassing some 13 Mb of the long arm of chromosome 1, the RCA does not contain an even distribution of CCPs. In fact all CCPs can be found within one of two distinct blocks, located at the telomeric and centromeric ends of the cluster, we refer to these as the α and β blocks respectively. Both blocks are about 500 kb in length and are characterised by extensive imperfect genomic duplication and degeneracy generated by insertions and deletions (indels) such as those created through domain duplication/deletion and retroviral insertion. CR1 is encoded within the RCAα block, where it has been duplicated as part of a segment along with MCP-Like, to form CR1-L and MCP (or vice versa). Segment A (CR1 and MCP-L) and B (CR1-L and MCP) are located next to each other and encompass ∼350 kb of the α block.16
As demonstrated some time ago within the MHC,17–19 and more recently throughout the entire genome,20–22 genomic blocks are normally defined by gene and segmental duplications while also exhibiting other characteristics such as suppression of recombination and an increased frequency of copy number, indel and single nucleotide polymorphisms.23 A consequence of recombination suppression is that haplotypes of these regions are likely to have been inherited faithfully over many generations (ie, ancestral haplotypes (AH)) and will therefore be important in defining complex genetic interactions.24
We recently reported a novel haplotyping approach, capable of interrogating the genetically complex RCAα block, by utilising the genomic duplication of CR1 and MCP (segment A and segment B).25 The assay uses the principles of the Genomic Matching Technique,23 used by us, and other groups, to interrogate the highly complex MHC region for the identification of donor/recipient matches prior to bone marrow transplantation.26–29 More recently the technique has been extended to forensic applications30 and used within the RCA complex on chromosome 125 and the class II region of the canine MHC.31 The technique involves the specific amplification, with a single primer pair, of multiple complex geometric elements, all linked and located within duplicated segments. The conceptual basis behind the technique is that duplicated elements make excellent haplospecific markers as they have evolved in line with the many other changes that have also occurred on each haplotype.
The aim of this study was to evaluate RCAα block haplotypes, as defined by the Genomic Matching Technique (GMT), in relation to both disease susceptibility and Ro/La autoantibody responses in patients with pSS. Specifically we hypothesised that genetic variation in the RCAα, in combination with HLA, would influence the diversification of the Ro/La autoantibody response.
MATERIALS AND METHODS
Study participants
Ninety-eight Caucasian controls and 115 Caucasian patients with pSS from the South Australian Sjögren syndrome research registry were included in the study. All patients met the revised 2002 American-European consensus research classification criteria for pSS,6 and controls were recruited from the same population base as the patients. Anti-Ro/La autoantibody specificity was determined by enzyme-linked immunosorbent assay (RELISA ANA Screening System, Immuno Concepts NA, Sacramento, California, USA) using recombinant Ro60 and La proteins, as part of standard diagnostic procedure. Sera from patients with anti-La were further tested by counterimmunoelectrophoresis (CIEP)32 to confirm whether or not anti-La antibodies detected by enzyme-linked immunosorbent assay were able to be detected by this method. HLA typing of patients with pSS (serological HLA-B and molecular DRB1) was performed by the Transplantation Laboratory, Australian Red Cross Blood Service, SA Division. Molecular DRB1 typing of the controls was performed by Conexio Genomics (Applecross, WA, Australia). The study was approved by the Human Ethics Committee of The Queen Elizabeth and Royal Adelaide Hospitals and all patients gave informed, written consent.
RCAα block haplotyping
Haplotypes of the RCAα block were obtained using the GMT assays as previously described.25 Briefly, two separate polymerase chain reaction reactions using primer sets CR1MCP5&6 and CR1MCP11&12 were performed on each genomic DNA sample. Each primer set was designed to amplify a duplicated element, located within the CR1 region of both segments (segment A containing CR1 and MCP-Like and segment B containing CR1-Like and MCP). The resulting mix of polymerase chain reaction products were used to define the haplotypic variation within the RCAα block. The polymerase chain reaction products were separated on the basis of size on a Corbett Research GS-3000 automated gel analysis system (Corbett Research (Australia), Mortlake, NSW, Australia). Haplotype assignment and nomenclature are as previously described.25
Statistical analysis
Contingency table analysis of haplotype and genotype frequencies in patients with pSS versus controls was performed by multivariate logistic regression using both additive and dominant allele coding. Associations were further reported as odds ratios (OR) with 95% confidence intervals (CI) obtained by back transformation of the regression coefficients.
