Abstract
The G2 DNA damage and slowing of S-phase checkpoints over mitosis function through tyrosine phosphorylation of NIMXcdc2 in Aspergillus nidulans. We demonstrate that breaking these checkpoints leads to a defective premature mitosis followed by dramatic rereplication of genomic DNA. Two additional checkpoint functions, uvsB and uvsD, also cause the rereplication phenotype after their mutation allows premature mitosis in the presence of low concentrations of hydroxyurea. uvsB is shown to encode a rad3/ATR homologue, whereas uvsD displays homology to rad26, which has only previously been identified in Schizosaccharomyces pombe. uvsBrad3 and uvsDrad26 have G2 checkpoint functions over mitosis and another function essential for surviving DNA damage. The rereplication phenotype is accompanied by lack of NIMEcyclinB, but ectopic expression of active nondegradable NIMEcyclinB does not arrest DNA rereplication. DNA rereplication can also be induced in cells that enter mitosis prematurely because of lack of tyrosine phosphorylation of NIMXcdc2 and impaired anaphase-promoting complex function. The data demonstrate that lack of checkpoint control over mitosis can secondarily cause defects in the checkpoint system that prevents DNA rereplication in the absence of mitosis. This defines a new mechanism by which endoreplication of DNA can be triggered and maintained in eukaryotic cells.
INTRODUCTION
Checkpoint pathways have been identified that respond to damaged or incompletely replicated DNA to prevent cell cycle progression and to subsequently allow DNA repair or the completion of replication (Elledge, 1996; Nurse, 1997; Weinert, 1997, 1998). The regulation of tyrosine 15 phosphorylation of the cdc2 kinase plays a crucial role in the control of mitotic entry. During interphase, cdc2 associates with its cyclin partner cyclin B (Evans et al., 1983; Booher et al., 1989), but the complex is kept inactive by phosphorylation at tyrosine 15 of cdc2 by Wee1/Mik1/Myt1 inhibitory tyrosine kinases (Gould and Nurse, 1989; Lundgren et al., 1991; Mueller et al., 1995). At the G2–M transition, cdc2/cyclin B is rapidily activated by tyrosine 15 dephosphorylation carried out by the cdc25 phosphatase (Russell and Nurse 1986; Gould and Nurse, 1989). The inability to phosphorylate the inhibitory tyrosine residue of cdc2 results in premature mitosis in many model systems (Gould and Nurse 1989; Broek et al., 1991; Krek and Nigg, 1991; Hayles et al., 1994; Blasina et al., 1997), including Aspergillus nidulans (Ye et al., 1997b), but not in Saccharomyces cerevisiae (Amon et al., 1992; Sorger and Murray, 1992). Lack of tyrosine phosphorylation of cdc2 abolishes the slowed S-phase and G2 DNA damage checkpoints in A. nidulans, which prevents entry into mitosis in the presence of incompletely replicated or damaged DNA, respectively (Ye et al., 1996, 1997b). This mechanism of delaying mitotic entry is conserved in Schizosaccharomyces pombe and mammalian systems (Jin et al., 1996; Rhind et al., 1997), as are many of the upstream regulators of these pathways.
In A. nidulans, BIME has also been demonstrated to play a role in the control of the S-phase checkpoint (Ye et al., 1996). bimE is an anaphase-promoting complex 1 (APC1) homologue and was originally identified as being required for exit from mitosis (Morris, 1976). Although the absence of cdc2 tyrosine phosphorylation is sufficient to allow mitosis in the presence of low concentrations of hydroxyurea (HU), these cells are not able to overcome a complete S-phase arrest in the presence of high concentrations of HU (Ye et al., 1996). However, lack of cdc2 tyrosine phosphorylation in combination with compromised BIME function is sufficient to overcome this S-phase arrest (Ye et al., 1996). This appears to be regulated through the mitosis-promoting NIMA kinase, because lack of cdc2 tyrosine phosphorylation allows the accumulation of NIMA protein during S-phase arrest, and inactivation of BIME leads to the activation of NIMA by phosphorylation (Ye et al., 1996).
In addition to checkpoint systems that ensure the completion of DNA replication or DNA repair before entry into mitosis, cells also have mechanisms that restrict DNA replication to occurring only once per cell cycle (Stillman, 1996; Wuarin and Nurse, 1996). This single round of replication is followed by mitosis, resulting in the segregation of the duplicated chromosomal DNA. Only once mitosis is completed can cells enter S-phase and replicate DNA again. Components of the cell cycle regulatory machinery are involved in maintaining this temporal order, which ensures the maintenance of genome ploidy (Hayles et al., 1994; Moreno and Nurse, 1994; Nasmyth, 1996; Nishitani and Nurse, 1997).
We were interested in identifying upstream regulators of tyrosine phosphorylation of the A. nidulans NIMXcdc2 kinase. Many DNA damage-sensitive mutants have been identified in A. nidulans (Jansen, 1970; Kafer and Mayor, 1986; Kafer and Chae, 1994; Kafer and May, 1997; Zhao and Kafer, 1992; Osman et al., 1993; Yoon et al., 1995; van Heemst et al., 1997; Han et al., 1998). Of these, uvsH encodes a DNA repair gene with homology to S. cerevisiae RAD18 (Yoon et al., 1995), uvsC is an S. cerevisiae RAD51 homologue (van Heemst et al., 1997), uvsI is an S. cerevisiae REV3 homologue (Han et al., 1998), and uvsF displays homology to DNA replication factor C (Kafer and May, 1997). In addition to having roles in the DNA damage repair response, some of these DNA damage-sensitive mutants are likely to have roles in checkpoint regulation. To identify these genes, we screened the known A. nidulans DNA damage-sensitive mutants for sensitivity to low concentrations of HU. Strains which are sensitive to both HU and DNA damage are likely candidates for having roles in checkpoint responses to G2-DNA damage and slowed S-phase. Here we describe the identification and complementation of two of these genes. uvsB encodes a rad3 homologue, whereas uvsD displays sequence and structural similarity to rad26 and is likely to be the first identified rad26 homologue. Similar to mutations that prevent tyrosine 15 phosphorylation of NIMXcdc2, mutations in uvsB or uvsD result in loss of checkpoint regulation in response to G2-DNA damage or prolonged S-phase. Moreover, loss of checkpoint regulation in these mutants in the presence of low concentrations of HU leads to a dramatic rereplication phenotype characterized by highly polyploid nuclei. Cells displaying the rereplication phenotype have low NIMXcdc2 kinase activity because of loss of NIMEcyclinB, but ectopic expression of nondegradable NIMEcyclinB does not prevent DNA rereplication even though NIMXcdc2 kinase activity is maintained at a high level. We propose that loss of checkpoint regulation over mitosis can secondarily cause defects in mechanisms that prevent DNA rereplication in the absence of mitosis.
MATERIALS AND METHODS
A. nidulans Strains and General Techniques
A. nidulans strains used in this study were R153 (pyroA4; wA3); GR5 (pyrG89; pyroA4; wA3); ΔankAwee1 (ΔankAwee1; pyrG89; pyr4+; pyroA4; wA3); FRY20-1 (nimXcdc2AF; pyroA4; pyrG89; wA3); AT27 (nimXcdc2AF; nimA5; pyroA1; riboA2; wA3); SO54 (nimA5, wA2); AT158 (uvsD308; nimA5; riboA1, fwA); AT136 (uvsB505; nimA5; nicA2); AT103 (uvsD308; pyrG89; riboA1, pyroA4 wA3); AT107 (uvsB505; pyrG89; pyroA4; chaA1); A329 (uvsH4; adE20; biA1; methG1; pyroA4; wA3); A826 (uvsB505; choA1; biA1; chaA1);A574 (uvsD308; riboA1; biA1; chaA1); AT33–1 (bimE7; nimXcdc2AF; pyroA4; pabaA1; pyrG89); and AT214 (nimXcdc2AF + alcA::nimEcyclin BΔD; pyroA4; pyrG89 wA3). AT214 was generated by transformation of FRY20-1 with the plasmid p122 containing a version of nimEcyclin B, which does not contain the destruction box and which is under control of the alcA promoter (alcA::nimEcyclin BΔD). Plasmid p122 was a kind gift from Dr. Matthew O’Connell (Peter MacCallum Cancer Institute, Melbourne, Victoria, Australia). The genotypes of strains listed in Figure 1 are available at http://www.kumc.edu/research/fgsc/nidlist.html. Media and general techniques for culture of A. nidulans, DAPI staining for chromosome mitotic index, protein extraction, immunoprecipitation, protein kinase assays, and Western blotting were as previously described (Osmani et al., 1987, 1991a,b, 1994; Oakley and Osmani, 1993; Ye et al., 1995). Measurements of relative DNA content were made as described by May et al. (1992) using an Eclipse E800 (Nikon, Tokyo, Japan) microscope equipped with a digital camera system. The data collected were analyzed with Phase 3 Imaging Systems software (Media Cybernetics, Silver Spring, MD), and the values presented represent the average fluorescence intensity for single nuclei (subtracting background) in 12 cells for each strain.
