Abstract
The peroxisomal matrix proteins involved in many important biological metabolism pathways in eukaryotic cells are encoded by nucleal genes, synthesized in the cytoplasm and then transported into the organelles. Targeting and import of these proteins depend on their two peroxisomal targeting signals (PTS1 and PTS2) in sequence as we have known so far. The vectors of the fluorescent fusions with PTS, i.e., green fluorescence protein (GFP)-PTS1, GFP-PTS2 and red fluorescence protein (RFP)-PTS1, were constructed and introduced into Magnaporthe oryzae Guy11 cells. Transformants containing these fusions emitted fluorescence in a punctate pattern, and the locations of the red and green fluorescence overlapped exactly in RFP-PTS1 and GFP-PTS2 co-transformed strains. These data indicated that both PTS1 and PTS2 fusions were imported into peroxisomes. A probable higher efficiency of PTS1 machinery was revealed by comparing the fluorescence backgrounds in GFP-PTS1 and GFP-PTS2 transformants. By introducing both RFP-PTS1 and GFP-PTS2 into Δmgpex6 mutants, the involvement of MGPEX6 gene in both PTS1 and PTS2 pathways was proved. In addition, using these transformants, the inducement of peroxisomes and the dynamic of peroxisomal number during the pre-penetration processes were investigated as well. In summary, by the localization and co-localization of PTS1 and PTS2, we provided a useful tool to evaluate the biological roles of the peroxisomes and the related genes.
Keywords: Peroxisomal targeting signal (PTS), Peroxisomal localization, MGPEX6 gene, Magnaporthe oryzae
INTRODUCTION
Peroxisomes, the single-membrane bound organelles, are ubiquitous in various eukaryotic cells (Elgersma and Tabak, 1996; Gould and Valle, 2000; Hettema et al., 1999). These organelles contain more than 50 different enzymes and control many important metabolic processes, including β-oxidation of fatty acids, synthesis of ether lipids, generation and degradation of reactive oxygen species (van den Bosch et al., 1992; Johnson and Olsen, 2001), and some specialized functions (Müller et al., 1991; Elgersma and Tabak 1996). In human, the defects in peroxisome arouse serious disorders [peroxisome biogenesis disorders (PBDs)], such as the Zellweger syndrome and the infantile Refsum’s disease (Gould and Valle, 2000). So far, studies on peroxisomes have mainly concentrated on a few yeast species, Arabidopsis thaliana and some mammalian systems (Subramani, 1998).
The peroxisomal matrix proteins are encoded by nucleal genes, synthesized in the cytoplasm, and then transported into the organelles (Lazarow and Fujiki, 1985). Targeting and import of these proteins depend on the peroxisomal targeting signals (PTSs) in sequence. PTS falls into at least two categories: PTS1 and PTS2 (Hettema et al., 1999). PTS1, located at the C-terminal of proteins with conserved tripeptide SKL (serine-lysine-leucine) or its derivative (S/C/A-K/R/H-L), is the most abundant one among all known peroxisomal matrix proteins (Purdue and Lazarow, 2001). And PTS2, mainly at the N-terminal of proteins with a consensus sequence [(R)-(A/L/Q/I)-X5-(H)-(I/L/F)], is found in a few instances (Kato et al., 1996). Studies in yeast and mammals revealed about 30 proteins involved in peroxisome biogenesis. These proteins were called peroxins and their coding genes were written as PEX. The components of peroxins involved in PTS1 and PTS2 pathways are different. For instance, the PTS1 receptor is Pex5p, while the PTS2 receptor is Pex7p (Subramani, 1998).
Despite all that, the peroxisome biogenesis is still an incompletely understood process. Compared with that in yeast, mammals or plants, less research has been done in filamentous fungi, especially in the phytopathogenic species. Magnaporthe oryzae, a well-known ascomycete which causes rice blast, was a model plant fungal pathogen. Research on the infection processes of this fungus including conidium germination, appressorium formation (Howard et al., 1991; Howard and Ferrari, 1989) and penetration (Rossman et al., 1990; Bourett and Howard, 1992) is becoming a hot point. In recent years, the peroxisome has been revealed to have important roles in the phytopathogenic fungi. The disruption of PEX6 blocked the fungal penetration and destroyed the virulence completely in M. oryzae and Colletotrichum lagenarium (Kimura et al., 2001; Ramos-Pamplona and Naqvi, 2006; Wang et al., 2007). The Woronin bodies, a special class of peroxisomes involved in occlusion of septal pores, are also necessary for pathogenicity of M. oryzae (Soundararajan et al., 2004). Some peroxisome targeted genes, such as ICL1, PTH2 and MFP1, were also proved crucial for plant fungal pathogens (Wang et al., 2003; 2007; Bhambra et al., 2006). Characterization of PEX6 and the enzymes localized in peroxisomal matrix revealed the primary function of peroxisome in fungal pathogenicity. However, to explore more peroxins involved in PTS1 or PTS2 pathway and peroxisome-containing enzymes, to investigate the roles of these proteins, and to learn the dynamic change of peroxisomes in the host infection of M. oryzae, will bring out more important messages about peroxisome biogenesis and pathogenicity mechanism in fungal pathogens.