RESULTS
RCAα block haplotype diversity
We have previously demonstrated substantial polymorphism in GMT RCAα block haplotypes.25 More than 20 haplotypes have been defined, although the majority are rare. In the current study of 213 Caucasians (pSS and controls combined), there were three relatively common haplotypes (AH1, AH2 and AH3 as designated by McLure et al25 each with a frequency of >10%. These three haplotypes combined accounted for 56% of the total haplotypes in the sample. There were a further 14 haplotypes with a frequency between 1 and 3%. These frequencies were considered too low to be informative given the study sample sizes and were therefore combined for analysis purposes. RCAα block genotypes were in Hardy–Weinberg equilibrium in both patients with pSS and controls (p = 0.93, p = 0.21 respectively, exact test).
RCAα block haplotypes are associated with Ro/La autoantibody-positive primary Sjögren syndrome
CR1 haplotype frequencies were significantly different between patients with pSS and controls (global χ2 = 14.6, df = 3, p = 0.002, table 1). Both AH1 (OR 2.1, 95% CI 1.3, 3.3) and AH3 (OR 2.4, 95% CI 1.3, 4.4) were significantly increased in pSS relative to controls implying an association between both of these haplotypes and susceptibility to pSS. However, this association was primarily attributable to the Ro/La autoantibody-positive subgroup of patients with pSS (n = 96, p = 0.0003) and was not evident in the relative minority of patients with pSS who were autoantibody negative (n = 19, p = 0.68).
Table 1. RCAα haplotype frequencies in patients with pSS compared with controls.
| Haplotype | pSS(2N = 230) | Controls(2N = 196) | OR (95% CI) | p Value |
| AH1 | 80 (34.8%) | 46 (23.5%) | 2.1 (1.3,3.3)* | 0.002 |
| AH2 | 25 (10.9%) | 27 (13.8%) | 1.3 (0.6,2.1) | 0.75 |
| AH3 | 38 (16.5%) | 19 (9.7%) | 2.4 (1.3,4.4) | 0.006 |
| Other | 87 (37.8%) | 104 (53.1%) | 1 | |
| Global test: χ2 = 14.6, df = 3, p = 0.002 | ||||
There were three common RCAα block ancestral haplotypes, AH1, AH2 and AH3. “Other” haplotypes, with a frequency between 1 and 3%, were combined for analysis purposes. Both AH1 and AH3 frequencies were increased in patients with pSS relative to controls. This association was attributable to the Ro/La autoantibody-positive subgroup of patients with pSS and was not evident in the minority of patients with pSS who were autoantibody negative (2N = 38).
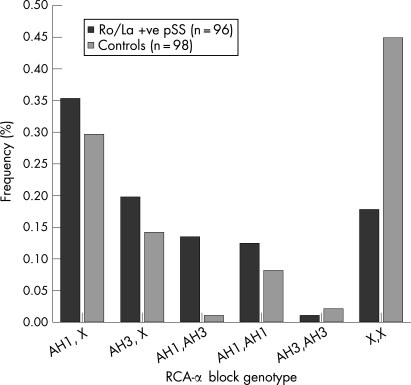
With the exception of the relatively rare AH3 homozygotes, the frequencies of all genotypes carrying either AH1 or AH3 were increased in Ro/La autoantibody-positive patients with pSS compared with controls (fig 1). This was accompanied by a striking concomitant decrease in the frequency of all other (X,X) genotypes. There was significant departure from an additive or allele dose model (p = 0.043) by logistic regression analysis, and the data were most consistent with both AH1 and AH3 exerting dominant genotypic effects. Under the dominant model, AH1 and AH3 appeared to be independently associated with autoantibody-positive pSS as there was no evidence of any interaction (p = 0.258). The odds ratios for the dominant genotypic association with autoantibody-positive pSS were 3.8 (95% CI 2.0, 7.3, p = 0.0001) for AH1 and 4.1 (95% CI 1.9, 8.7, p = 0.0003) for AH3. The association for the compound heterozygous AH1,AH3 genotype was predicted by multiplication of these odds ratios.