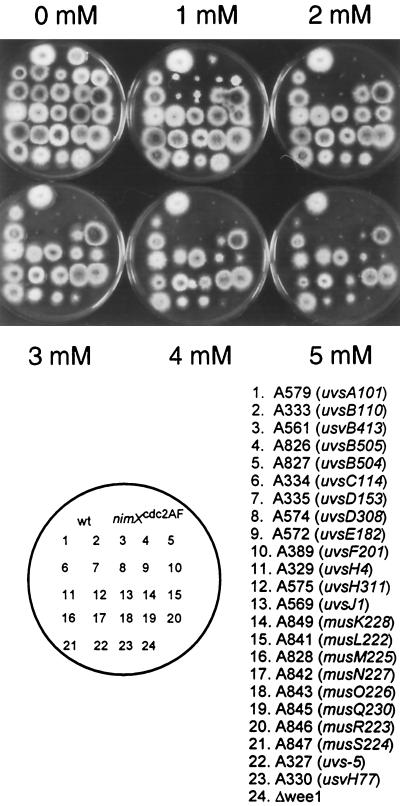
Figure 1.
Mutations in uvsB and uvsD cause marked HU sensitivity. The indicated strains were replica plated onto media containing varying concentrations of HU. After incubation plates were photographed.
Library Construction, Aspergillus nidulans Transformation, and Complementation of uvsB505 and uvsD308
Genomic DNA isolated from GR5 was partially digested with AluI to generate blunt-ended gDNA fragments of an average size of ∼7 kb. The gDNA (14 μg) was then ligated to an adaptor (28 μg) in a total volume of 40 μl at 16°C overnight in the presence of 5 mM spermidine. The sequence of the 5′ phosphorylated adaptor was as follows: 5′-ATCCGGCACGAG-3′ and 5′-CTCGTGCCG-3′. After ligation, the DNA was precipitated, and adaptor-ligated genomic DNA (gDNA) was separated from unligated linkers by agarose gel electrophoresis. Adaptor-ligated gDNA (4–20 kb) was excised from the gel and purified by freeze–thawing and ethanol precipitation (Qian and Wilkinson, 1991). Adaptor-ligated gDNA was then ligated into pRG3 (Waring et al., 1989), which had been digested with BamHI and partially filled by incubation with Taq DNA polymerase (Perkin-Elmer, Norwalk, CT) in the presence of 25 μM dGTP for 2 min at 72°C. Ligated DNA was transformed into Epicurian Coli XL10-Gold Ultracompetent cells (Stratagene, La Jolla, CA) or TOP10 Electocomp Escherichia coli cells (Invitrogen, San Diego, CA), and transformants were selected by resistance to ampicillin. Ampicillin-resistant colonies (7.7 × 105) were obtained, 80% of which contained inserts of an average insert size of 6.7 kb, giving >170 genomic equivalents assuming a genome size of 2.3 × 104 kb (Timberlake, 1978). Aliquots of the primary library were stored in Luria–Bertani medium containing 20% glycerol at −70°C. The remaining cells were grown overnight at 37°C, and plasmid library DNA was isolated using the alkaline lysis procedure followed by purification on a cesium chloride gradient using standard procedures (Maniatis et al., 1982).
Complementation of uvsB505 and uvsD308 alleles was carried out by library transformation of AT107 and AT103, respectively, using standard techniques (Oakley and Osmani, 1993) selecting for transformants that were complemented for sensitivity to HU and methyl methane sulfonate (MMS). Single-copy integration was confirmed by Southern blotting, and plasmids were recovered from complemented strains (Osmani et al., 1987). Recovered plasmids were retransformed into the original mutant strains, and complementing plasmids were sequenced. We confirmed homologous integration at the uvsB locus by two-step gene replacement (Osmani et al., 1987) and for uvsD by sequencing the mutant uvsD308 allele. To obtain the coding sequence for uvsB and uvsD, rapid amplification of cDNA ends (RACE)-PCR was performed using the Marathon cDNA amplification kit (Clontech, Cambridge, United Kingdom), and the 5′ and 3′ RACE-PCR products were sequenced.
Targeted Disruption of uvsB and uvsD
Targeted disruption of uvsB was performed using standard techniques (Osmani et al., 1994) by transforming GR5 with a plasmid containing a 6153-bp internal fragment of uvsB genomic DNA. Transformants were able to grow in the absence of uridine and uracil and contained the above plasmid integrated homologously at uvsB. This leads to a duplication of uvsB with one copy lacking its 3′ end and the other lacking its 5′ end. The 3′-deleted version lacks 1815 bp of 3′ coding sequence, including the kinase domain, and also its normal termination and processing sequences. The 5′-deleted version lacks a promoter and 800 bp of the 5′ coding sequence. A similar strategy was use to disrupt uvsD using a 1512-bp internal fragment. Homologous integration disrupts uvsD generating a 3′-deleted version lacking 465 bp of coding sequence and normal termination and processing sequences and a 5′-deleted version lacking 407 bp of 5′ coding sequence and its promoter. Disruptions were confirmed by Southern blotting.
Sensitivity Test to UV Irradiation and nimA5 Block Release
Nondividing and dividing cells were tested for sensitivity to UV irradiation as previously described using a microprocessor controlled UV cross-linker (FBUVXL-1000; Fischer Biotech, Pittsburgh, PA; 254 nm) (Ye et al., 1997b). Entry into mitosis after MMS (0.02%) treatment at the nimA5 arrest point was determined as previously described (Ye et al., 1997b).
RESULTS
uvsB and uvsD Mutants Are Sensitive to Both DNA Damage and Prolonged S-Phase
We have previously demonstrated that checkpoint regulation over entry into mitosis is regulated through tyrosine phosphorylation of the NIMXcdc2 kinase in response to G2 DNA damage or prolonged S-phase (Ye et al., 1996, 1997b). To identify upstream regulators of checkpoint regulation of NIMXcdc2 tyrosine phosphorylation, we screened known DNA damage-sensitive mutants (Jansen, 1970; Kafer and Mayor, 1986; Kafer and Chae, 1994; Kafer and May, 1997; Zhao and Kafer, 1992; Osman et al., 1993; Yoon et al., 1995; van Heemst et al., 1997; Han et al., 1998) for sensitivity to the DNA replication inhibitor HU (Figure 1). Of the 18 genes tested, all four alleles of uvsB and both alleles of uvsD were highly sensitive to low concentrations of HU, whereas other genes displayed no or only limited sensitivity (Figure 1). All alleles of uvsB and uvsD displayed similar sensitivity to HU, consistent with the previous finding that they belong to the same complementation group (Kafer and Mayor, 1986). Interestingly, uvsB and uvsD mutant strains displayed similar sensitivity to HU as a strain (nimXcdc2AF), which contains a single copy of NIMXcdc2 that is nonphosphorylatable on the inhibitory tyrosine and threonine residues (Figure 1). In addition, a strain in which the major NIMXcdc2 tyrosine kinase ANKAwee1 has been deleted (ΔankAwee1) was less sensitive to HU than the uvsB, uvsD and nimXcdc2AF mutant strains (Figure 1). These results are consistent with uvsB and uvsD playing roles in checkpoint regulation in response to both DNA damage and prolonged S-phase and suggest that they may function in the pathway leading to tyrosine phosphorylation of NIMXcdc2.
uvsB and uvsD Mutants Are Defective in the G2 DNA Damage and Prolonged S Phase Checkpoints over Mitosis
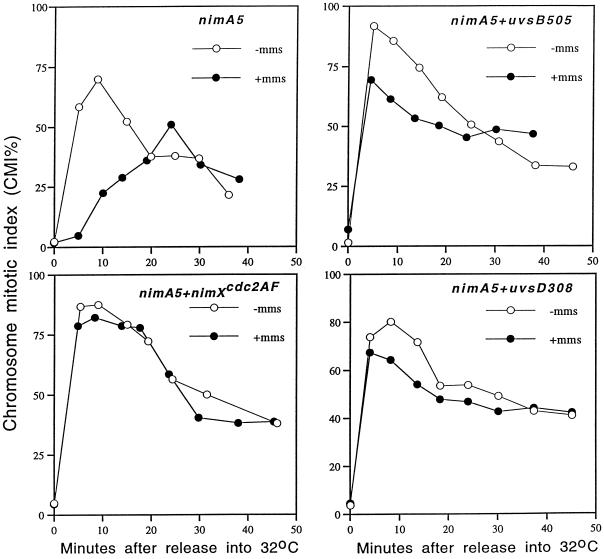
To confirm that the sensitivity of uvsB and uvsD mutants to DNA damage was due to loss of checkpoint control over entry into mitosis, we used strains that also carried the nimA5 temperature-sensitive mutation. Cells arrested in G2 at the nimA5 arrest point were either treated or not treated with 0.02% MMS to elicit DNA damage. Cells were then released in the absence of MMS to the permissive temperature and entry into mitosis followed by determining the chromosome mitotic index at time points after release. Under these conditions, entry into mitosis is delayed 15 min in the presence of DNA damage in the nimA5 mutant (Figure 2). In contrast, uvsB505 + nimA5 and uvsD308 + nimA5 double mutants failed to arrest in response to G2 DNA damage and entered mitosis with similar kinetics as when MMS was not included and like the nimXcdc2AF + nimA5 double mutant in the presence of MMS (Figure 2). These data suggest that the sensitivity to DNA damage of uvsB and uvsD mutants is due to premature entry into mitosis with damaged DNA.