Fluorescent localization is a useful strategy to investigate the organelle biogenesis and is also used in research of peroxisome. By introducing the fluorescent fusions with PTS into the fungal cells, the fluorescence indicates the locations of peroxisomes in transformants. Mutations in PEX genes usually result in the mislocalization of peroxisomal matrix proteins. Therefore, the fluorescent fusion with PTS is also useful to study PEX genes by the fluorescence pattern in the related mutants (Mano et al., 2006; Stasyk et al., 2006). The misallocation of PTS1 protein resulted from MGPEX6 disruption has been already demonstrated in M. oryzae (Ramos-Pamplona and Naqvi, 2006; Wang et al., 2007). To learn more factors in peroxisome biogenesis and the roles of peroxisome in fungal pathogenicity, we constructed vectors with the fusions of GFP or RFP with PTS1 or PTS2 signal, and introduced them into M. oryzae cells. By the fluorescence in these transformants, the dynamic of peroxisomal number in the pre-penetration processes was observed, and the defect in PTS2 pathway of Δmgpex6 mutant was also investigated in this paper.
MATERIALS AND METHODS
Strains and growth conditions
M. oryzae wild-type was Guy11 (Notteghem and Silue, 1992); Δmgpex6 mutant (Wang et al., 2007) was a kind gift from Dr. Zhengyi Wang (Talbot’s Laboratory, Exeter University, UK). All M. oryzae strains used were grown routinely on complete medium (CM) (Talbot et al., 1993) using routine procedures (Wang et al., 2005). Escherichia coli strain DH5α and Agrobacterium tumefaciens strain AGL1 were cultured on Luria-Bertani (LB) medium in 37 °C and 28 °C, respectively.
Vector construction
The GFP (green fluorescence protein) ORF (open reading frame) without PTS was amplified from pEGFP (Clontech, USA) using the forward primer GFP-Xb and the reverse primer GFPstop-Bm. The GFP ORF with PTS1 signal (GFP-PTS1, GFPA) was amplified using the forward primer GFP-Xb and the reverse fusion primer GFP1-Bm that contains a tripeptide SKL encoding serial (underlined in Table 1). The GFP ORF with PTS2 signal (GFP-PTS2, GFPB) was amplified using the forward fusion primer GFP2-Xb, which contains a PTS2 encoding region (underlined) derived from N-terminal of Saccharomyces cerevisiae thiolase gene (GeneID: 854646), and the reverse primer GFPstop-Bm. The RFP (red fluorescence protein) ORF without PTS was amplified from pDsRED (Clontech, USA) using the forward primer RFP-Xb and the reverse primer RFPstop-Bm. The RFP ORF with PTS1 signal (RFP-PTS1, RFPA) was amplified using the forward primer RFP-Xb and the reverse fusion primer RFP1-Bm that contains SKL encoding serial. XbaI and BamHI sites were added respectively to 5′- and 3′-terminals of each of above amplificons (marked in box in Table 1). The promoter of MPG1 gene (Talbot et al., 1993) was used to express the fusions. The fragment of MPG1 promoter was amplified from M. oryzae Guy11 genome using the forward primer MProF-Pv and the reverse primer MProR-Xb, with PvuI and XbaI sites introduced into 5′- and 3′-terminals respectively. The BAR gene resistant to glufosinate-ammonium and the NPTII gene resistant to G418 were used as the selection markers. The fragment of the BAR gene under the control of trpC promoter (PtrpC) was amplified from pBARKS1 (Pall and Brunelli, 1993) using the forward primer BAR-Xh1 and the reverse primer BAR-Xh2. The NPTII under PtrpC was amplified from pBSTrp-Neo (a reconstructed vector by introducing PtrpC and NPTII respectively into the XhoI-EcoRI and EcoRI-PstI sites of the pBluscript) using the primers NEO-Xh1 and NEO-Xh2. XhoI sites were introduced into each terminal of amplificon of selection marker. All amplificons were cloned into pGEM-T easy vector (Promega, USA) and sequenced to ensure the accuracy.
Table 1.