Figure 1. RCAα ancestral haplotype (AH) 1 and AH3 genotype frequencies in patients with primary Sjögren syndrome who were Ro/La autoantibody positive compared with controls. There was a significant departure from an additive or allele dose model (p = 0.043) by logistic regression analysis, and the data are most consistent with AH1 and AH3 exerting dominant effects of similar size. Under the dominant model, AH1 and AH3 appear to be independently associated with autoantibody-positive primary Sjögren syndrome, ie, with multiplicative risks.
Epistatic interaction between the RCAα block and the major histocompatibility complex in susceptibility to Ro/La autoantibody-positive pSS
An association between both HLA-DRB1*0301 (DR3) and HLA-DRB1*1501 (DR15) and autoantibody-positive pSS is well established in Caucasians.4 5 Therefore it was of interest to examine relationships between MHC and RCAα block associations with autoantibody-positive pSS.
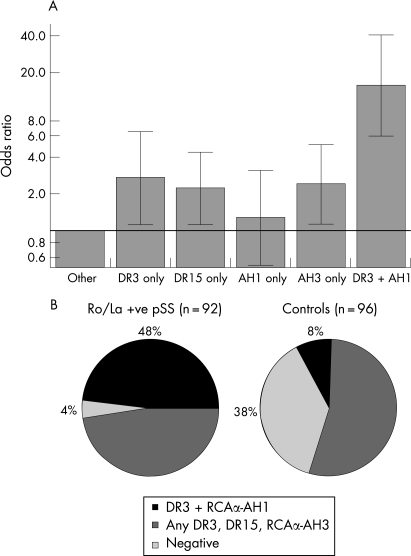
Analysis of the cross-classification of HLA-DR3, DR15 and RCAα AH1 and AH3 genotypic combinations in autoantibody-positive patients with pSS (n = 92) versus controls (n = 96) was performed by multivariate logistic regression using dominant coding and all two-factor interaction terms were evaluated. The odds ratios from this analysis are illustrated in fig 2A. HLA DR3, DR15 alleles and the RCAα AH3 haplotype were independent risk factors for autoantibody-positive pSS, with similar effect sizes. Therefore, the association for compound heterozygous genotypes was predicted by multiplication of these odds ratios. However, there was a disproportionately increased frequency of patients with pSS who carried both HLA DR3 and RCAα AH1 (interaction term, p = 0.021) demonstrating an epistatic relationship.33 In the absence of RCAα AH1, HLA DR3 was still associated with Ro/La-positive pSS (OR 2.7, p = 0.025), although the association was greatly enhanced by the presence of AH1 in the same individual (OR 15.7, p = 10−8). Conversely, the RCAα AH1 association was not associated with genetic susceptibility to pSS in the absence of DR3 (OR = 1.3, p = 0.6). Moreover, this epistatic genotypic combination of HLA DR3 and RCAα AH1 was the largest genetic risk factor for autoantibody-positive pSS. It was highly specific for autoantibody-positive pSS as it was present in 44 of 92 (48%) patients with pSS who were Ro/La autoantibody positive compared with only eight of 96 (8%) controls (fig 2B). The majority of patients with pSS who did not carry this epistatic genetic risk factor carried any combination of DR3/DR15/AH3, the other risk genotypes identified in the analysis. Therefore, only four of 92 (4%) autoantibody-positive patients with pSS did not carry any risk genotypes compared with 36 of 96 (38%) of controls.
Figure 2. Epistatic interaction between the MHC and RCAα block in Ro/La autoantibody-positive patients with primary Sjögren syndrome (pSS). (A) Odds ratios (y-axis, logarithmic scale) derived by logistic regression for the cross-classification of HLA DR3, DR15 and RCAα AH1, AH3 genotypic combinations (dominant coding) in Ro/La autoantibody-positive patients with pSS (n = 92) relative to controls (n = 98). The vertical bars represent 95% confidence intervals, and the horizontal line represents an odds ratio of 1 (no effect). HLA DR3, DR15 alleles and the RCAα AH3 haplotype were independent risk factors for autoantibody-positive pSS (ie, multiplicative risks), but there was an epistatic interaction between HLA DR3 and RCAα AH1 (interaction term p = 0.021). The genotypic combination of HLA DR3 and RCAα AH1 was the greatest genetic risk factor for autoantibody-positive pSS (OR 15.7, p = 10−8), but in the absence of DR3, there was no effect of RCAα AH1. (B) Pie chart depicting the relative proportions of risk genotypes. The HLA DR3–RCAα AH1 epistatic combination was present in 48% of autoantibody-positive patients with pSS compared with 8% of controls. The majority of other patients with pSS carried any combination of HLA DR3, DR15 and RCAα AH3.
