Abstract
Agrobacterium tumefaciens-mediated transformation (ATMT) system was assessed for conducting insertional mutagenesis in Penicillium digitatum, a major fungal pathogen infecting post-harvest citrus fruits. A transformation efficiency of up to 60 transformants per 106 conidia was achieved by this system. The integration of the hph gene into the fungal genome was verified by polymerase chain reaction (PCR) amplification and sequencing. These transformants tested were also shown to be mitotically stable. Southern blot analysis of 14 randomly selected transformants showed that the hph gene was randomly integrated as single copy into the fungal genome of P. digitatum. Thus, we conclude that ATMT of P. digitatum could be used as an alternatively practical genetic tool for conducting insertional mutagenesis in P. digitatum to study functional genomics.
Keywords: Penicillium digitatum, Agrobacterium tumefaciens-mediated transformation (ATMT), Hygromycin B resistance gene, Insertional mutagenesis
INTRODUCTION
Several species in genus Penicillium are the causative agents of a number of post-harvest fruits and vegetables, and some species in this genus are important microbes for industries and environments. Penicillium digitatum and P. italicum causing green and blue molds, respectively, are the most economically important pathogens to post-harvest citrus and ubiquitous to all citrus growing regions. They cause a rapid breakdown of fruit punctured or bruised during harvesting and packing operations, usually resulting in 20% to 30% of the harvested citrus loss during storage, transportation and marketing in the case of no post-harvest fungicide treatment (Zhu et al., 2006). To control the infection, sodium o-phenylphenate, thiabendazole and imazalil have been commonly used as fungicides (Smilanick et al., 2006; Zhang, 2007), and some new fungicides, such as pyrimethanil, azoxystrobin and fludioxonil, have been approved for citrus post-harvest treatments for decay control (Zhang, 2007). However, the control efficiencies are often negatively affected by the emergences of the fungicide-resistant biotypes of these fungi (Holmes and Eckert, 1999).
Although it was known that the pectic enzymes (polygalacturonase (PG) and cellulase) produced by P. digitatum are responsible for the maceration of orange-rind tissue (Bush and Codner, 1968), and that a plant hormone ethylene produced by both this fungus and citrus is involved in susceptibility of citrus fruits to P. digitatum, virtually little has been known about the molecular mechanism of pathogenesis. For better understanding the genetic bases of pathogenicity and fungicide-resistance of this fungus, therefore, to facilitate the design of new strategies to control this disease, a readily accessible genetic tool to generate and tag mutants and recover the corresponding mutant genes is essentially prerequisite.
Here, we describe the procedure for the genetic transformation of conidia of P. digitatum by applying Agrobacterium tumefaciens-mediated transformation (ATMT) system using the hygromycin B resistant (hph) gene as a selected marker. To our best knowledge, this is the first report of the genetic transformation of P. digitatum by ATMT. The described protocol will be not only useful for identification and analysis of fungal genes required for pathogenicity and fungicide-resistance in P. digitatum, but also a reference for other species in Penicillium genus.
MATERIALS AND METHODS
Strains, plasmids, chemicals and culture conditions
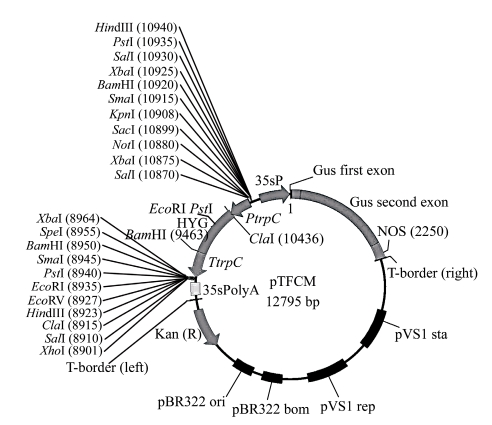
Penicillium digitatum strain Pd01, a single spore strain from an infected citrus fruit in a local storage house (Zhu et al., 2006), was used throughout this work. Fungal strains used for transformation and genomic DNA isolation were grown in a potato dextrose agar (PDA) at 25 °C in dark. Agrobacterium tumefaciens AGL1 strain was kindly provided by Dr. Zhonghua Ma (Zhejiang University, China) and was cultured in yeast extract peptone (YEP) (An et al., 1988), minimal medium (MM) (Hooykaas et al., 1979), and induction medium (IM) (Bundock et al., 1995), respectively, at 28 °C. Escherichia coli strain DH5α was cultured in Luria Bertani (LB) (Sambrock et al., 1989) at 37 °C. Plasmid pTFCM containing the T-DNA border repeat sequence and the hph driven by the Aspergillus nidulans trpC promoter and terminated by trpC terminator was generously given by Dr. Daohong Jiang (Huazhong Agricultural University, China) and maintained in E. coli (Fig.1).