Figure 2.
uvsB and uvsD mutants fail to arrest in response to DNA damage in G2. The four indicated nimA5 strains were arrested in G2 at 42°C before downshift to 32°C to allow synchronous release into mitosis. Cells were released with or without MMS-induced DNA damage as indicated, and the chromosome mitotic index was determined in >200 cells per time point after staining DNA of fixed cells with DAPI.
We next investigated whether the sensitivity of uvsB and uvsD mutants to low concentrations of HU was due to premature entry into mitosis using a wild-type and a nimXcdc2AF mutant strain as controls. Conidiospores were germinated in the presence or absence of 6 mM HU, and the chromosome mitotic index was determined at time points after germination. Consistent with previous studies, entry into mitosis was markedly delayed in the wild-type strain in the presence of HU, but the nimXcdc2AF mutant strain, which cannot be negatively regulated by tyrosine phosphorylation, entered mitosis 1 h earlier than the wild-type strain in the presence of HU (Ye et al., 1996; our unpublished results). Similarly, uvsB and uvsD mutants germinated in the presence of 6 mM HU entered mitosis 30 min earlier than the wild-type strain germinated under the same conditions. Thus, like mutations that impair tyrosine phosphorylation of NIMXcdc2, mutations in uvsB and uvsD lead to sensitivity to low concentrations of HU, at least in part because of loss of the S-phase checkpoint over entry into mitosis. Supporting this, the lethality of uvsB and uvsD mutants germinated in the presence of 6 mM HU could be rescued if premature entry into mitosis was prevented by arresting cells in G2 at the nimA5 arrest point followed by release to the permissive temperature in the absence of HU (our unpublished results).
uvsB and uvsD Also Have a Function That Is Independent of Tyrosine Phosphorylation of NIMXcdc2
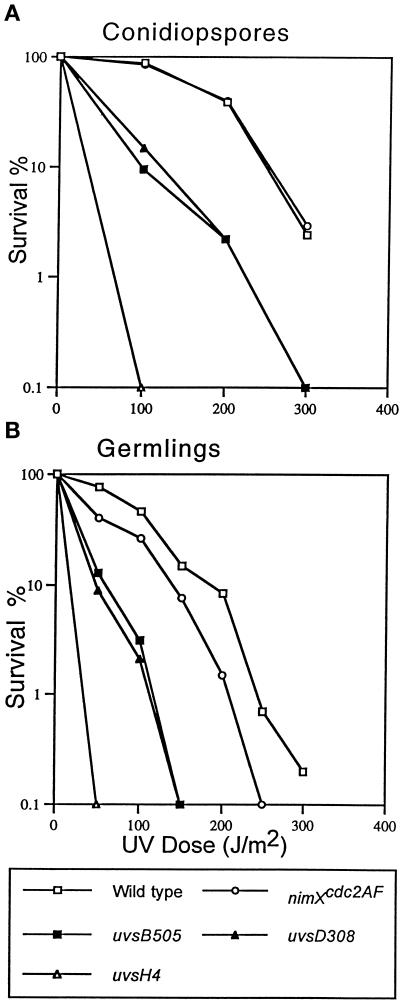
Although the nimXcdc2AF mutant is sensitive to DNA damage elicited during the cell cycle, remarkably this strain is no more sensitive than a wild-type strain when quiescent conidiospores are subjected to UV irradiation (Ye et al., 1997b). In contrast, previous studies have demonstrated that uvsB and uvsD mutant conidiospores are highly sensitive to DNA damage (Kafer and Mayor, 1986). To directly compare the sensitivity of these mutants to DNA damage, we determined their viability after UV irradiation, using the DNA damage repair-deficient uvsH4rad18 mutant (Kafer and Mayor, 1986; Yoon et al., 1995) and wild-type strains as controls. uvsB and uvsD mutants were more sensitive to UV irradiation than the nimXcdc2AF mutant when either conidiospores or germlings were irradiated (Figure 3, A and B). As shown previously (Ye et al., 1997a), nimXcdc2AF mutant conidiospores displayed sensitivity to UV irradiation similar to that of the wild-type strain, but uvsB505 and uvsD308 mutant conidiospores displayed significant sensitivity to UV irradiation (Figure 3A). These data indicate that uvsB and uvsD have functions in response to DNA damage that are independent of tyrosine phosphorylation of NIMXcdc2. In addition, uvsB505 and uvsD308 mutants are not as sensitive as the repair deficient uvsH4rad18 strain (Figure 3A), suggesting that these mutants are not completely DNA damage repair deficient.
Figure 3.
Differential UV sensitivity of strains deficient in the G2 DNA damage checkpoint. Conidiospores (250 per plate, two plates per strain) of wild-type and nimXcdc2AF, uvsB505, uvsD308, and uvsH4 mutant strains were spread on YAG plates and either immediately UV irradiated as indicated (A) or incubated at 32°C for 4.5 h to allow spore germination and then UV irradiated (B). After irradiation the plates were incubated at 32°C for 2 d to allow colony formation. The percent survival after DNA damage by UV irradiation is expressed as the percentage of colonies produced in the absence of treatment.
uvsB Is a rad3 Homologue, and uvsD Displays Homology to rad26
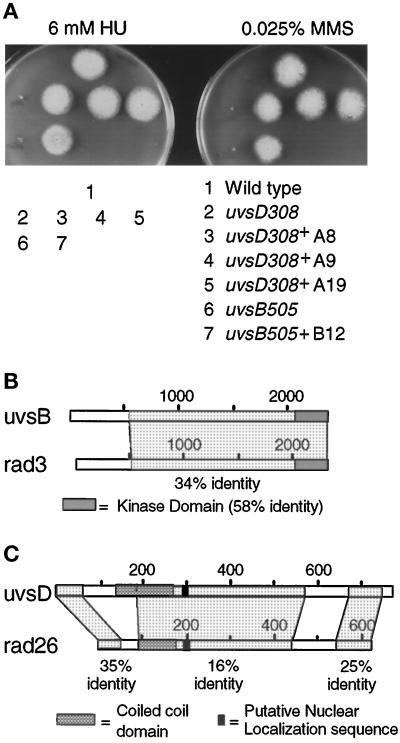
We cloned uvsB and uvsD by complementation of the HU sensitivity of the uvsB505 and uvsD308 alleles using an A. nidulans plasmid-based genomic DNA library. Positive transformants were also fully complemented for sensitivity to MMS (Figure 4A). Single-copy integration was confirmed by Southern blotting, and plasmids were recovered from complemented strains (Osmani et al., 1987). Recovered plasmids were retransformed into the original mutant strains, and complementing plasmids were sequenced. We confirmed homologous integration at the uvsB locus by two-step gene replacement (Osmani et al., 1987). A database search revealed extensive homology of uvsB to the S. pombe checkpoint rad gene rad3; however, the genomic sequence failed to identify any large open reading frames. To obtain the coding sequence for uvsB, RACE-PCR was performed, and sequencing of the 5′ and 3′ RACE-PCR products identified a single large open reading frame of 7365 bp coding for a 2454-amino-acid protein assuming that the first in-frame methione is used for translational initiation. Comparison of the gDNA and the cDNA identified a coding region of 8768 bp containing 25 introns, which are present throughout the coding region (GenBank accession number AF178850). This is a remarkably high number of introns given that the S. pombe rad3 gDNA sequence consists of a single open reading frame (Bentley et al., 1996). Sequence comparison confirmed that uvsB is a rad3 homologue with 34% overall identity (Figure 4B) and indicated that it also displays high homology to other members of this family, including the human ATM (22%) and ATR (28%) genes. UVSB is most homologous to rad3 in its kinase domain, where it displays a higher level of homology to rad3 (58% identity) than does the next closest rad3 homologue, ATR (53% identity; Figure 4B) (Bentley et al., 1996).
Figure 4.
Identification of uvsB as a rad3 homologue and uvsD as a rad26-like gene. (A) Growth of the wild-type and the uvsB505 and uvsD308 mutant strains and the complemented transformants uvsD308+ A8, A9, A19, and uvsB505+ B12, as indicated 3 d after inoculation at 32°C. (B) Schematic diagram showing the kinase domain of UVSB and the region of homology to rad3. (C) Schematic diagram of UVSD showing regions of homology with rad26, including a coiled coil domain followed by a putative nuclear localization sequence as indicated.
