Abstract
Quantitative trait loci (QTLs) analysis has been used to examine natural variation of phenotypes in the mouse somatosensory cortex, hippocampus, cerebellum, and amygdala. QTL analysis has also been utilized to map and identify genes underlying anatomical features such as muscle, organ, and body weights. However, this methodology has not been previously applied to identification of anatomical structures related to gustatory phenotypes. In this study, we used QTL analysis to map and characterize genes underlying tongue size, papillae number, and papillae area. In a set of 43 BXD recombinant inbred (RI) mice (n = 111) and 2 parental strains (C57BL/6J and DBA/2J; n = 7), we measured tongue length, width, and weight. In a subset of 23 BXD RI mice and the parental mice, we measured filiform and fungiform papillae number and fungiform papillae area. Using QTL linkage analysis (through WebQTL), we detected 2 significant and noninteracting QTLs influencing tongue length on chromosomes 5 and 7. We also found a significant QTL on chromosome 19 underlying fungiform papillae area and a suggestive QTL on chromosome 2 linked to fungiform papillae number. From these QTLs, we identified a number of candidate genes within the QTL intervals that include SRY-box containing gene, nebulin-related anchoring protein, and actin-binding LIM protein 1. This study is an important first step in identifying genetic factors underlying tongue size, papillae size, and papillae number using QTL analysis.
Keywords: BXD, C57BL/6J, DBA/2J, filiform papillae, fungiform papillae, genes, interval mapping, QTL analysis, taste buds, tongue
Introduction
Quantitative trait loci (QTL) analysis is an effective way of relating positions in the genome with variation in a phenotype (Williams 1998). QTL analysis has been applied to map variation of phenotypes in the central nervous system, such as somatosensory cortical barrel field size (Li et al. 2005); hippocampal structure, volume, and cell number (Lu et al. 2001; Peirce et al. 2003); and volume and cell number in basolateral amygdala (Mozhui et al. 2007). In addition, QTL analysis has also been applied to map and characterize genes underlying anatomical variation such as muscle weight (Lionikas et al. 2005) and body and organ weights in mice (Neuschl et al. 2007) and pigs (Zhang et al. 2007). Although QTL analysis has also been used to characterize gustatory behaviors (Phillips et al. 1994; Bachmanov et al. 1997; Blizard et al. 1999; Inoue, Beauchamp, and Bachmanov 2004; Nelson et al. 2005), it has not been used to study the genetic control of variation in tongue size or the size, number, and distribution of papillae. Such a quantitative genetic approach exploiting natural variation has the potential to uncover gene variants that influence gustatory development and behavior (Barlow 2000; Nosrat et al. 2004; Mistretta and Liu 2006; Krimm 2007).
The tongue contains several types of epithelial structures, termed papillae. Among these, fungiform, foliate, and vallate papillae all contain taste buds and are known to be involved in taste perception. Another class of papillae, filiform, are numerous and found on the anterior tongue; these papillae do not possess taste buds and are reported to be involved in tactile sensation (Bradley 1971; Miller and Smith 1984; Finger and Simon 2000). Fungiform number has been implicated in taste behavior in mice (Miller and Whitney 1989) and humans (Bartoshuk et al. 1994). Therefore, fungiform and filiform papillae were chosen as representative phenotypes to be studied.
QTL analysis has been greatly aided by examining recombinant inbred (RI) strains of mice created from progenitor strains (C57BL/6J and DBA/2J). The resulting inbred strains, known as the BXD panel, possess a set of homozygous genotypes at each locus, with parental alleles segregated among the strains (Taylor 1989; Peirce et al. 2004). To date, 89 strains have been genotyped at a density of over 13 000 single nucleotide polymorphisms (SNPs) allowing high precision mapping of chromosomal, loci influencing phenotypes (Peirce et al. 2004; also see www.genenetwork.org).
In the present study, we measured tongue size in 43 BXD RI strains of mice and papillae size and numbers in 23 strains as a first step in identifying chromosomal loci and candidate genes that might underlie anatomical variation involved in taste and oromotor function. We identified 2 significant QTLs for tongue length on chromosomes 5 and 7, a suggestive QTL on chromosome 2 for fungiform papillae number, and a significant QTL on chromosome 19 for fungiform papillae area. We highlight a number of genes in these intervals that we regard as interesting candidates.
Materials and methods
Animals
In this study, we utilized a total of 118 mice, which included 43 BXD RI strains (n = 111, mean = 2.58 mice per strain) and 2 parental strains (C57BL/6J and DBA/2J, n = 7). A nearly equal numbers of males and females were used. All animals were examined between 37 and 57 days of age (mean ± standard error of the mean = 45.00 ± 0.30). Tongue dimension measurements were collected from all 43 BXD RI strains, whereas papillae measurements were collected from 23 (n = 55, 2.39 mice per strain) of the total 43 BXD RI strains. Both tongue dimension and papillae measurements were also collected from the parental strains. All strains were inbred (>F20) at the time of measurement except 4: BXD55 (F18), BXD56 (F14), BXD80 (F19), and BXD83 (F17). In addition, 3 of the strains (BXD71, BXD77, and BXD95) contained individuals both below and above the 20-generation plateau.