Primers used in this paper
The fragment of PtrpC-BAR or PtrpC-NPTII was inserted between two XhoI sites of pCAMBIA1300 (CAMBIA) to generate p1300BAR or p1300NPT. The fragment of MPG1 promoter was then inserted into the PvuI-XbaI site of p1300BAR and p1300NPT respectively to generate p1300BM and p1300NM. The GFP, GFPA and GFPB were introduced respectively into p1300BM to generate p1300BMGFP, p1300BMGFPA and p1300BMGFPB. The RFP and RFPA were introduced respectively into p1300NM to generate p1300NMRFP and p1300NMRFPA by XbaI-BamHI digestion. All the primers used were listed in Table 1.
Fungal transformation
The inducible medium (IM) preparation and Agrobacterium tumefaciens-mediated transformation (ATMT) were processed as described by Mullins et al.(2001). CM plates containing corresponding antibiotics (300 μg/ml glufosinate-ammonium or 800 μg/ml G418) were used to select transformants.
Nucleic acid manipulations and Southern blot
Genomic DNA was extracted from mycelia collected from 2-day-old CM shaken at 150 r/min at 28 °C using a CTAB (hexadecyltrimethylammonium bromide) procedure described by Talbot et al.(1993). Gel electrophoresis, restriction enzyme digestion and fragment cloning were carried out using standard procedures (Sambrook et al., 1989). Genomic DNA from different strains was digested, size-separated on a 0.7% (w/v) agarose gel and transferred to a nylon membrane. Probes labeling and DNA hybridization were processed by using the DIG high prime DNA labeling and detection starter kit I (Roche, Germany).
Fluorescence microscopy
Fluorescence of conidia, hyphae and appressoria was observed using the Leica SP2 Confocal System (Leica, Germany). The objective used was a 63× Plan-Apochromat (numerical aperture, 1.4) oil immersion lens. GFP was imaged with 488 nm wavelength laser excitation and 505~530 nm band pass emission filter. RFP was imaged with 558 nm laser excitation and 580~600 nm filter. An Olympus-BX51 fluorescence microscope was also used for primary observation.
Inducement and counting of peroxisomes
To induce the peroxisomal proliferation, conidia from GFPA and RFPA transformants were allowed to form the hyphae in CM liquid shaken at 28 °C darkness for 48 h. The generated mycelia were washed for 3 times in sterilized water and then transferred into CM, minimal medium containing 6 mmol/L glucose (GM) or sucrose (SM), 2 g/L olive oil (OM) or Tween 80 (TM), respectively. After cultured for 24 h, the peroxisomes in the hyphae were observed and imaged under the fluorescence microscope.
To analyze the dynamic of peroxisomal number during pre-penetration processes, conidia of GFPA and RFPA transformants were harvested from a 10-day-old CM plate and allowed to form appressoria on inducible membrane. The cells incubated for 0, 2, 4, …, 24 h were observed and imaged. The numbers of peroxisomes in more than 100 conidia (or with germ tubs and appressoria) for each time point were counted and statistically compared.
RESULTS
Binary vectors, fungal transformation and validation of transformants
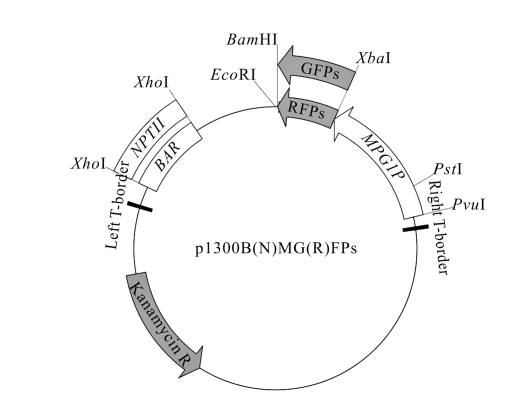
The structures of 1300BMGFP, p1300BMGFPA, p1300BMGFPB, p1300NMRFP and p1300NMRFPA are shown in Fig.1. The expression cassettes of fluorescence proteins and the selection markers were both enclosed between two T-borders to ensure the expression in the transformants.
Fig. 1.