Sequencing of uvsD gDNA and RACE-PCR products identified an open reading frame of 2377 bp containing a single intron and coding for a 778-amino-acid protein (GenBank accession number AF180367). We confirmed integration at the uvsD locus by sequencing the mutant allele in the complemented strain, as we were unable to perform a two-step gene replacement because the complemented strain failed to undergo a self-cross. This identified a single point mutation substituting a stop codon instead of the glutamine at codon number 237 (CAG→TAG). Database searches identified UVSD as having highest homology to rad26 (Figure 4C). UVSD and rad26 both contain a coiled coil domain followed by a putative nuclear localization sequence (Figure 4C). Given the similar phenotypes of uvsD and rad26 mutants and the sequence and structural homologies of these genes, uvsD is likely to be the first identified rad26 homologue.
rad3 and rad26 are nonessential genes in S. pombe (Jimenez et al., 1992; Al-Khodairy et al., 1994); however, the S. cerevisiae rad3 homologue MEC1 plays an essential role in budding yeast (Kato and Ogawa, 1994). To determine whether uvsB and uvsD have essential functions in A. nidulans, we performed targeted gene disruption of the respective genes (see MATERIALS AND METHODS; Osmani et al., 1994). Disruptions were confirmed by Southern blotting, and strains were analyzed for sensitivity to HU and MMS (our unpublished results). The resulting ΔuvsB and ΔuvsD strains were viable and displayed sensitivity to HU and MMS similar to that of the respective mutant alleles (our unpublished results). Thus, similar to S. pombe rad3 and rad26, uvsB and uvsD are apparently nonessential genes that are involved in checkpoint regulation over G2 DNA damage and prolonged S-phase.
Deregulation of cdc2 Kinase Activity during Prolonged S Phase Leads to an Abnormal Mitosis and Subsequent Over-Replication of DNA
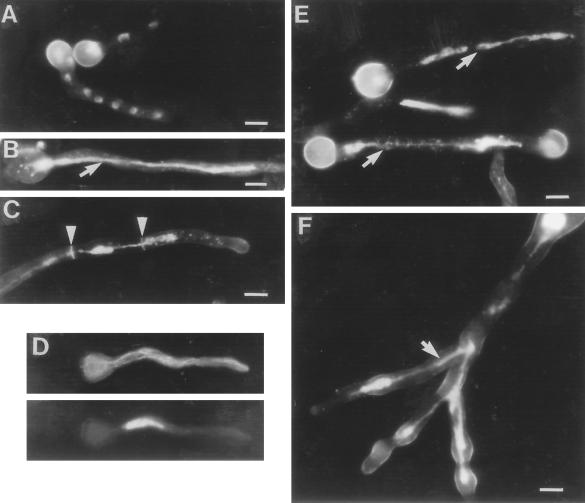
Phenotypically, uvsBrad3 and uvsDrad26 mutants are similar to strains that are unable to tyrosine phosphorylate NIMXcdc2. To investigate the phenotype of premature entry into mitosis, we germinated the ΔankAwee1 strain in the presence of 6 mM HU and examined germlings by DAPI staining to visualize DNA (Figure 5, B–D). Under these conditions, wild-type strains delay entry into mitosis (Ye et al., 1996), but nuclear division and migration occur normally (Figure 5A). In contrast, the ΔankAwee1 germlings displayed striking, abnormal DNA morphologies consisting of polyploid nuclei, which became highly stretched as hyphal growth continued (Figure 5, B and C). Failure to segregate DNA is expected for cells prematurely entering mitosis from S-phase; however, the over-replication of DNA was completely unexpected. To confirm that cells displaying over-replicated DNA were in interphase and not undergoing mitosis, we examined the microtubule network by immunofluorescent staining at 1-h intervals after germination of the ΔankAwee1 strain in 6 mM HU. Cells with polyploid nuclei always displayed interphase patterns of microtubule staining (Figure 5D), and mitotic spindles were never observed after the initial premature mitosis, even though these cells continued to replicate their DNA, resulting in the formation of polyploid nuclei. The nimXcdc2AF mutant strain is more sensitive to low concentrations of HU than the ΔankAwee1 strain (Ye et al., 1996). Examination of germlings of a nimXcdc2AF strain germinated in the presence of 6 mM HU revealed that in marked contrast to the wild-type strain (Figure 5A), these cells displayed massive nuclei, which were often highly stretched in both germlings (Figure 5E) and hyphae (Figure 5F). The nuclei of the ΔankAwee1 and nimXcdc2AF strains germinated in low concentrations of HU are clearly polyploid, suggesting that over-replication of DNA is occurring in these cells in the absence of mitosis. This can clearly be seen in Figure 5F, in which the DNA of one nucleus has been extensively stretched into the three separate branched hyphae. Because cells over-replicating their DNA do not undergo mitosis, this dramatic stretching cannot be due to mitotic forces but rather may be the result of the action of nud genes, which function to position individual nuclei within the cytoplasm (Morris et al., 1998). We therefore induced rereplication in a nudC3 + nimXcdc2AF double mutant at the restrictive temperature for nudC3. Although the rereplication phenotype was still observed, no stretching of nuclei occurred (our unpublished results). This demonstrates that the nuclear stretching is the result of attempted migration of a single nucleus. In extreme examples (Figure 5F), large polyploid nuclei were found stretched into several different hyphal branches. Clearly such multidirectional stretching cannot be the result of abortive mitosis.
Figure 5.
DNA rereplication in strains that are unable to tyrosine phosphorylate NIMXcdc2 in response to a slowed S-phase. (A–C) DAPI staining for DNA of wild-type cells 11 h after germination in 6 mM HU (A), a ΔankAwee1 germling 11 h after germination in the presence of 6 mM HU (B), and a ΔankAwee1 germling 15 h after germination. Arrowheads indicate septa stained by Calcofluor. (D) Tubulin staining of a ΔankAwee1 germling 10 h after germination in 6 mM HU. The top panel shows tubulin staining, and the lower panel shows DAPI staining of DNA in the same germling. (E) DAPI staining of nimXcdc2AF mutant germlings grown in 6 mM HU for 11 h. (F) Hyphae after growth of a nimXcdc2AF strain for 14 h in 6 mM HU. Arrows indicate stretched DNA (B, D, and F). Bars, 5 μm.
Septum formation was often deregulated in mutants displaying the rereplication phenotype and was observed more frequently and in shorter germlings compared with wild-type strains germinated under the same conditions (Figure 5C; our unpublished results). Moreover, septation often occurred in the absence of nuclear division in these mutants, resulting in a cut-like phenotype (Figure 5C).
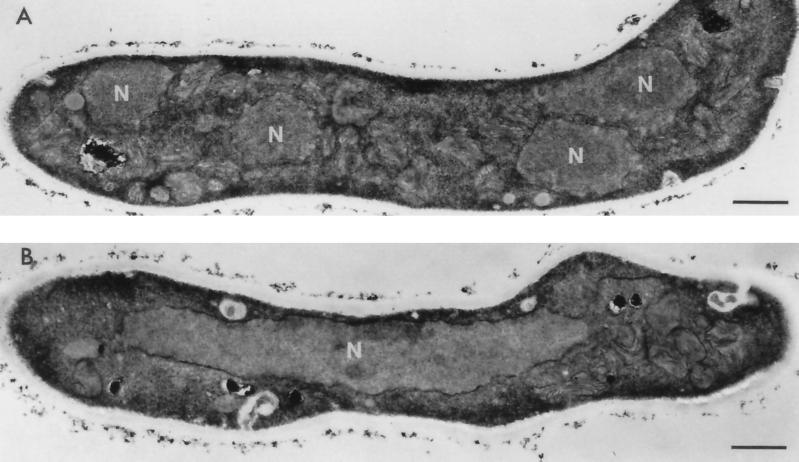
To further examine the morphology of the nuclei in cells displaying the rereplication phenotype, germlings of the ΔankAwee1 strain were grown for 10 h in the absence or presence of 6 mM HU and subjected to electron microscopy. In the absence of HU nuclear division occurred normally, with four distinct nuclei being apparent in the cell shown in Figure 6A. In striking contrast, cells grown in HU displayed abnormal, giant nuclei, which were polyploid (Figure 6B), demonstrating that DNA replication had continued in the absence of an effective mitosis, and that the DNA is maintained in a single nuclear membrane.
Figure 6.
Electron micrographs of ΔankAwee1 control and over-replicating germlings. (A) Germling germinated in the absence of HU for 10 h. (B) Germling germinated in the presence of 6 mM HU for 10 h displaying a massive polyploid nucleus. N, nuclei. Bar, 1 μm.