Fig. 1.
Physical map of the transformation plasmid pTFCM
HYG: the hygromycin B resistant gene; PtrpC and TtrpC: promoter and terminator of Aspergillus nidulans, respectively
Sensitivity of P. digitatum to hygromycin B
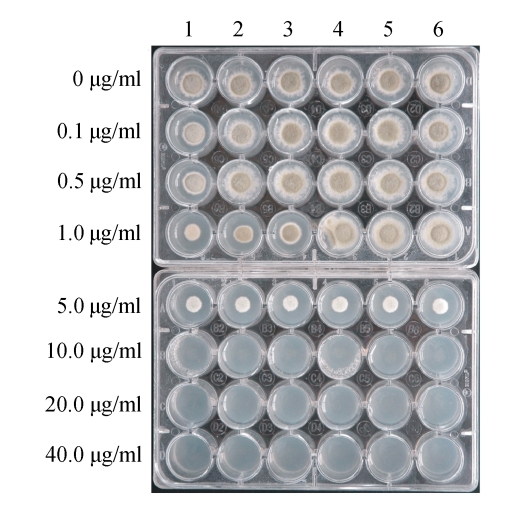
Six wild strains of P. digitatum isolated from local infected citrus were randomly selected for the evaluation of their sensitivities to hygromycin B. The canidial suspensions were prepared from the cultures grown in PDA for 7 d and diluted to 106 per ml. Two microliters of those suspensions were added to PDA medium with hygromycin B at various concentrations, which were allocated in a 24-well cell growth plate (Fig.2), and incubated at 25 °C. The growth was determined by scoring the formation of colony after 5~6 d. Culture with each hygromycin B concentration was replicated thrice and the experiment was repeated twice.
Fig. 2.
Assessment of sensitivity of Penicillium digitatum to hygromycin B
Lanes 1~6: 6 randomly selected P. digitatum wild strains. The concentrations of hygromycin B are given on the left
A. tumefaciens-mediated transformation
Initially, plasmid pTFCM was transformed into A. tumefaciens AGL1 strain by a heat-shock method (Bowyer, 2001). Cells of A. tumefaciens AGL1 strain carrying plasmid pTFCM were grown in MM supplemented with kanamycin (50 µg/ml) at 28 °C for 2 d, and then were collected by centrifugation at 3000 r/min, washed once with fresh IM, and finally diluted to OD 660 (optical density at 660 nm)=0.15 with IM with/without 200 µmol/L acetosyringone (IM+AS/IM−AS). After growing for an additional 6 h, the bacterial cells were mixed with an equal volume of conidial suspensions with concentrations of 1×104 to 1×107 per ml. The 200 µl mixture was spread onto a chemistry analysis qualitative filter paper that was plated on agar plate containing the co-cultivation medium (the same as IM+AS or IM−AS, but the glucose concentrations was 5 mmol/L instead of 10 mmol/L). After co-cultivation at 25 °C for 2 d, the filters were transferred to PDA medium containing 75 µg/ml hygromycin B and 50 µg/ml cefoxitin to select fungal transformants and to kill A. tumefaciens cells. After an additional incubation for 3~4 d, individual colonies were transferred to hygromycin B-containing PDA.
Mitotic stability of transformants
To determine the mitotic stability of inserted T-DNA in the transformants, putative transformants were cultured on PDA for five successive generations (7 d for a generation) in the absence of hygromycin B, and thereafter, the transformants were tranferred to PDA with 100 µg/ml hygromycin B.