The structures of p1300BMGFP, p1300BMGFPA, p1300BMGFPB, p1300NMRFP and p1300NMRFPA. GFPs include the GFP expression cassettes without PTS (GFP), with PTS1 (GFPA), and with PTS2 (GFPB); RFPs include the RFP expression cassettes without PTS (RFP) and with PTS1 (RFPA). BAR and NPTII represent the BAR and NPTII cassettes respectively; and MPG1P is the MPG1 promoter
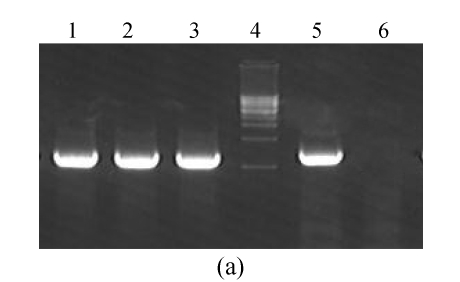
The GFP, GFPA and GFPB were introduced into M. oryzae Guy11 cells, respectively. From glufosinate-ammonium-containing media, 20 strains for each type of transformants were selected. Polymerase chain reaction (PCR) method and Southern blot were used to check the DNA integration and their copies. Except for 4 of them, all 60 selected strains contained the GFP fragment, and most of which were single-copy. The frequency of the single-copy integration was 83%. The results of transformants checking are partially shown in Fig.2. By similar means with G418 as the selection agent, the RFPA was introduced into a GFPB transformant to generate the peroxisome co-localized strains. As a control, RFP was also introduced to a GFP transformant.
Fig. 2.
PCR (a) and Southern blot (b) to check the GFP transformants. (a) GFP-Xb and GFPstop-Bm were used to amplify from the templates of p1300BMGFP (Lane 1, as positive control) and genomic DNAs of GFP (Lane 2), GFPA (Lane 3), GFPB (Lane 5) transformants and Guy11 strain (Lane 6, as negative control). Lane 4 is a 1-kb DNA marker; (b) The p1300BMGFP (Lane 1, as positive control) and genomic DNAs of GFP (Lanes 2 and 3), GFPA (Lanes 4 and 5), GFPB (Lane 6) transformant and Guy11 stain (Lane 7, as negative control) were digested with EcoRI and hybridized with GFP fragment as a probe
Peroxisomal co-localization of PTS1 and PTS2 fusions
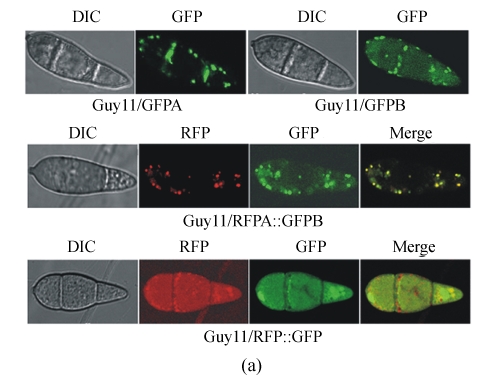
The single-copied transformants were primarily examined under the fluorescence microscope. The ones that express the fluorescence protein in high level and stably during subculture were chosen and observed under the laser confocal microscope. Under the control of MPG1 promoter, all GFP or RFP fusions were expressed in a high level. The fluorescence in conidia was stronger than that in other tissues. The fluorescence of GFPA, GFPB and RFPA showed a punctate pattern, comparing with that of GFP and RFP without PTS which dispersed in the whole cytoplasm (Fig.3a). In conidia, the fluorescence emerged more frequently at the periphery than the central region. In the co-transformed strains of RFPA and GFPB, the locations of red and green fluorescence were coincident strictly. These data indicate that the fluorescence proteins were transported from the cytoplasm into the peroxisomes exactly because of the existence of either PTS1 or PTS2.
Fig. 3.
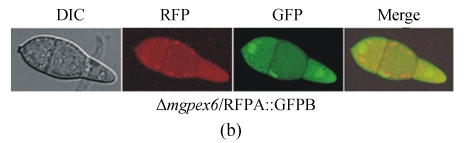
Fluorescence of the GFP, GFPA, GFPB, RFP and RFPA fusions. Conidia from an 8-day-old CM plate of each type of transformants were checked and recorded by the Leica SP2 Confocal System. (a) The transformants derived from wild type Guy11; (b) The transformants derived from Δmgpex6 mutant
Besides the dominant punctate fluorescence, the dispersed fluorescent backgrounds were also visible in conidia of GFPB transformants, which were stronger than those in conidia of GFPA transformants. The fluorescent backgrounds were considered from the fusion proteins remained in the cytoplasm which still had not been imported into peroxisomes immediately, so more protein residues would produce more cytoplasmic backgrounds. However, it was deducible that GFPB was expressed in equal amount as GFPA under the same promoter. Therefore, we concluded that the transport efficiency of PTS2 machinery was lower than that of PTS1 machinery.
Analysis of PTS2 pathway in Δmgpex6 mutant
To test the capability of these fusions to investigate gene functions, and also to learn the effect of MGPEX6 disruption to PTS2 pathway, we introduced the GFPB and RFPA into a Δmgpex6 mutant (Wang et al., 2007). The import of RFPA was blocked in Δmgpex6 mutant, and the red fluorescence dispersed in the whole cytoplasm. Similarly, the green fluorescence of GFPB also showed to be cytoplasmic (Fig.3b). These results indicate that, besides the PTS1, the PTS2 pathway in peroxisomal import was also blocked by the invalidity of MGPEX6.