Genetic associations with Ro/La autoantibody subsets in primary Sjögren syndrome
Of 115 patients with pSS, 19 (16%) were negative and 97 (84%) positive for anti-Ro/La autoantibodies. Seropositive Ro+La patients by enzyme-linked immunosorbent assay were further subdivided into non-precipitating La, ie, Ro+La (ppt–), or precipitating, ie, Ro+La (ppt+), on the basis of a precipitin line formed by anti-La antibodies on CIEP. Therefore, in addition to a seronegative subset, seropositive patients with pSS were classified into one of three serological subsets: anti-Ro alone (19 of 115 = 16%), anti-Ro+La(ppt−) (22 of 115 = 19%) and anti-Ro+La(ppt+) (55 of 115 = 46%). These subgroups are characterised by increasing titre and a more polyclonal autoantibody response in addition to higher rheumatoid factor and IgG levels.4 32
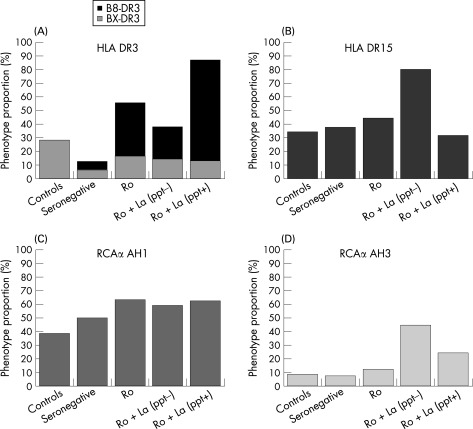
We, and others4 5 have previously demonstrated that HLA-DR3 and DR15 frequencies differ between autoantibody subsets, and this question is also of interest in relation to the RCAα haplotypes. The phenotypic prevalences of DR3, DR15 and RCAα AH1 and AH3 in controls, seronegative pSS and the three pSS autoantibody subsets are displayed in fig 3. While the data are difficult to interpret definitively because of the relatively small sample sizes in three of the serological subsets, some inferences may be drawn.
Figure 3. Phenotypic prevalence of HLA DR3 (A), HLA-DR15 (B), RCAα AH1 (C) and RCAα AH3 (D) haplotypes by diversification of the Ro/La autoantibody response within primary Sjögren syndrome (pSS). There were 115 patients with pSS in the study: 19 were seronegative, 19 with anti-Ro only, 22 with anti-Ro+La (ppt−) and 55 with anti-Ro+La (ppt+). The increased prevalence of HLA DR3 in autoantibody-positive pSS is largely B8-DR3 (the prevalence of B8-DR3 in the controls is not known) and there are genetic differences between the pSS autoantibody subgroups.
The increased prevalence of HLA DR3 in autoantibody-positive pSS is largely B8-DR3, which is carried on the 8.1 ancestral haplotype. B8-DR3 is most strongly associated with the polyclonal Ro+La (ppt+) autoantibody response, whereas, as previously reported,4 DR15 is almost exclusively associated with the restricted Ro+La (ppt−) response. The RCAα AH1 prevalence, although elevated, is relatively constant in autoantibody-positive patients. The basis for the epistatic interaction between RCAα AH1 and DR3 is most likely restricted to the 8.1 ancestral haplotype, rather than other DR3 containing haplotypes, and may exert a primary influence on the autoantibody response in pSS. Interestingly, 8.1 ancestral haplotype only one, rather than two or more C4 genes and is therefore associated with a relative C4 deficiency.34 However, there are clearly other genes on the MHC 8.1 haplotype that contribute additionally to regulation of the autoantibody response. The RCAα AH3 haplotype has the highest prevalence in patients with anti-La and may therefore potentiate diversification of the autoantibody response.
The genes for C2 are also in the extended MHC region and type 1 C2 deficiency is encoded within the 18.1 haplotype, which carries B18-DR15. There was no evidence of any epistatic interaction between RCAα block haplotypes and DR15. Further, only four B18-DR15 (from a total of 52 DR15) haplotypes were observed in patients with pSS and, as expected, there were no associations.