In fission yeast rad3 and rad26 are thought to function in checkpoint control through a pathway that regulates tyrosine phosphorylation of cdc2 (Al-Khodairy and Carr, 1992; Furnari et al., 1997; Uchiyama et al., 1997; Lindsay et al., 1998; Martinho et al., 1998). Given that strains unable to tyrosine phosphorylate NIMXcdc2 entered mitosis early in the presence of low concentrations of HU and subsequently over-replicated their DNA, we were interested in determining whether the same occurred in uvsBrad3 and uvsDrad26 mutants. We examined uvsB505rad3 and uvsD308rad26 germlings grown in the presence of 6 mM HU for 12 h by DAPI staining and observed similar over-replication phenotypes as in the ΔankAwee1 and nimXcdc2AF mutants under these conditions (our unpublished results; Figure 7).
Figure 7.
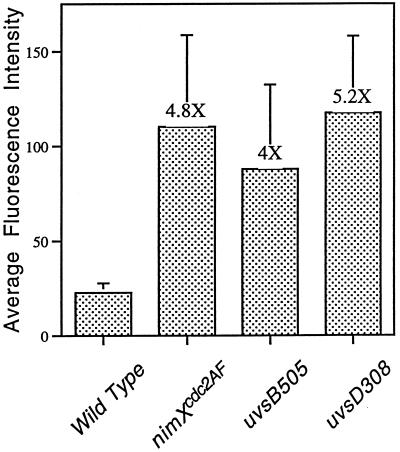
Comparison of the relative DNA content of cells undergoing DNA replication. Average fluorescence intensity of nuclei in 12 cells (6 cells from two independent experiments) of the indicated strains germinated in the presence of 6 mM HU for 12 h. Error bars represent the SD observed for each strain.
To estimate the ploidy of cells that had undergone over-replication of DNA, we measured the relative nuclear fluorescence of nuclei in cells from wild-type, nimXcdc2AF, uvsB505, and uvsD308 strains germinated for 12 h in the presence of 6 mM HU (Figure 7). A time point of 12 h was chosen for this experiment, even though over-replication continued to occur, because DNA subsequently became highly stretched, making measurements difficult. Even at 12 h, the nuclei of nimXcdc2AF, uvsB505, and uvsD308 germlings were clearly polyploid displaying 4.8, 3.9, and 5.2 times, respectively, the DNA content of wild-type nuclei when grown in the presence of 6 mM HU (Figure 7).
The rereplication phenotype described above may be a consequence of entry into mitosis before the completion of DNA replication, which would subsequently cause defects in DNA segregation as observed in Figure 5. To determine whether cells displaying over-replicated and incompletely segregated DNA had undergone an abnormal mitosis, we germinated the ΔankAwee1 mutant in 6 mM HU for 6.5 h and determined the average spindle length of cells undergoing the first mitosis. In comparison with an average spindle length of 2.3 μm for normal wild-type cells undergoing their first mitosis, the mean spindle length of ΔankAwee1 germlings grown in 6 mM HU was only 1.4 μm. This was largely because of the failure of the ΔankAwee1 cells to elongate their spindles in the presence of HU with no spindles >4 μm being observed in this strain compared with the wild-type strain, in which 17% of spindles were >4 μm long. Together, these data strongly suggest that cells entering mitosis before the completion of DNA replication undergo an abnormal mitosis leading to the failure of DNA segregation and the subsequent over-replication of DNA.
Combination of Impaired APC Function and the nimXcdc2AF Mutation Results in the Rereplication Phenotype in the Absence of HU
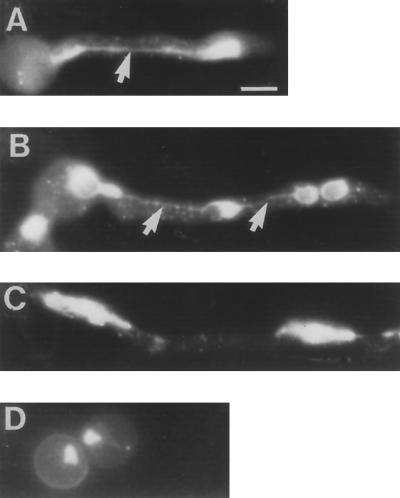
The APC1 homologue BIME has been previously demonstrated to play a role in S-phase checkpoint regulation (Ye et al., 1996, 1997b). Specifically, the bimE7APC1 mutation can override the S-phase arrest induced by 100 mM HU in a nimXcdc2AF mutant by a mechanism that leads to the activation of the NIMA kinase. Similarly, the bimE7APC1 mutation also negates S-phase arrest induced by inactivation of the mini chromosome maintenance protein nimQmcm2 at the restrictive temperature (James et al., 1995; Ye et al., 1997b). In both of these cases, it is a combination of compromised APC function and lack of cdc2 tyrosine phosphorylation that overrides the S-phase arrest (Ye et al., 1996, 1997b). The ability of bimE7APC1 to allow entry into mitosis of a nimXcdc2AF mutant arrested in S-phase by 100 mM HU at 32°C (the permissive temperature for bimE7APC1) suggests that the mutant BIMEAPC1 is not completely functional in this mutant at 32°C. We have previously observed that the nimXcdc2AF + bimE7APC1 double mutant is synthetically lethal and grows poorly at 32°C. This is likely to be a consequence of the unregulated entry of cells into mitosis during S-phase, which is analogous to what occurs in uvsB, uvsD and nimXcdc2AF mutants grown in the presence of low concentrations of HU. With this in mind, we examined the phenotype of the nimXcdc2AF + bimE7APC1 double mutant germinated at 32°C by DAPI staining. These cells either contained nuclei that displayed incomplete DNA segregation (Figure 8, A and B), or polyploid nuclei (Figure 8C). These phenotypes are almost identical to those of uvsB, uvsD and nimXcdc2AF mutants grown in the presence of low concentrations of HU. These data suggest that the bimE7APC1 mutation in combination with lack of tyrosine phosphorylation of NIMXcdc2 can promote premature entry into mitosis at varying stages of S-phase and that rereplication seen in previous experiments is not a consequence of the presence of HU. Interestingly, if the nimXcdc2AF + bimE7APC1 double mutant strain was germinated in low concentrations of HU at the restrictive temperature for bimE7APC1, cells arrested with a BIM phenotype (Figure 8D) instead of the rereplication phenotype observed in the nimXcdc2AF mutant under these conditions. Thus, completely inhibiting APC function prevents rereplication of DNA by arresting cells in mitosis.
Figure 8.
nimXcdc2AF + bimE7APC1 double mutant cells display polyploid nuclei. DAPI staining of cells showing representative nuclear morphologies observed in the double mutant germlings are shown. (A) 12-h-old germling with a highly stretched large nucleus (17% of cells). (B) Germling showing incomplete DNA segregation indicated by the arrows (50% of cells). (C) Part of a hyphae showing large abnormal nuclei (33% of cells). (D) Double mutant spores 12 h after germination at 42°C, showing the typical mitotic block phenotype of the bimE7 mutation with highly condensed DNA. Arrows indicate stretched nuclei. Bar, 5 μm.
Mitotic Kinase Activities Are Decreased in Cells Undergoing DNA Rereplication
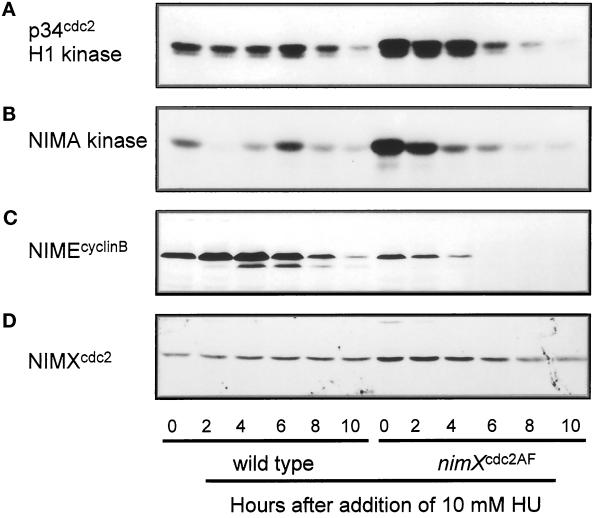
The rereplication phenotype appears to be a consequence of over-replication of DNA after premature entry into mitosis. In S. pombe, cells deleted for cdc13cyclinB (Hayles et al., 1994) or overexpressing the cdc2/cdc13cyclin B kinase inhibitor rum1 (Moreno and Nurse, 1994), undergo multiple rounds of replication without mitosis, suggesting that rereplication of DNA may be induced by the failure to accumulate cdc2 kinase activity during S-phase. In A. nidulans, the kinase activities of both NIMXcdc2 and NIMA are required for entry into mitosis (Osmani et al., 1991a; Ye and Osmani 1997). To examine these kinase activities in cells undergoing rereplication, we performed kinase assays on extracts prepared from log phase cultures of a wild-type and a nimXcdc2AF strain at various time points after addition of 10 mM HU (Figure 9). Under these conditions, the rereplication phenotype is evident in the nimXcdc2AF strain after the first premature mitosis, and large polyploid nuclei are clearly evident by 10 h. As expected, NIMXcdc2 kinase activity was considerably higher compared with wild-type cells before the addition of HU, because negative regulation of this kinase by tyrosine phosphorylation cannot occur in this mutant (Figure 9A). However, NIMXcdc2 kinase activity decreased markedly by 6 h after addition of HU in the nimXcdc2AF strain (Figure 9A). NIMA kinase levels also decreased during this period, consistent with NIMA being activated downstream of NIMXcdc2 (Figure 9B, Ye et al., 1995). This decrease in NIMXcdc2 kinase activity was not due to a decrease in levels of NIMXcdc2 protein but rather the decrease in levels of its cyclin partner NIMEcyclinB (Figure 9C). In contrast, the activity of the mitotic kinases fluctuated after addition of HU to wild-type cells, which undergo a normal, albeit delayed, mitosis under these conditions (Figure 9, A and B). These data indicate that over-replication of DNA occurs in the presence of low NIMXcdc2 kinase activity in the nimXcdc2AF strain after premature mitotic entry in the presence of incomplete DNA replication.