Peroxisomal numbers under different conditions and numerical dynamic during appressorium formation
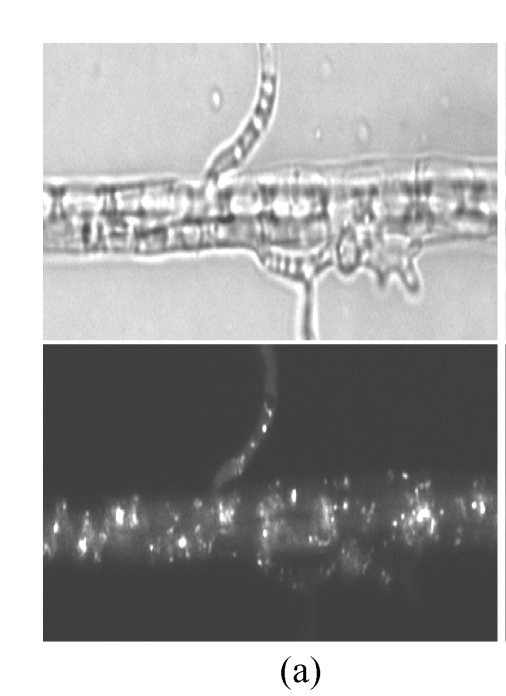
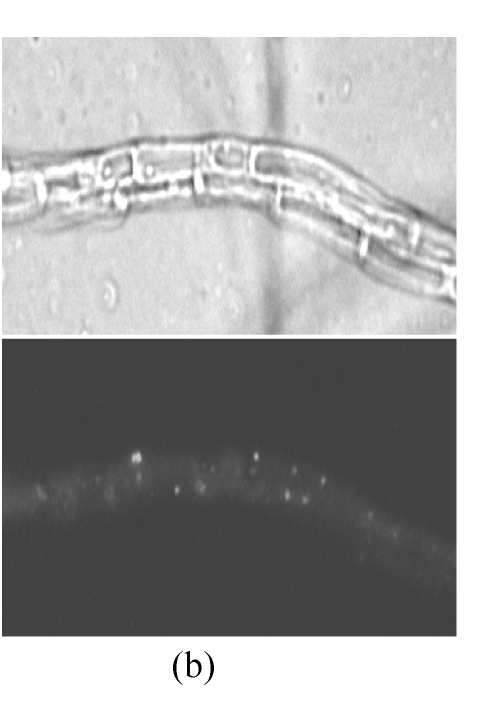
Fluorescent localization made it viable to survey the numbers of peroxisomes in cells. We compared the peroxisomal numbers in the hyphae induced in different culture media. The CM, 2 media with carbohydrate (GM and SM) and 2 with lipid as sole carbon source (OM and TM), were used to treat the hyphae of GFPA transformants. After induced in lipid media, the number of peroxisomes increased significantly, comparing with that unchanged on CM, GM or SM (Fig.4). Lipid degradation is one of the main metabolisms in peroxisome. These findings indicate that the number of peroxisomes was correlated to the intensity of the lipid metabolism in M. oryzae cells.
Fig. 4.