DISCUSSION
The GMT haplotyping approach has been used in this study to identify AH of the RCAα block and their associations with pSS, a systemic autoimmune disease. Duplication and copy number variation reveal more genetic diversity than single nucleotide polymorphisms,19 35 yet for most haplotyping assays, interpreting the results and defining haplotypes in these regions is difficult. The GMT assay is useful in interrogating complex genomic regions, such as the MHC and RCA blocks, as it requires and utilises these features to efficiently define the genomic polymorphism and AH within the region. The GMT RCAα block haplotyping has revealed extensive haplotypic polymorphism in this region (which also includes CR1-L, MCP and MCP-L genes), with more than 20 AH defined,25 although the majority are rare.
In this study we report that relatively frequent RCAα block haplotypes, AH1 and AH3, are associated with pSS, an autoimmune disease with a high prevalence of antinuclear Ro/La autoantibodies, and which shares both clinical and genetic susceptibility overlap with SLE.
The RCAα block contains CR1, CR1-L, MCP and MCP-L genes, and possibly also CR2, although the boundaries of the block have not been precisely defined. Erythrocyte CR1 in humans provides an important mechanism for the non-inflammatory clearance of immune complexes opsonised with C3a and C4b, and genetic variation in CR1 expression and/or function may influence both the predisposition to autoantibody-mediated disease (the clearance hypotheses)36 and self-perpetuating autoimmune reactivity resulting from prolonged, immune complex-mediated, type I interferon production.8 Although an acquired loss of erythrocyte CR1 expression has been observed in diseases such as SLE and pSS, the phenotypic high/low CR1 expression polymorphism is not associated with SLE,11 although the genetic basis for this is not well defined. CR1 expression is downregulated by immune complexes and upregulated by interferon γ in SLE,37 and in the mouse, CR1/CR2 expression (encoded by the same gene) is upregulated by BAFF.38 Therefore, genetic differences in the compound regulation of CR1 expression are likely to be strongly influenced by the complex inflammatory disease milieu.
In addition to clearance of immune complexes, CR1 and CR2 play a direct, instructive role in setting the threshold for B cell responses to antigen39 and potentially, the loss of B cell tolerance,40 a fundamental step in the pathogenesis of autoimmune disease. Further, cross-linking of MCP (CD46) with the TCR on naïve CD4+ T cells induces regulatory T cells,41 which are critical in the balance between autoimmunity and tolerance.42
Finally, complement control proteins may function as receptors for ligands other than complement components. The CR2 receptor is of particular interest in this regard, and a recent study has demonstrated an association between a common CR2 single nucleotide polymorphism based haplotype and SLE.43. CR2 is not only a functional receptor for interferon α,44 but is also the Epstein–Barr virus (EBV) receptor. There has long been a strongly suspected link between EBV infection and systemic autoimmunity. Intriguingly, molecular mimicry between the EBNA-1 epitope of the EBV virus and the Ro autoantigen has been demonstrated in initiation of the anti-Ro response in patients who ultimately developed SLE.45
Similar to HLA haplotypes, RCAα block haplotypes exert an influence on Ro/La autoantibody responses in patients with pSS. Importantly, there was an epistatic interaction between RCAα AH1 and HLA B8-DR3 in patients with pSS with Ro/La autoantibodies, which is indicative of a biological relationship between gene products of these haplotypes and disease pathways. The most likely basis for this epistasis is an interaction between the CR1 receptor and C4, one of its ligands. The genes for C4 are in the extended MHC region. HLA B8-DR3 and a relative C4 insufficiency (C4A*Q0,C4B*1)34 are both part of the 8.1 ancestral haplotype. The genetic structure of the C4 region is itself complex and highly polymorphic with both allelic and copy number variation of C4A and C4B genes.46 Genetic variation in C4 is known to contribute to disease susceptibility associated with MHC haplotypes47–49, but other MHC genes are also likely to be involved.34 48 In addition to systemic diseases such as pSS and SLE, the MHC 8.1 haplotype is associated with a number of organ-specific autoimmune diseases such as type 1 diabetes mellitus, Hashimoto’s thyroiditis, Graves’ disease, myasthenia gravis and multiple sclerosis. Evaluation of RCAα/MHC interactions in these diseases, and their association with susceptibility, autoantibody production and clinical phenotype would clearly be of great interest.