Analysis of transformants
Genomic DNA of P. digitatum was isolated as described by Raeder and Broda (1985). Polymerase chain reaction (PCR) was used to confirm the presence of T-DNA by amplifying an internal 0.6 kb region of the hph gene of T-DNA using primers hph1 (5′-TTC GAT GTA GGA GGG CGT GGA T-3′) and hph2 (5′-CGC GTC TGC TGC TCC ATA CAA G-3′) (Irie et al., 2001), whereas Southern blots were performed to determine the frequency and randomness of T-DNA integration. PCR reaction consisted of approximately 0.1~2 µg of genomic DNA, 2.5 U Taq DNA polymerase, 10 µl 10× polymerase buffer, 1.5 µmol/L MgCl2, 200 µmol/L dNTP and 0.5 µmol/L each primer, adding ddH2O to 100 µl. The cycling conditions were as follows: an initial denaturation of 5 min at 94 °C, followed by 35 cycles of 45 s for denaturation at 92 °C, 1 min for annealing at 60 °C, and 1 min for polymerization at 72 °C, with a final extension of 72 °C for 10 min (Malonek and Meinhardt, 2001). Amplified products were compared to a product amplified from plasmid pTFCM on 0.5% (w/v) agarose gel.
For Southern blot analysis genomic DNA (60~100 µg) was digested with KpnI for 8 h, fragments were separated on a 0.5% (w/v) agarose gel, and transferred to a Nylon membrane (Millipore, Billerica, MA, USA). The probes were labelled by PCR using the primers of hph1 and hph2 with the PCR DIG probe synthesis kit I (Roche, Mannheim, Germany), according to the manufacturer’s instruction manual. Aqueous hybridizations were performed at 42 °C overnight and detected using the DIG Luminescent Detection Kit (Roche) according to the manufacturer’s instruction.
Analysis of regions flanking T-DNA insertions in transformants
Thermal asymmetric interlaced PCR (TAIL-PCR) (Mullins et al., 2001; Rooney et al., 2001) was used to identify genomic DNA sequences flanking T-DNA insertions for 14 randomly selected transformants. Genomic DNA was used as template in successive reactions using left border-specific primers LB1, LB2, LB3 and right border-specific primers RB1, RB2, RB3, and arbitrary degenerate primers AD1, AD2 and AD3, as described by Rooney et al.(2001) and Sambrock et al.(1989). The sequences of primers were listed in Table 1.
Table 1.
Sequences of the primers used for this research
| Primer name | Nucleotide sequence (5′ to 3′) | Tm (°C) |
| hph1 | TTCGATGTAGGAGGGCGTGGA | 58 |
| hph2 | CGCGTCTGCTGCTCCATACAAG | 58 |
| LB1 | GGGTTCCTATAGGGTTTCGCTCATG | 64 |
| LB2 | CATGTGTTGAGCATATAAGAAACCCT | 60 |
| LB3 | GAATTAATTCGGCGTTAATTCAGT | 56 |
| RB1 | GGCACTGGCCGTCGTTTTACAAC | 64 |
| RB2 | AACGTCGTGACTGGGAAAACCCT | 62 |
| RB3 | CCCTTCCCAACAGTTGCGCA | 61 |
| AD1 | WGTGNAGWANCANAGA | 45 |
| AD2 | NGTCGASWGANAWGAA | 47 |
| AD3 | NTCGASTWTSGWGTT | 45 |
RESULTS AND DISCUSSION
Sensitivity of P. digitatum to hygromycin B
To determine whether the hph gene could be used as a selection marker during the transformation, the sensitivities of 6 randomly selected wild isolates of P. digitatum were evaluated in PDA medium with various concentrations of hygromycin B before the transformation experiment. As shown in Fig.2, the growth of all isolates of P. digitatum was completely inhibited in media with hygromycin B up to 10 µg/ml. The formation of colonies did not occur even on Day 10 of inoculation. No significant difference of sensitivity was found among the tested isolates. Thus, hygromycin B was considered to be a suitable marker to select putative transformants in ATMT. To exclude the unstable transformants, PDA containing 75 µg/ml hygromycin B was routinely used in transformation experiment.
A. tumefaciens-mediated transformation in P. digitatum
To determine if acetosyringone (AS) was essential component for ATMT in P. digitatum, transformations were conducted in AS-present liquid IM for pre-cultivation of A. tumefaciens followed by AS-present solid IM for co-cultivation with P. digitatum (104~107 per ml), and the same was done for AS-absent IM. The result revealed that the hygromycin B-resistant colonies only recovered when AS was present in media for both pre- and co-cultivation, followed by growing in PDA medium containing hygromycin B for 4 d in 3 independent transformation experiments (Table 2). The number of resistant colonies generated ranged from 35 to 60 per 106 conidia in 3 independent experiments (Table 2). Transformation efficiency obviously decreased, when the conidial concentration decreased from 1×106 to 1×104 or 1×105 conidia per ml. However, increasing the conidial concentration from 1×106 to 1×107 per ml did not significantly increase the transformation efficiency.