Peroxisomal numbers increase in lipid media. Hyphae from GFPA transformants were induced in (a) OM (olive oil media) and (b) GM (glucose media) respectively, and observed under fluorescence microscope. The top, bright image; the bottom, fluorescent image
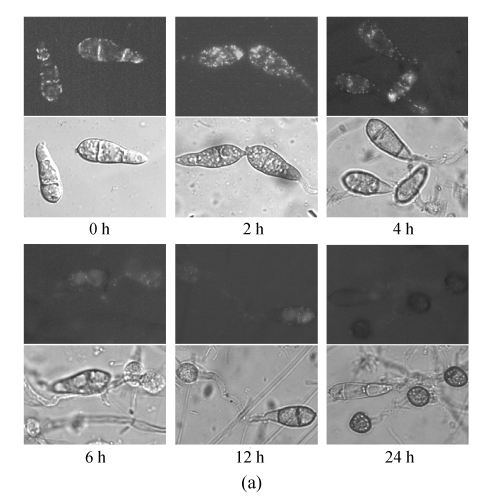
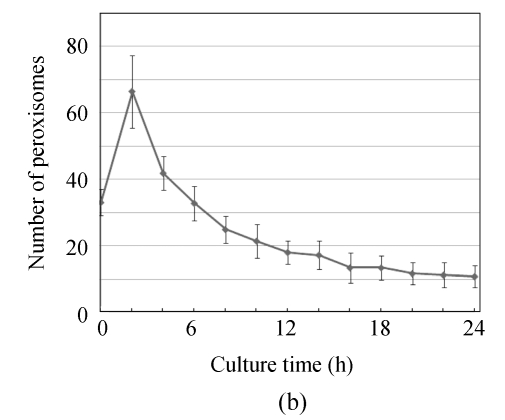
Considering that the number of peroxisomes reflected the lipid metabolism, we investigated the dynamic of peroxisomal number during the appressoria differentiation. In the conidia incubated for 0 h, many peroxisomes were observed clearly, indicating that the peroxisomal metabolism was required in the basic life activities in conidia (Fig.5). Incubated in 27 °C dewiness, the conidia were activated and prepared for the germination. The number of peroxisomes increased rapidly in this stage. To 2 h, along with germination, the total number of peroxisomes in conidia and germ tubes reached a peak. Then, the peroxisomes began to decrease and flowed with the cytoplasm along the germ tubes into the infant appressoria. To 6 h, the number of peroxisomes went back to the level of 0 h and decreased continuously in the later time. To 12 h, typical appressoria formed. Only a few of peroxisomes were found in conidia and appressoria. And up to 24 h, all the cytoplasms in the conidia were transferred into the appressoria. The conidia and germ tubes no longer contained any peroxisomes, and in the appressoria, peroxisomes were hardly found either.
Fig. 5.
Numerical dynamic of peroxisome during appressorium differentiation. Conidia from GFPA transformants were allowed to form appressoria on hydrophobic membrane; (a) Fluorescence of different time points (the top, fluorescent image; the bottom, bright image); (b) Numbers of peroxisomes (fluorescent puncta) of different time points
DISCUSSION AND CONCLUSION
Fluorescent localization is an effective strategy in the study of peroxisome biogenesis in yeast, mammals and plants (Mano et al., 2006; Stasyk et al., 2006). In M. oryzae, the fluorescent fusion with PTS1 has already been used to analyze the gene functions (Maggio-Hall and Keller, 2004; Ramos-Pamplona and Naqvi, 2006; Wang et al., 2007). For different purposes, the serials used as PTS1 and the promoters to control the fusion expression were different in these studies. For instance, the 164-aa (amine acid) residues at the C-terminal of FoxA were used to construct the RFP-PTS1 in Aspergillus nidulans (Maggio-Hall and Keller, 2004). And this fusion could also be imported into peroxisomes of M. oryzae (Wang et al., 2007). By searching in whole genome of M. oryzae, we found that SKL is most frequent PTS1 in M. oryzae. The SKL was therefore selected as PTS1 in this work. MPG1 is a gene expressed in a high level in conidia, hyphae and appressoria of M. oryzae (Talbot et al., 1993). Using of the MPG1 promoter ensured the expression of fusion in these tissues. We also tried to use the promoters of gpdA and trpC in this work but had not gotten an ideal result.
The application of PTS2 is less than that of PTS1 in the peroxisomal localization (Hayashi et al., 2005) and has yet not been reported in phytopathogenic fungi. However, the PTS2 fusions are indispensable for analysis of the factors in PTS2 pathway. Therefore, we constructed the GFP-PTS2 and RFP-PTS1 and co-localized them with PTS1 fusions. To ensure that the PTS2 is intact and to avoid that the fusion is too large, the 17 amine acids serial at N-terminal of thiolase in S. cerevisiae (GeneID: 854646), a well-defined PTS2 (Glover et al., 1994), was used. In the transformed cells of PTS1 or PTS2 fusions, the fluorescence showed a punctate pattern and mainly emerged in periphery of cells, which is consistent with the locations of peroxisomes in former studies. Further, the locations of PTS1 and PTS2 fluorescence overlapped exactly. These data indicate that SKL and the N-terminal serial of thiolase in S. cerevisiae were both efficient PTS in M. oryzae.
Most known peroxisomal matrix proteins were imported by PTS1 pathway, and only the seldom by PTS2 pathway. We therefore suppose that the efficiencies of PTS1 and PTS2 machineries may be different. To clarify this point, we compared the fluorescence in GFPA and GFPB transformants. Stronger fluorescence backgrounds were found in the GFPB transformants than in GFPA. The backgrounds were considered from the fusion proteins, which were expressed too much to be imported immediately and thus remained in the cytoplasm. However, it was deducible that the amount of GFPB expressed in the cells was the same as that of GFPA under the same promoter. Therefore, we concluded that the PTS1 machinery was more effective than PTS2 machinery. PEX6 gene encodes an AAA-type ATPase involved in peroxisome biogenesis with unclear function details (Yahraus et al., 1996). The involvement of PEX6 in PTS1 pathway and pathogencity processes were already demonstrated in M. oryzae and C. lagenarium. To clarify the contributions of PEX6 to PTS2 pathway and test the capability to apply the fusions in functional gene analysis, the localizations of both GFPA and RFPB in Δmgpex6 mutant were investigated. The data indicate that the invalidity of MGPEX6 blocked both the PTS1 and PTS2 pathways in M. oryzae.