In addition to demonstrating the utility of the GMT approach for interrogating polymorphism in complex genomic regions, this study has also demonstrated that normal population variation in the RCAα block contributes substantially to susceptibility to systemic autoimmune disease. This is therefore direct evidence of the importance of regulation of complement activation in disease pathogenesis. The differing functional effects of the RCAα block haplotypes have not yet been defined and will be the focus of future research. These are likely to be pleiotropic and may well involve further epistatic genetic interactions. Although the task of unravelling these effects will be highly complex, we predict they will provide important insights into underlying autoimmune disease mechanisms.
Acknowledgments
The authors thank Professor Tom Gordon and Ms Dimi Beroukas from the Department of Allergy, Immunology and Arthritis, Flinders Medical Centre, for CIEP autoantibody screening of patient sera. We also gratefully acknowledge the support of the Arthritis Foundation of Australia and the CYO Immunogenetics Research Foundation. Genetic Technologies Ltd provided infrastructural support.
Footnotes
Competing interests: The CY O’Connor Village has a financial interest in Genetic Technologies Ltd, which has filed a provisional patent application, including RCAα block typing (PCT/AU2006001232, “Identification of ancestral haplotypes and uses thereof”).
REFERENCES
- 1.Manoussakis MN, Georgopoulou C, Zintzaras E, Spyropoulou M, Stavropoulou A, Skopouli FN, et al. Sjögren’s syndrome associated with systemic lupus erythematosus: clinical and laboratory profiles and comparison with primary Sjögren’s syndrome. Arthritis Rheum 2004;50:882–91 [DOI] [PubMed] [Google Scholar]
- 2.Rigby RJ, Dawkins RL, Wetherall JD, Hawkins BR. HLA in systemic lupus erythematosus: influence on severity. Tissue Antigens 1978;12:25–31 [PubMed] [Google Scholar]
- 3.Reinertsen JL, Klippel JH, Johnson AH, Steinberg AD, Decker JL, Mann DL. B-lymphocyte alloantigens associated with systemic lupus erythematosus. N Engl J Med 1978;299:515–18 [DOI] [PubMed] [Google Scholar]
- 4.Rischmueller M, Lester S, Chen Z, Champion G, Van Den Berg R, Beer R, et al. HLA class II phenotype controls diversification of the autoantibody response in primary Sjögren’s syndrome (pSS). Clin Exp Immunol 1998;111:365–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottenberg JE, Busson M, Loiseau P, Cohen-Solal J, Lepage V, Charron D, et al. In primary Sjögren’s syndrome, HLA class II is associated exclusively with autoantibody production and spreading of the autoimmune response. Arthritis Rheum 2003;48:2240–5 [DOI] [PubMed] [Google Scholar]
- 6.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis 2002;61:554–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon TP, Bolstad AI, Rischmueller M, Jonsson R, Waterman SA. Autoantibodies in primary Sjögren’s syndrome: new insights into mechanisms of autoantibody diversification and disease pathogenesis. Autoimmunity 2001;34:123–32 [DOI] [PubMed] [Google Scholar]
- 8.Nordmark G, Alm GV, Ronnblom L. Mechanisms of disease: primary Sjögren’s syndrome and the type I interferon system. Nat Clin Pract Rheumatol 2006;2:262–9 [DOI] [PubMed] [Google Scholar]
- 9.Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. Annu Rev Immunol 2004;22:431–56 [DOI] [PubMed] [Google Scholar]
- 10.Uko G, Dawkins RL, Kay P, Christiansen FT, Hollingsworth PN. CR1 deficiency in SLE: acquired or genetic? Clin Exp Immunol 1985;62:329–36 [PMC free article] [PubMed] [Google Scholar]
- 11.Nath SK, Harley JB, Lee YH. Polymorphisms of complement receptor 1 and interleukin-10 genes and systemic lupus erythematosus: a meta-analysis. Hum Genet 2005;118:225–34 [DOI] [PubMed] [Google Scholar]