Table 2.
Efficiency of Agrobacterium tumefaciens-mediated genetic transformation of Penicillium digitatum
| No. | +AS/−AS* | Number of transformants from suspensions at different conidial concentrations |
Mitotically stable transformants (%) | |||
| 1×104 ml−1 | 1×105 ml−1 | 1×106 ml−1 | 1×107 ml−1 | |||
| 1 | +AS | 5 | 21 | 48 | 52 | 97.6 |
| −AS | 0 | 0 | 0 | 0 | ||
| 2 | +AS | 3 | 19 | 35 | 43 | 100 |
| −AS | 0 | 0 | 0 | 0 | ||
| 3 | +AS | 3 | 15 | 60 | 61 | 97.1 |
| −AS | 0 | 0 | 0 | 0 | ||
+AS: AS-present medium; −AS: AS-absent medium
Analysis of fungal transformants
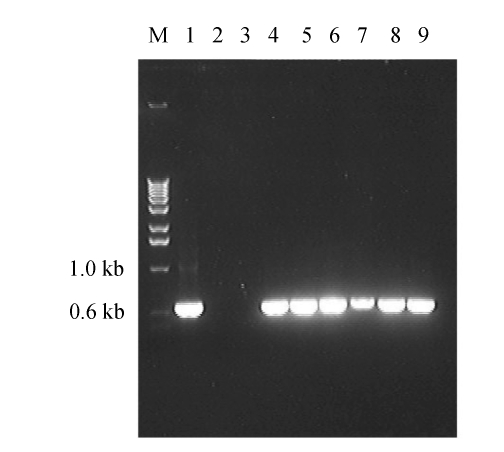
To confirm the genetic stability of the transformants, 6 randomly selected putative transformants were grown in hygromycin B-free PDA for 5 successive generations (7 d for one generation), then transferred to hygromycin B (100 µg/ml)-containing PDA. The results show that no discernible difference for hygromycin resistance was found between the transformants grown on hygromycin B-free medium and those grown on hygromycin-containing medium, suggesting that these transformants were mitoticaly stable. The stability of the putative transformants was also confirmed by PCR detection in presence of hph gene (Fig.3). As shown in Fig.3, the expected 0.6-kb fragments were detected in all the 6 transformants randomly chosen, but not in parental strain Pd01. The presence of hph gene in these transformants was further confirmed by subsequent sequencing.
Fig. 3.
PCR analysis of transformed P. digitatum was confirmed by amplifying the hph gene
Lane 1: pTFCM plasmid; Lane 2: wild strain Pd01; Lane 3: water; Lanes 4~9: transformants T1, T2, T3, T4, T5 and T6, respectively; M: the molecular markers
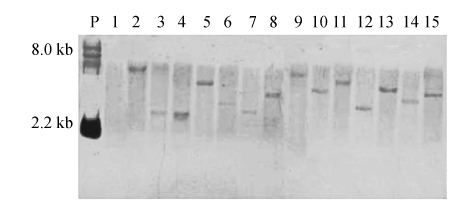
The copy number of T-DNA integration in P. digitatum was determined by Southern hybridization using partial of the hph gene as a probe. Genomic DNA isolated from 14 randomly selected transformants was digested with KpnI for 8 h. All of the analyzed transformants produced a single hybridizing fragment (Fig.4), suggesting a single T-DNA insertion. Out of the 14 transformants, 12 were detected to have distinct sizes (Fig.4), suggesting unique insertion positions. Two of the transformants (Lanes 3 and 4) produced hybridizing fragments of approximately the same size. But, sequencing demonstrated that they were inserted into the different genomic positions.
Fig. 4.