Figure 9.
Loss of NIMXcdc2 and NIMA kinase activities and proteolysis of NIMEcyclinB in nimXcdc2AF mutant cells after addition of 6 mM HU. HU was added to log phase cultures of either a wild-type or the nimXcdc2AF mutant strain to the final concentration of 10 mM, and samples were collected at the time intervals indicated. NIMXcdc2 (A) and NIMA (B) kinase activities were assayed using histone H1 or β-caesin as substrate. NIMEcyclin B (C) and NIMXcdc2 (D) protein levels were determined by Western blotting.
DNA Over-Replication Continues in the Presence of High NIMXcdc2 Kinase Activity
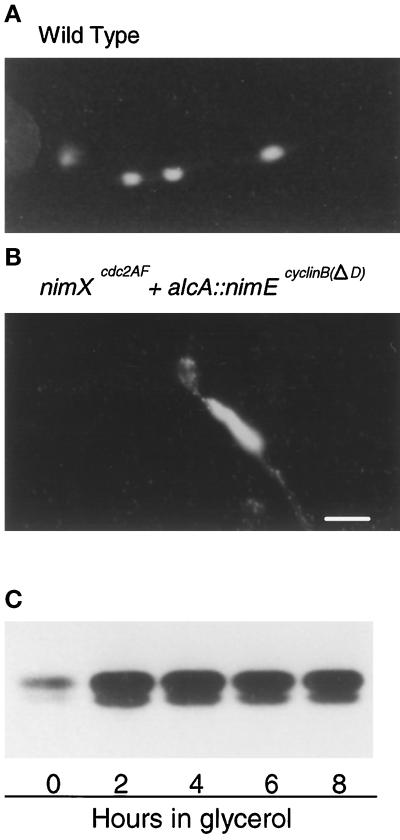
Given that DNA over-replication can be induced in S. pombe if cyclin B/cdc2 kinase activity is eliminated (Hayles et al., 1994; Moreno and Nurse, 1994), we next determined whether maintaining a high level of NIMXcdc2 kinase activity could prevent the rereplication phenotype we observe in the nimXcdc2AF mutant strain. To do this, we used a nimXcdc2AF strain that also contained a nondegradable form of NIMEcyclin B under the control of the inducible alcA promoter. Cells grown on coverslips in minimal acetate medium (repressing) were allowed to undergo the first premature mitosis in the presence of 6 mM HU, after which expression of nondegradable of NIMEcyclinB was allowed in glycerol medium. Under these conditions, cells still clearly displayed the rereplication phenotype characterized by large polyploid nuclei (Figure 10B). To follow NIMXcdc2 kinase activity biochemically after induction of nondegradable NIMEcyclin B, cells were grown in liquid culture to an early log phase before the addition of 6 mM HU for 4 h to cause premature mitosis. DAPI staining of aliquots of these HU-treated cells indicated that they did not display the rereplication phenotype at this time (our unpublished results). Expression of nondegradable NIMEcyclin B was then allowed by medium change, and samples were analyzed at the indicated times for NIMXcdc2 kinase activity (Figure 10C) and by DAPI staining. These cells still displayed the rereplication phenotype (our unpublished results), even though NIMXcdc2 kinase activity was maintained at a high level (Figure 10C). This indicates that loss of NIMXcdc2 kinase activity is not essential for continuing over-replication of DNA. Together with the experiments described earlier, this suggests that the rereplication phenotype occurs as a result of loss of checkpoint regulation over entry into mitosis because of the inability to tyrosine phosphorylate NIMXcdc2, but that the subsequent over-replication of DNA occurs independently of NIMXcdc2 activity.
Figure 10.
Maintaining high levels of NIMXcdc2 kinase activity in the nimXcdc2AF mutant fails to prevent DNA over-replication. The nimXcdc2AF + alcA::nimEcyclin BΔD strain, which contains a nondegradable (D box minus) nimEcyclin B under control of the alcA promoter, and a wild-type strain were first germinated in minimal medium containing acetate (repressing) in the presence of 6 mM HU. Cells were allowed to undergo the first premature mitosis, which occurs after 9 h in minimal medium. Expression of nimEcyclin B ΔD was then allowed by transferring cells to glycerol-containing medium. DAPI staining of representative cells of (A) the wild-type and (B) nimXcdc2AF + alcA::nimEcyclin BΔD strains after 7 h growth in glycerol-containing medium. Please note that when grown in minimal medium A. nidulans cells are much thinner than those shown in nutrient-rich medium (Figure 5). (C) p34cdc2 H1 kinase activity was isolated from the nimXcdc2AF + alcA::nimEcyclin BΔD strain at the indicated times after transfer to glycerol-containing medium. Bar, 5 μm.
DISCUSSION
Here we demonstrate that A. nidulans uvsB and uvsD play roles in checkpoint regulation over entry into mitosis in response to slow S-phase and G2 DNA damage. These genes display homology to two members of the checkpoint rad gene family of S. pombe, rad3 and rad26, respectively. rad3 and rad26 are thought to function in checkpoint control through a pathway that regulates cdc2 tyrosine phosphorylation (Al-Khodairy and Carr, 1992; Furnari et al., 1997; Uchiyama et al., 1997; Lindsay et al., 1998; Martinho et al., 1998). Homology between uvsD and rad26 exists throughout the coding region of the gene and includes a coiled coil domain followed by a putative nuclear localization sequence. Given these sequence and structural homologies and the similar phenotypes caused by mutations of these genes, uvsD is likely to be the first identified rad26 homologue. uvsB is a member of the rad3/ATR/ATM/MEC1/TEL1 family of proteins displaying highest homology to rad3 in S. pombe. The human ATM (ataxia–telangiectasia mutated) gene is involved in G2 checkpoint control and when mutated leads to cancer predisposition (Savitsky et al., 1995), emphasizing the importance of cell cycle checkpoint control and normal health in humans.
Over-replication of DNA is a consequence of the loss of coordination between S-phase and mitosis. Maintenance of an ordered progression of S-phase and mitosis is thought to be carried out by a replication-licensing system in which a replication-licensing factor binds chromatin early in the cell cycle, is removed from chromatin as DNA replicates, and is unable to rebind replicated chromatin until after the following mitosis (Blow, 1993; Chong et al., 1995). Here we demonstrate that in A. nidulans, the initiation of mitosis before the completion of DNA replication results in an abnormal mitosis, after which cells subsequently undergo over-replication of DNA without ever attempting mitosis again. Quantitation of the average nuclear fluorescence intensity indicated that the nuclei of cells displaying the rereplication phenotype contained greater than four times the DNA of wild-type nuclei by 12 h germination in 6 mM HU. That over-replication occurs without cells attempting another mitosis is supported by the absence of mitotic spindles in all cells displaying polyploid nuclei. The rereplication phenotype was demonstrated in uvsB and uvsD mutant strains as well as in strains that were unable to tyrosine phosphorylate NIMXcdc2. It is therefore likely that any breakdown in checkpoint regulation that allows cells prematurely into a defective mitosis will subsequently break the regulation that normally maintains ploidy in A. nidulans.