- 12.Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, et al. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity 2001;15:775–85 [DOI] [PubMed] [Google Scholar]
- 13.Boackle SA, Holers VM. Role of complement in the development of autoimmunity. Curr Dir Autoimmun 2003;6:154–68 [DOI] [PubMed] [Google Scholar]
- 14.Boackle SA, Culhane KK, Brown JM, Haas M, Bao L, Quigg RJ, et al. CR1/CR2 deficiency alters IgG3 autoantibody production and IgA glomerular deposition in the MRL/lpr model of SLE. Autoimmunity 2004;37:111–23 [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Jiang N, Deppong C, Singh J, Dolecki G, Mao D, et al. A role for the Cr2 gene in modifying autoantibody production in systemic lupus erythematosus. J Immunol 2002;169:1587–92 [DOI] [PubMed] [Google Scholar]
- 16.McLure CA, Williamson JF, Stewart BJ, Keating PJ, Dawkins RL. Indels and imperfect duplication have driven the evolution of human Complement Receptor 1 (CR1) and CR1-like from their precursor CR1 alpha: importance of functional sets. Hum Immunol 2005;66:258–73 [DOI] [PubMed] [Google Scholar]
- 17.Zhang WJ, Degli-Esposti MA, Cobain TJ, Cameron PU, Christiansen FT, Dawkins RL. Differences in gene copy number carried by different MHC ancestral haplotypes. Quantitation after physical separation of haplotypes by pulsed field gel electrophoresis. J Exp Med 1990;171:2101–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokunaga K, Zhang WJ, Christiansen FT, Dawkins RL. The genomic structure of two ancestral haplotypes carrying C4A duplications. Immunogenetics 1991;34:247–51 [DOI] [PubMed] [Google Scholar]
- 19.Alper CA, Larsen CE, Dubey DP, Awdeh ZL, Fici DA, Yunis EJ. The haplotype structure of the human major histocompatibility complex. Hum Immunol 2006;67:73–84 [DOI] [PubMed] [Google Scholar]
- 20.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature 2001;411:199–204 [DOI] [PubMed] [Google Scholar]
- 21.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet 2001;29:229–32 [DOI] [PubMed] [Google Scholar]
- 22.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–9 [DOI] [PubMed] [Google Scholar]
- 23.Gaudieri S, Longman-Jacobsen N, Tay GK, Dawkins RL. Sequence analysis of the MHC class I region reveals the basis of the genomic matching technique. Hum Immunol 2001;62:279–85 [DOI] [PubMed] [Google Scholar]
- 24.Dawkins R, Leelayuwat C, Gaudieri S, Tay G, Hui J, Cattley S, et al. Genomics of the major histocompatibility complex: haplotypes, duplication, retroviruses and disease. Immunol Rev 1999;167:275–304 [DOI] [PubMed] [Google Scholar]
- 25.McLure CA, Williamson JF, Smyth LA, Agrawal S, Lester S, Millman JA, et al. Extensive genomic and functional polymorphism of the complement control proteins. Immunogenetics 2005;57:805–15 [DOI] [PubMed] [Google Scholar]
- 26.Tay GK, Witt CS, Christiansen FT, Charron D, Baker D, Herrmann R, et al. Matching for MHC haplotypes results in improved survival following unrelated bone marrow transplantation. Bone Marrow Transplant 1995;15:381–5 [PubMed] [Google Scholar]
- 27.Witt C, Sayer D, Trimboli F, Saw M, Herrmann R, Cannell P, et al. Unrelated donors selected prospectively by block-matching have superior bone marrow transplant outcome. Hum Immunol 2000;61:85–91 [DOI] [PubMed] [Google Scholar]
- 28.Grosse-Wilde H, Ketheesen N, Christiansen FT, Ottinger HD, Ferencik S, Tay G, et al. The genome matching technique (GMT): a new tool for selecting unrelated marrow donors. Charron D, ed. 12th International Histocompatibility Workshop and Conference. Paris: EDK, 1997:589–91 [Google Scholar]
- 29.Kitcharoen K, Witt CS, Romphruk AV, Christiansen FT, Leelayuwat C. MICA, MICB, and MHC beta block matching in bone marrow transplantation: relevance to transplantation outcome. Hum Immunol 2006;67:238–46 [DOI] [PubMed] [Google Scholar]