Southern blot analysis of genomic DNA of transformants of Penicillium digitatum
DNA was digested with KpnI and probed with hph. Lane 1: parental strain Pd01; Lanes 2~15: 14 randomly selected transformants; P: the plasmid pTFCM digested with HindIII
TAIL-PCR was used to isolate the genomic DNA of P. digitatum flanking the site of T-DNA insertion with a left or right border-specific primer in combination with a degenerate primer. Three rounds of PCR were performed and border region amplification was enriched using three different and increasingly internal left and right border primers (LB1, LB2, LB3 or RB1, RB2, RB3) in combination with a degenerate primer (AD1, AD2 or AD3). Products from the third round of TAIL-PCR were sequenced if the predicted sizes shift between the second and third rounds of amplification had occurred. Sequence analysis of the 12 TAIL-PCR products suggests that the insertion of T-DNA within the host genome appears to be a random event (Table 3). The host DNA sequences flanking the insertion sites showed no significant similarity to each other or to the sequences available in databases.
Table 3.
Sequence analysis of T-DNA junctions
| No. | Left border | Right border |
| 0 | CCGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGACAGGGATATATT |
| 1 | ttagacctatgacgtgtaGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAattatccgttgtgttgac |
| 2 | actaccttcttttacttggtGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAccggaagatgtcgatg |
| 3 | aggaccgagaggaggGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAacggcaaacaacccg |
| 4 | cgacccatacccgttagGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGActcttaaaaggtcttcc |
| 5 | agagctcgaactatagtGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGActgtcgaactgtccac |
| 6 | gtggatactgaagagtcGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAcaggggggctctgga |
| 7 | ctctttcccttactacagGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAgacggaatcattgcc |
| 8 | aggctgccacgaaggGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAtcgatcagcgttacag |
| 9 | cccgttatcttgacctaaGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAgcgacaggaatcttttc |
| 10 | cttgtctagtttattataGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAacataggccatcaaca |
| 11 | ctacccgtggtataacGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAagaaggacgaagaga |
| 12 | aactcgtgctgattgggGTCCTATATAACACCACATTTGtttaactg- | -ccgccttcagtttaaactatcagtgtttGAacgttgaggctggaag |
Nos. 1~12: partial nucleotide sequences of TAIL-PCR products obtained from 12 randomly selected transformants were aligned; No. 0: the left and the right border sequences of T-DNA from plasmid pTFCM
CONCLUSION
The efficient transformation system is an essential tool for gene manipulation and studying the functional genomics of P. digitatum, a most important pathogen of post-harvest citrus worldwide. The transformation via protoplast was successfully used for the P. digitatum in several studies (Nakaune et al., 1998; 2002; Hamamoto et al., 2001). The ATMT via germinated conidia could avoid the preparation of protoplast, which is usually expensive and time-consuming (Song et al., 2004).
Random integration of a single copy into the fungal genome is another important character for functional genomic study (Morioka et al., 2006). In this study, the hph was demonstrated to be randomly integrated in P. digitatum genome with single copy by Southern blot and sequencing of 14 randomly selected transformants. In addition, the efficiency of transformation of P. digitatum could be achieved up to 60 transformants per 106 conidia by ATMT. These results make ATMT a very feasible alternative for gene manipulation of this economically important fungus.
Acknowledgments
We gratefully acknowledge Dr. Xiaoguang Liu (University of Texas A & M, USA) for critical reading of the manuscript. We would particularly like to thank Dr. Zhonghua Ma (Zhejiang University, China) and Dr. Daohong Jiang (Huazhong Agricultural University, China) for providing the strain of Agrobacterium tumefaciens and plasmid pTFCM.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30571236), the Modern Agriculture (Citrus) Technology System (MATS) of China, and the Science and Technology Department of Zhejiang Province, China (No. 2007C22007)
Selected Papers of 2008 International Symposium on Fungal Diversity, Oct. 16~19, Hangzhou, China. The symposium is supported by the National Natural Science Foundation of China (No. 30671351)