Although over-replication of DNA does not occur as a result of entry into mitosis before the completion of DNA replication or G2 DNA repair in S. pombe, over-replication can be induced under certain circumstances in this organism. Mutations in S. pombe cut1 and cut2 lead to a failure of sister chromatid separation, and when coupled with the cdc11 mutation to prevent cytokinesis, cells over-replicate their DNA, resulting in the formation of polyploid nuclei (Creanor and Mitchison, 1990; Uzawa et al., 1990; Funabiki et al., 1996). In A. nidulans, mutations in the cut1 homologue bimB and in the kinesin-like protein bimC lead to the failure of mitosis and nuclear division, but cells still replicate their DNA and form polyploid nuclei (Enos and Morris 1990; May et al., 1992). In contrast to the nimXcdc2AF, uvsB and uvsD mutants, however, bimB and bimC mutants attempt multiple abortive mitosis after the failure of the initial mitosis (Enos and Morris 1990; May et al., 1992). In mutants lacking bimB and bimC function, cells progress through the cell cycle, but their replicated DNA is not segregated because of mechanical defects during mitosis. Such cells may delay in a mitotic-like state but fail to segregate DNA, exit mitosis, and resume DNA synthesis. They then reenter mitosis, fail to segregate DNA, and enter another round of DNA replication. Such repeated rounds of the cell cycle subsequently generate large polyploid nuclei. In contrast, premature mitosis induced by lack of uvsB or uvsD or tyrosine phosphorylation of NIMXcdc2 prevents progression through the first premature mitosis, but cells never again attempt mitosis, presumably because of lack of mitotic kinase activities. Indeed, loss of cdc2 kinase activity has also been associated with over-replication in S. pombe. Notably, severely impairing cdc2 kinase activity by deletion of the cdc13cyclinB gene (Hayles et al., 1994) or by overexpression of the cdc2/cdc13cyclinB kinase inhibitor rum1 (Moreno and Nurse, 1994) leads to the formation of polyploid cells. Endoreplication is also known to occur naturally in certain plant, Drosophila, and mammalian cells (Grafi and Larkins; Williams and Jackson, 1982; Sauer et al., 1995; Datta et al., 1996; Weiss et al., 1998). Interestingly, similar to S. pombe, endoreplication appears to be associated with a loss of cdc2 kinase activity in terminally differentiating megakaryocytes (Datta et al., 1996) and in maize endosperm (Grafi and Larkins, 1995). This suggests that low cdc2 kinase activity may allow over-replication of DNA.
We have demonstrated that NIMXcdc2 kinase activity decreases as a result of NIMEcyclinB proteolysis in the nimXcdc2AF mutant under conditions that induce over-replication of DNA. However, maintaining high kinase activity by expressing a nondegradable form of NIMEcyclinB after premature mitosis failed to abolish over-replication of DNA. Thus, over-replication can still occur in the presence of high NIMXcdc2 kinase activity if cells are first allowed to undergo a premature mitosis. Further understanding of the rereplication phenotype described here should provide information on how DNA replication and successful exit from mitosis are interlinked and how some cells normally break this relationship to naturally produce polyploid cells.
Over-replication can also be induced in S. pombe by overexpression of the replication initiator cdc18 (Nishitani and Nurse, 1997; Greenwood et al., 1998). Interestingly, cells overexpressing a version of cdc18 in which consensus CDK phosphorylatable sites have been mutated to alanine continue to over-replicate their DNA, even in the presence of high levels of cdc2 kinase activity (Jallepelli et al., 1997). Furthermore, phosphorylation of these consensus CDK sites promotes wild-type cdc18 ubiquitin-dependent degradation (Jallepelli et al., 1997). This suggests that over-replication in S. pombe induced by lack of cdc2 activity may be due to defects in the degradation of cdc18. In addition to cdc18, proteolysis of rum1 (Kominami and Toda, 1997) and Xenopus wee1 (Michael and Newport, 1998) are also thought to be involved in progression into mitosis from S-phase. Our observation of over-replication in the bimE7APC1 + nimXcdc2AF double mutant further implicates APC/C function in checkpoint control over mitotic entry. The observed loss of cyclin B protein in the nimXcdc2AF mutant undergoing rereplication suggests that the APC/C is active in these cells. It will be of interest to determine whether an A. nidulans cdc18 homologue plays a role in the rereplication phenotype described in this study and to define the precise role of proteolysis in maintaining the temporal order of S-phase and mitosis.
Our data also indicate that uvsBrad3 and uvsDrad26 have another role in response to DNA damage, which is independent of NIMXcdc2. Specifically, UV irradiation of quiescent conidiospores causes a marked loss of viability of uvsB and uvsD mutants, but nimXcdc2AF conidiospores are no more sensitive than wild type. Germinating conidiospores do not enter the cell cycle directly, because they need to break dormancy and get their metabolism up and running. This delay presumably gives cells time to repair damaged DNA. The high UV sensitivity of uvsB and uvsD mutant conidiospores suggests that they have roles in the DNA damage response other than the regulation of NIMXcdc2 tyrosine phosphorylation. rad3 and rad26 are thought to function early in the DNA damage response and may also play roles in the initiation of DNA repair. Supporting this, the S. cerevisiae uvsBrad3 homologue MEC1 is involved in the transcriptional activation of genes involved in DNA repair (Huang et al., 1998).
In conclusion, we have identified A. nidulans uvsBrad3 and uvsDrad26 as rad3 and rad26 homologues, respectively. The finding that, like in S. pombe, uvsDrad26 functions in both the G2 DNA damage and prolonged S-phase checkpoints suggests functional conservation of this checkpoint gene. The isolation of uvsDrad26 as a potential homologue of S. pombe rad26 may enable human rad26-like genes to be identified to determine whether this class of checkpoint function is conserved in the same manner as uvsBrad3. We further show that in A. nidulans, loss of checkpoint control over mitotic initiation leads to a defective mitosis followed by over-replication of genomic DNA. This defines a new mechanism by which endoreplication of DNA can be triggered. Finally, the dramatic rereplication phenotype we have defined will enable us to screen easily for further mutations causing defective checkpoint control over mitosis.
ACKNOWLEDGMENTS
We thank Dr. L.Ellis and Dr. P.Ramos (W.M. Keck Center for Genome Informatics, Institute of Biosciences and Technology, Texas A&M University, College Station, TX) for providing the ΔankAwee1 strain, Dr. Etta Kafer (Institute of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, British Columbia, Canada) for helpful discussions, and Dr. Matthew O’Connell for providing the plasmid p122 containing alcA inducible nimEcyclinBΔD. This work was supported by National Institutes of Health grant GM-42564.
REFERENCES
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJF, Lehmann AR, Carr AM. Identification of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;4:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A, Surana U, Muroff I, Nasmyth K. Regulation of p34cdc28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae. Nature. 1992;355:368–371. doi: 10.1038/355368a0. [DOI] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;23:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Blasina A, Paegle S, Mc Gowan CH. The role of inhibitory phosphorylation of cdc2 following DNA replication block and radiation-induced damage in human cells. Mol Cell Biol. 1997;8:1013–1023. doi: 10.1091/mbc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ. Preventing rereplication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Alfa CE, Hyams JS, Beach DH. The fission yeast cdc2/cdc13/suc1 protein kinase: regulation of catalytic activity and nuclear localization. Cell. 1989;58:485–497. doi: 10.1016/0092-8674(89)90429-7. [DOI] [PubMed] [Google Scholar]
- Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S-phase on mitosis. Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani HM, Khoo C-Y, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–424. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Creanor J, Mitchison JM. Continued DNA synthesis after a mitotic block in the double mutant cut1 cdc11 of the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1990;96:435–438. doi: 10.1242/jcs.96.3.435. [DOI] [PubMed] [Google Scholar]
- Datta NS, Williams JL, Caldwell J, Curry AM, Ashcraft EK, Long MW. Novel alterations in CDK1/cyclin B1 kinase complex formation occur during the acquisition of a polyploid DNA content. Mol Cell Biol. 1996;7:209–223. doi: 10.1091/mbc.7.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing and identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Evans T, Rosenthal ET, Youngblom Y, Distel D, Hunt T. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell. 1983;33:389–396. doi: 10.1016/0092-8674(83)90420-8. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. EMBO J. 1996;15:6617–6628. [PMC free article] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- Gould KL, Nurse P. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature. 1989;342:39–45. doi: 10.1038/342039a0. [DOI] [PubMed] [Google Scholar]
- Grafi G, Larkins B. Endoduplication in maize endosperm: involvement of M-phase-promoting factor inhibition and induction of S-phase-related kinases. Science. 1995;269:1262–1264. doi: 10.1126/science.269.5228.1262. [DOI] [PubMed] [Google Scholar]