- 30.Laird R, Dawkins RL, Gaudieri S. Use of the genomic matching technique to complement multiplex STR profiling reduces DNA profiling costs in high volume crimes and intelligence led screens. Forensic Sci Int 2005;151:249–57 [DOI] [PubMed] [Google Scholar]
- 31.McLure CA, Kesners PW, Lester S, Male D, Amadou C, Dawkins JR, et al. Haplotyping of the canine MHC without the need for DLA typing. Int J Immunogenet 2005;32:407–11 [DOI] [PubMed] [Google Scholar]
- 32.Beer RG, Rischmueller M, Coates T, Purcell AW, Keech CL, McCluskey J, et al. Nonprecipitating anti-La(SS-B) autoantibodies in primary Sjögren’s syndrome. Clin Immunol Immunopathol 1996;79:314–18 [DOI] [PubMed] [Google Scholar]
- 33.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet 2002;11:2463–8 [DOI] [PubMed] [Google Scholar]
- 34.Candore G, Lio D, Colonna Romano G, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev 2002;1:29–35 [DOI] [PubMed] [Google Scholar]
- 35.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carroll MC. A protective role for innate immunity in systemic lupus erythematosus. Nat Rev Immunol 2004;4:825–31 [DOI] [PubMed] [Google Scholar]
- 37.Arora V, Mondal AM, Grover R, Kumar A, Chattopadhyay P, Das N. Modulation of CR1 transcript in systemic lupus erythematosus (SLE) by IFN-gamma and immune complex. Mol Immunol 2007;44:1722–8 [DOI] [PubMed] [Google Scholar]
- 38.Gorelik L, Cutler AH, Thill G, Miklasz SD, Shea DE, Ambrose C, et al. Cutting edge: BAFF regulates CD21/35 and CD23 expression independent of its B cell survival function. J Immunol 2004;172:762–6 [DOI] [PubMed] [Google Scholar]
- 39.Fearon DT, Locksley RM. The instructive role of innate immunity in the acquired immune response. Science 1996;272:50–3 [DOI] [PubMed] [Google Scholar]
- 40.Carroll MC. The complement system in B cell regulation. Mol Immunol 2004;41:141–6 [DOI] [PubMed] [Google Scholar]
- 41.Kemper C, Verbsky JW, Price JD, Atkinson JP. T-cell stimulation and regulation: with complements from CD46. Immunol Res 2005;32:31–44 [DOI] [PubMed] [Google Scholar]
- 42.Paust S, Cantor H. Regulatory T cells and autoimmune disease. Immunol Rev 2005;204:195–207 [DOI] [PubMed] [Google Scholar]
- 43.Wu H, Boackle SA, Hanvivadhanakul P, Ulgiati D, Grossman JM, Lee Y, et al. Association of a common complement receptor 2 haplotype with increased risk of systemic lupus erythematosus. Proc Natl Acad Sci USA 2007;104:3961–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asokan R, Hua J, Young KA, Gould HJ, Hannan JP, Kraus DM, et al. Characterization of human complement receptor type 2 (CR2/CD21) as a receptor for IFN-alpha: a potential role in systemic lupus erythematosus. J Immunol 2006;177:383–94 [DOI] [PubMed] [Google Scholar]
- 45.McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med 2005;11:85–9 [DOI] [PubMed] [Google Scholar]
- 46.Blanchong CA, Chung EK, Rupert KL, Yang Y, Yang Z, Zhou B, et al. Genetic, structural and functional diversities of human complement components C4A and C4B and their mouse homologues, Slp and C4. Int Immunopharmacol 2001;1:365–92 [DOI] [PubMed] [Google Scholar]
- 47.Christiansen FT, Dawkins RL, Uko G, McCluskey J, Kay PH, Zilko PJ. Complement allotyping in SLE: association with C4A null. Aust N Z J Med 1983;13:483–8 [DOI] [PubMed] [Google Scholar]
- 48.Christiansen FT, Zhang WJ, Griffiths M, Mallal SA, Dawkins RL. Major histocompatibility complex (MHC) complement deficiency, ancestral haplotypes and systemic lupus erythematosus (SLE): C4 deficiency explains some but not all of the influence of the MHC. J Rheumatol 1991;18:1350–8 [PubMed] [Google Scholar]
- 49.Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 2007;80:1037–54 [DOI] [PMC free article] [PubMed] [Google Scholar]