Lipid degradation is one of the main metabolisms in peroxisomes, and the number of peroxisomes reflects the strength of lipid metabolism. It is reported that the peroxisomes proliferated when the hyphae or conidia were induced in lipid and some homological chemicals in yeast and mammals (Gray and La DeIglesia, 1984; Veenhuis et al., 1987). Our data indicate that peroxisomes were also inducible in Magnaporthe grisea. In the prophase of infection, the lipids in conidia degraded rapidly to supply the substance and energy required for penetration (Weber et al., 2001). In the present study, we found that the number of peroxisomes changed constantly in this process, which was consistent with the lipid degradation in conidia. After induction, the lipid began to degrade and the peroxisome started to proliferate. And to 2 h, the number reached a peak. Along with the germination and the appressorium differentiation, the lipid degradation became slower, the redundant peroxisomes began to be disassembled, and the number decreased gradually. And in the 24-h mature appressoria, the lipids were degraded almost completely and converted to glycerol, all peroxisomes and other organelles were disassembled, and the punctate fluorescence indicating the peroxisomes was hardly found. The dynamic of peroxisomal number gave an intuitional proof to the lipid metabolism during the pre-penetration processes.
In summary, by PTS1 and PTS2 localization and co-localization, we provide a useful tool to analyze the location, number and numerical dynamic of peroxisome, which is helpful to study the metabolism and related genes in peroxisome biogenesis, proliferation and degradation.
Footnotes
Project supported by the National Natural Science Foundation of China (Nos. 30671351 and 30810033) and the Natural Science Foundation of Zhejiang Province of China (No. Y306638)
Selected Papers of 2008 International Symposium on Fungal Diversity, Oct. 16~19, Hangzhou, China. The symposium is supported by the National Natural Science Foundation of China (No. 30671351)
References
- 1.Bhambra GK, Wang ZY, Soanes DM, Wakley GE, Talbot NJ. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea . Molecular Microbiology. 2006;61(1):46–60. doi: 10.1111/j.1365-2958.2006.05209.x. [DOI] [PubMed] [Google Scholar]
- 2.Bourett TM, Howard RJ. Actin in penetration pegs of the fungal rice blast pathogen, Magnaporthe grisea . Protoplasma. 1992;168(1-2):20–26. doi: 10.1007/BF01332647. [DOI] [Google Scholar]
- 3.Elgersma Y, Tabak HF. Proteins involved in peroxisome biogenesis and functioning. Biochim Biophys Acta Rev Biomembr. 1996;1286(3):269–283. doi: 10.1016/s0304-4157(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 4.Glover JR, Andrews DW, Subramani S, Rachubinski RA. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo . J Biol Chem. 1994;269(10):7558–7563. [PubMed] [Google Scholar]
- 5.Gould SJ, Valle D. Peroxisome biogenesis disorders: genetics and cell biology. TIG. 2000;16(8):340–345. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- 6.Gray RH, La DeIglesia FA. Quantitative microscopy comparison of peroxisome proliferation by the lipid-regulating agent gemfibrozil in several species. Hepatology. 1984;4(3):520–530. doi: 10.1002/hep.1840040328. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in arabidopsis. J Biol Chem. 2005;280(15):14829–14835. doi: 10.1074/jbc.M411005200. [DOI] [PubMed] [Google Scholar]
- 8.Hettema EH, Distel B, Tabak HF. Import of proteins into peroxisomes. Biochimica et Biophysica Acta. 1999;1451:17–34. doi: 10.1016/s0167-4889(99)00087-7. [DOI] [PubMed] [Google Scholar]
- 9.Howard RJ, Ferrari MA. Role of melanin in appressorium function. Experimental Mycology. 1989;13(4):403–418. doi: 10.1016/0147-5975(89)90036-4. [DOI] [Google Scholar]
- 10.Howard RJ, Ferrari MA, Roach DH, Money NP. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci. 1991;88(24):11281–11284. doi: 10.1073/pnas.88.24.11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson TL, Olsen L. Building new models for peroxisome biogenesis. Plant Physiol. 2001;127(3):731–739. doi: 10.1104/pp.127.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato A, Hayashi M, Kondo M, Nishimura M. Targeting and processing of a chimeric protein with the N-terminal presequence of the precursor to glyoxysomal citrate synthase. Plant Cell. 1996;8(9):1601–1611. doi: 10.1105/tpc.8.9.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura A, Takano Y, Furusawa I, Okuno T. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium . Plant Cell. 2001;13(8):1945–1957. doi: 10.1105/tpc.13.8.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1(1):489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 15.Maggio-Hall LA, Keller NP. Mitochondrial β-oxidation in Aspergillus nidulans . Molecular Microbiology. 2004;54(5):1173–1185. doi: 10.1111/j.1365-2958.2004.04340.x. [DOI] [PubMed] [Google Scholar]