References
- 1.An G, Ebert PR, Mitra A, et al. Binary Vectors. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Great Britain: Kluwer Academic Publishers; 1988. pp. 1–19. [Google Scholar]
- 2.Bowyer P. DNA-mediated Transformation of Fungi. In: Talbot N, editor. Molecular and Cellular Biology of Filamentous Fungi. Oxford: Oxford Univ. Press; 2001. pp. 33–46. [Google Scholar]
- 3.Bundock P, Dulk-Ras A, Beijersbergen AGM, Hooykaas PJJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Ssaccharomyces cerevisiae . EMBO J. 1995;14(13):3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush DA, Codner RC. The nature of macerating factor of Penicillium digitatum saccardo. Phytochemistry. 1968;7(5):863–869. doi: 10.1016/S0031-9422(00)84844-X. [DOI] [Google Scholar]
- 5.Hamamoto H, Nawata O, Hasegawa K, Nakaune R, Lee YJ, Makizumi Y, Akutsu K, Hibi T. The role of the ABC transporter gene PMR1 in demethylation inhibitor resistance in Penicillium digitatum . Pest Biochem Physiol. 2001;70(1):19–26. doi: 10.1006/pest.2001.2530. [DOI] [Google Scholar]
- 6.Holmes GJ, Eckert JW. Sensitivity of Penicilium digitatum and P. italicum to postharvest citrus fungicides in California. Phytopathology. 1999;89(9):716–721. doi: 10.1094/PHYTO.1999.89.9.716. [DOI] [PubMed] [Google Scholar]
- 7.Hooykaas PJJ, Roobol C, Schilperoort RA. Regulation of the transfer of Ti-plasmids of Agrobacterium tumefaciens . J Gen Microbiol. 1979;110(1):99–109. [Google Scholar]
- 8.Irie T, Honda Y, Watanabe T, Kuwahara M. Efficient transformation of the filamentous fungus Pleurotus ostreatus using single-strand carrier DNA. Appl Microbiol Biotechnol. 2001;55(5):563–565. doi: 10.1007/s002530000535. [DOI] [PubMed] [Google Scholar]
- 9.Malonek S, Meinhardt F. Agrobacterium tumefaciens mediated genetic transformation of the phytopathogenic ascomycete Calonectria morganii . Curr Genet. 2001;40(2):152–155. doi: 10.1007/s002940100236. [DOI] [PubMed] [Google Scholar]
- 10.Morioka LRI, Furlaneto MC, Bogas AC, Pompermayer P, Duarte RTD, Vieira MLC, Watanabe MAE, Fungaro MHP. Efficient genetic transformation system for the ochratoxigenic fungus Aspergillus carbonarius . Curr Microbiol. 2006;52(6):469–472. doi: 10.1007/s00284-005-0402-6. [DOI] [PubMed] [Google Scholar]
- 11.Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S. Agrobacterium tumefaciens-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology. 2001;91(2):173–180. doi: 10.1094/PHYTO.2001.91.2.173. [DOI] [PubMed] [Google Scholar]
- 12.Nakaune R, Adachi K, Nawata O, Tomiyama M, Akutsu K, Hibi T. A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum . Appl Environ Microbiol. 1998;64(10):3983–3988. doi: 10.1128/aem.64.10.3983-3988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakaune R, Hamamoto H, Imada J, Akutsu K, Hibi T. A novel ABC transporter gene, PMR5, is involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum . Mol Genet Genomics. 2002;267(2):179–185. doi: 10.1007/s00438-002-0649-6. [DOI] [PubMed] [Google Scholar]
- 14.Raeder U, Broda P. Rapid prepraration of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1(1):17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x. [DOI] [Google Scholar]
- 15.Rooney PJ, Sullivan TD, Klein BS. Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism. Mol Microbiol. 2001;39(4):875–889. doi: 10.1046/j.1365-2958.2001.02300.x. [DOI] [PubMed] [Google Scholar]
- 16.Sambrock J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Smilanick JL, Mansour MF, Sorensen D. Pre- and postharvest treatments to control green mold of citrus fruit during ethylene degreening. Plant Dis. 2006;90(1):89–96. doi: 10.1094/PD-90-0089. [DOI] [PubMed] [Google Scholar]
- 18.Song AH, Li HY, Liu XH. Isolation and regeneration of protoplasts from Penicillium digitatum . Chin J Agric Biotechnol. 2004;1(3):197–202. doi: 10.1079/CJB200442. [DOI] [Google Scholar]
- 19.Zhang JX. The potential of a new fungicide fludioxonil for stem-end rot and green mold control on Florida citrus fruit. Postharvest Biol Technol. 2007;46(3):262–270. doi: 10.1016/j.postharvbio.2007.05.016. [DOI] [Google Scholar]
- 20.Zhu JW, Xie QY, Li HY. Occurrence of imazalil-resistant biotype of Penicillium digitatum in China and the resistant molecular mechanism. J Zhejiang Univ Sci A. 2006;7(2):362–365. doi: 10.1631/jzus.2006.AS0362. [DOI] [Google Scholar]