- Greenwood E, Nishitani H, Nurse P. Cdc18 can block mitosis by two independent mechanisms. J Cell Sci. 1998;111:3101–3108. doi: 10.1242/jcs.111.20.3101. [DOI] [PubMed] [Google Scholar]
- Han KY, Chae SK, Han DM. The uvsI gene of Aspergillus nidulans required for UV-mutagenesis encodes a homolog to REV3, a subunit of the DNA polymerase zeta of yeast involved in translesion DNA synthesis. FEMS Microbiol Lett. 1998;164:13–19. doi: 10.1111/j.1574-6968.1998.tb13061.x. [DOI] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Jallepelli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes & Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SW, Mirabito PM, Scaheri PC, Morris NR. The Aspergillus nidulans bimE (blocked-in-mitosis) gene encodes multiple cell cycle functions involved in mitotic checkpoint control and mitosis. J Cell Sci. 1995;108:3485–3499. doi: 10.1242/jcs.108.11.3485. [DOI] [PubMed] [Google Scholar]
- Jansen GJO. Abnormal frequencies of spontaneous mitotic recombination in uvsB and uvsC mutants of Aspergillus nidulans. Mutat Res. 1970;10:33–41. doi: 10.1016/0027-5107(70)90143-0. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Yucel J, Rowley R, Subramani S. The rad3+ gene of Schizosaccharomyces pombe is involved in multiple functions and in DNA repair. Proc Natl Acad Sci USA. 1992;89:4952–4956. doi: 10.1073/pnas.89.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Gu Y, Morgan DO. Role of inhibitory cdc2 phosphorylation in radiation-induced G2 arrest in human cells. J Cell Biol. 1996;134:963–970. doi: 10.1083/jcb.134.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer E, Chae S-K. Phenotypic and epistatic grouping of hypo- and hyper-rec mus mutants in Aspergillus. Curr Genet. 1994;25:223–232. doi: 10.1007/BF00357166. [DOI] [PubMed] [Google Scholar]
- Kafer E, May G. The uvsF gene region in Aspergillus nidulans codes for a protein with homology to DNA replication factor C. Gene. 1997;191:155–159. doi: 10.1016/s0378-1119(97)00052-8. [DOI] [PubMed] [Google Scholar]
- Kafer E, Mayor O. Genetic analysis of DNA repair in Aspergillus: evidence for different types of MMS-sensitive hyperrec mutants. Mutat Res. 1986;161:119–134. doi: 10.1016/0027-5107(86)90003-5. [DOI] [PubMed] [Google Scholar]
- Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek W, Nigg EA. Mutations of p34cdc2 phosphorylation sites induce premature mitotic events in HeLa cells: evidence for a double block to p34cdc2 kinase activation in vertebrates. EMBO J. 1991;10:3331–3341. doi: 10.1002/j.1460-2075.1991.tb04897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami K, Toda T. Fission yeast WD-repeat protein Pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator cdc18. Genes & Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsh EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Martinho RG, Lindsay HD, Flaggs G, DeMaggio AJ, Hoekstra MF, Carr AM, Bentley NJ. Analysis of rad3 and chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May GS, McGoldrick CA, Holt CL, Denison SH. The bimB3 mutation of Aspergillus nidulans uncouples DNA replication from the completion of mitosis. J Biol Chem. 1992;267:15737–15743. [PubMed] [Google Scholar]
- Michael WM, Newport J. Coupling of mitosis to the completion of S phase through cdc-34-mediated degradation of Wee1. Science. 1998;282:1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Morris NR. Mitotic mutants of Aspergillus nidulans. Genet Res. 1976;26:237–254. doi: 10.1017/s0016672300016049. [DOI] [PubMed] [Google Scholar]
- Morris NR, Efimov VP, Xiang X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998;8:467–470. doi: 10.1016/s0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- Mueller PR, Coleman TR, Kumagai A, Dunphy WG. Myt1: a membrane-associated kinase that phosphorylates cdc2 on both threonine-14 and tyrosine-15. Science. 1995;270:86–90. doi: 10.1126/science.270.5233.86. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Viewpoint: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. The cdc18 protein initiates DNA replication in fission yeast. Prog Cell Cycle Res. 1997;3:135–142. doi: 10.1007/978-1-4615-5371-7_11. [DOI] [PubMed] [Google Scholar]
- Nurse P. Checkpoints come of age. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Osmani SA. Cell cycle analysis using the filamentous fungus Aspergillus nidulans. In: Fantes P, Brooks R, editors. The Cell Cycle: A Practical Approach. New York: Oxford University Press; 1993. pp. 127–142. [Google Scholar]
- Osman F, Tomsett B, Strike P. The isolation of mutagen-sensitive nuv mutants of Aspergillus nidulans and their effects on mitotic recombination. Genetics. 1993;134:445–454. doi: 10.1093/genetics/134.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, McGuire SL, Osmani SA. Parallel activation of the NIMA and p34cdc2 cell cycle regulated protein kinases is required to initiate mitosis in A. nidulans. Cell. 1991a;67:283–291. doi: 10.1016/0092-8674(91)90180-7. [DOI] [PubMed] [Google Scholar]
- Osmani AH, O’Donnell K, Pu RT, Osmani SA. Activation of the NIMA protein kinase plays a unique role during mitosis that cannot be bypassed by absence of the bimE checkpoint. EMBO J. 1991b;10:2669–2679. doi: 10.1002/j.1460-2075.1991.tb07810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, van Peij N, Mischke M, O’Connell MJ, Osmani SA. A single p34cdc2 protein kinase (encoded by nimXcdc2) is required at G1 and G2 in Aspergillus nidulans. J Cell Sci. 1994;107:1519–1528. doi: 10.1242/jcs.107.6.1519. [DOI] [PubMed] [Google Scholar]
- Osmani SA, May GS, Morris NR. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol. 1987;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Wilkinson M. DNA fragment purification: removal of agarose 10 min after electrophoresis. BioTechniques. 1991;10:736–737. [PubMed] [Google Scholar]
- Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes & Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct roles of cyclin E/cdc2 kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes & Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfez S, Tagle DA, Ziv Y, Sartiel A, Collins FS, Shiloh Y, Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Murray AW. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28. Nature. 1992;355:365–368. doi: 10.1038/355365a0. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1663. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Timberlake WE. Low repetitive DNA content in Aspergillus nidulans. Science. 1978;202:773–775. doi: 10.1126/science.362530. [DOI] [PubMed] [Google Scholar]
- Uchiyama M, Galli I, Griffiths DJF, Wang TS-F. A novel allele of Schizosaccharomyces pombe rad26 defective in monitoring S-phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa S, Samejima I, Hirano T, Tanaka K, Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990;62:913–925. doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Swart K, Holub EF, van Dijk R, Offenberg HH, Goosen T, van den Broek HW. Cloning, sequencing, disruption and phenotypic analysis of uvsC, an Aspergillus nidulans homologue of yeast RAD51. Mol Gen Genet. 1997;254:654–664. doi: 10.1007/s004380050463. [DOI] [PubMed] [Google Scholar]
- Wuarin J, Nurse P. Regulating S phase: CDK, licencing and proteolysis. Cell. 1996;85:785–787. doi: 10.1016/s0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR. Shuttle vectors and inducible expression vectors using alcA in Aspergillus nidulans. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Weinert T. A DNA damage checkpoint meets the cell cycle engine. Science. 1997;277:1450–1451. doi: 10.1126/science.277.5331.1450. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- Weiss A, Herzig A, Jacobs H, Lehner CF. Continuous cyclin E expression inhibits progression through endoreduplication cycles in Drosophila. Curr Biol. 1998;12:239–242. doi: 10.1016/s0960-9822(98)70090-9. [DOI] [PubMed] [Google Scholar]
- Williams N, Jackson H. Kinetic analysis of megakaryocyte numbers and ploidy levels in developing colonies from mouse bone marrow cells. Cell Tissue Kinet. 1982;15:483–494. doi: 10.1111/j.1365-2184.1982.tb01571.x. [DOI] [PubMed] [Google Scholar]
- Ye XS, Fincher RR, Tang A, McNeal KK, Gygax SE, Wexler AN, Ryan KB, James SW, Osmani SA. Proteolysis and tyrosine phosphorylation of p34cdc2/cyclin B. J Biol Chem. 1997a;272:33384–33393. doi: 10.1074/jbc.272.52.33384. [DOI] [PubMed] [Google Scholar]
- Ye XS, Fincher RR, Tang A, O’Donnell K, Osmani SA. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 1996;15:3599–3610. [PMC free article] [PubMed] [Google Scholar]
- Ye XS, Fincher RR, Tang A, Osmani SA. The G2/M DNA damage checkpoint inhibits mitosis through Tyr15 phosphorylation of p34cdc2 in Aspergillus nidulans. EMBO J. 1997b;16:182–192. doi: 10.1093/emboj/16.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XS, Osmani SA. Regulation of p34cdc2/cyclin B H1 and NIMA kinases during the G2/M transition and checkpoint responses in Aspergillus nidulans. In: Meijer L, Guidet S, Philippe M, editors. Progress in Cell Cycle Research. Vol. 3. New York: Plenum Press; 1997. pp. 221–232. [DOI] [PubMed] [Google Scholar]
- Ye XS, Xu G, Pu PT, Fincher RR, McGuire SL, Osmani AH, Osmani SA. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 1995;14:986–994. doi: 10.1002/j.1460-2075.1995.tb07079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Leem BJ, Kang HS. The Aspergillus uvsH gene encodes a product homologous to yeast RAD18 and Neurospora UVS-2. Mol Gen Genet. 1995;248:174–178. doi: 10.1007/BF02190798. [DOI] [PubMed] [Google Scholar]
- Zhao P, Kafer E. Effects of mutagen-sensitive mus mutations on spontaneous mitotic recombination in Aspergillus. Genetics. 1992;130:171–728. doi: 10.1093/genetics/130.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]