- 16.Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. The Plant Journal. 2006;47(4):604–618. doi: 10.1111/j.1365-313X.2006.02809.x. [DOI] [PubMed] [Google Scholar]
- 17.Müller WH, van der Krift TP, Krouwer AJJ, Wosten HAB, van der Voort LHM, Smaal EB, Verkleij AJ. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum . EMBO J. 1991;10(2):489–495. doi: 10.1002/j.1460-2075.1991.tb07971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins ED, Romaine CP, Chen X, Geiser D, Raina R, Kang S. Agrobacterium tumefaciens-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology. 2001;91(2):173–180. doi: 10.1094/PHYTO.2001.91.2.173. [DOI] [PubMed] [Google Scholar]
- 19.Notteghem JL, Silue D. Distribution of the mating alleles of Magnaporthe grisea populations pathogenic on rice. Phytopathology. 1992;82(4):421–423. doi: 10.1094/Phyto-82-421. [DOI] [Google Scholar]
- 20.Pall ML, Brunelli JP. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet Newsl. 1993;40:59–62. [Google Scholar]
- 21.Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu Rev Cell Dev Biol. 2001;17(1):701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 22.Ramos-Pamplona M, Naqvi NI. Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Molecular Microbiology. 2006;61(1):61–75. doi: 10.1111/j.1365-2958.2006.05194.x. [DOI] [PubMed] [Google Scholar]
- 23.Rossman AY, Howard RJ, Valent B. Pyricularia grisea, the correct name for the rice blast disease fungus. Mycologia. 1990;82(4):509–512. doi: 10.2307/3760024. [DOI] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Soundararajan S, Jedd G, Li X, Ramos-Pamplon M, Chua NH, Naqvi NI. Woronin body function in Magnaporthe grisea is essential for efficient pathogenesis and for survival during nitrogen starvation stress. Plant Cell. 2004;16(6):1564–1574. doi: 10.1105/tpc.020677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stasyk OV, Stasyk OG, Mathewson RD, Farré JC, Nazarko VY, Krasovska OS, Subramani S, Cregg JM, Sibirny AA. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris . Autophagy. 2006;2(1):30–38. doi: 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- 27.Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiological Reviews. 1998;78(1):171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 28.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell. 1993;5(11):1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bosch H, Schutgens RBH, Wanders RJ, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61(1):157–197. doi: 10.1146/annurev.biochem.61.1.157. [DOI] [PubMed] [Google Scholar]
- 30.Veenhuis M, Mateblowski M, Kunau WH, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae . Yeast. 1987;3(2):77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- 31.Wang JY, Liu XH, Lu JP, Lin FC. Sequence analysis and expression pattern of MGTA1 gene in rice blast pathogen Magnaporthe grisea . J Zhejiang Univ Sci B. 2005;6(8):817–824. doi: 10.1631/jzus.2005.B0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang ZY, Thornton CR, Kershaw MJ, Li DB, Talbot NJ. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea . Molecular Microbiology. 2003;47(6):1601–1612. doi: 10.1046/j.1365-2958.2003.03412.x. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZY, Soanes DM, Kershaw MJ, Talbot NJ. Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. MPMI. 2007;20(5):475–491. doi: 10.1094/MPMI-20-5-0475. [DOI] [PubMed] [Google Scholar]
- 34.Weber RWS, Wakley GE, Thines E, Talbot NJ. The vacuole as central element of the lytic system and sink for lipid droplets in maturing appressoria of Magnaporthe grisea . Protoplasma. 2001;216(1-2):101–112. doi: 10.1007/BF02680137. [DOI] [PubMed] [Google Scholar]
- 35.Yahraus T, Braverman N, Dodt G, Kalish JE, Morrell JC, Moser HW, Valle D, Gould SJ. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 1996;15(12):2914–2923. [PMC free article] [PubMed] [Google Scholar]