All animals were maintained in the University of Tennessee Animal Facility at a temperature of 22 °C on a 12-h light/dark cycle with 35–40% humidity in a specific pathogen-free environment. Animals were given a diet of 5% fat Agway Prolab 3000 rat and mouse chow. All experimental procedures were performed in accordance with Principles of Laboratory Animal Care (NIH publication No. 86-23, revised 1985) and were approved by the Institutional Animal Care and Use Committee at the University of Tennessee Health Science Center. The Animal Care Facility is approved by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Tongue removal and dimension analysis
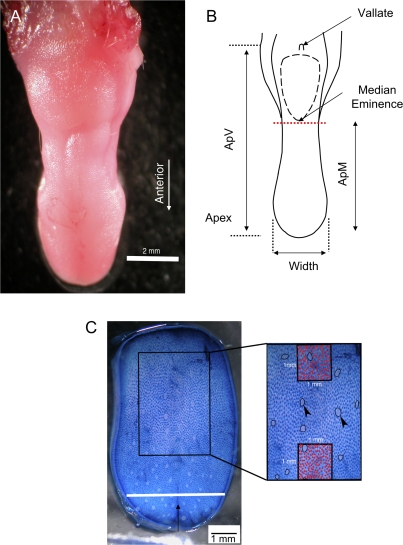
Each mouse was euthanized and weighed. The tongue was excised at the level of the trachea and placed on a glass slide. Three length measurements were made under a dissecting microscope using a millimeter ruler: apex to vallate (ApV), apex to median eminence (ApM), and tongue width. Using a millimeter scale, tongue width was measured at the point where the anterior tongue began to curve to form the tongue tip. All measurements were made by a single investigator (D.J.R.). Following these measurements, the anterior tongue was sectioned at the rostral-most point of the median eminence and weighed on an analytical balance (Figure 1). The anterior sectioned tongue was then stained with 0.5% methylene blue for approximately 90 s. Fungiform papillae were counted on both sides of the median fissure. To accomplish this, the tongue was placed on a glass slide and flattened slightly to visualize all fungiform papillae. Using a dissecting microscope, the fungiform papillae were then counted. The tongue was then placed on its ventral sides to count any fungiform papillae that were not observed when flattened on the glass slide. A digital photograph of the anterior tongue surface was then taken and used to measure fungiform area and filiform number.
Figure 1.
Mouse tongue morphology. (A) Photomicrograph of a mouse tongue. (B) Line drawing reconstruction of the tongue from (A) showing the location of the vallate papillae and median eminence. Measurements where ApV, ApM, and width were taken are indicated with solid-headed arrows. The dashed red line marks the location where the tongue was sectioned for weighing. (C) Photomicrograph of a methylene blue–stained tongue surface showing location of the region (rectangle) where fungiform and filiform papillae numbers were measured. Inset displays the 2 regions where filiform papillae (red dots) numbers were counted. Arrowheads show examples of fungiform papillae.
Papillae analysis
All images of the tongue surface were transferred to Adobe Photoshop for papillae measurements. A rectangle was then superimposed over the tongue surface and used to demarcate an area for filiform number and fungiform area measurement (Figure 1). The location and size of the rectangle were standardized in order to control for variation of tongue size. The rectangle was placed at 15% of the ApM length anterior to the median eminence. Lateral and vertical dimensions were both 60% of the overall width and ApM distance measurements, respectively. Fungiform papillae in the rectangle were outlined and their area measured using NIH ImageJ 1.33u. In addition, two 1-mm2 squares were placed medially at the anterior and posterior boundaries of the rectangle; filiform papillae number was counted within the squares. Filiform papillae counts were corrected for the presence of fungiform papillae within the squares using simple density and area calculations (filiform papillae number × area of fungiform).
Papillae asymmetry
We counted the number of filiform papillae in the anterior and posterior regions of the tongue. After correcting for the presence of fungiform papillae, anterior and posterior filiform papillae numbers were summed, resulting in a total filiform papillae number for each case. However, a paired t-test showed that there was a significant difference (P = 2.53 × 10−6) between the anterior and posterior filiform papillae, and the 2 are highly correlated (r = 0.85). Therefore, we combined anterior and posterior regions and conducted data analyses and QTL mapping on total filiform papillae number. Because this trend persisted in all mice studied, we only used the total filiform papillae number in the data analyses and QTL mapping.
There is no significant difference between the number of fungiform papillae on the right and left sides of the median fissure (P = 0.75). Thus, these 2 values were added to yield a total fungiform papillae number.
Data analysis and modeling
All measurements were entered in an Excel spreadsheet. Data analyses were performed using Data Desk 6.1 and Excel. ApM, ApV, width, and tongue weight were examined in 43 BXD RI strains. Filiform number and fungiform papillae number and area were examined in a subset (n = 23) of the 43 BXD RI strains. Parental strains were also measured. We utilized a linear model to test the effects of factors that may influence the trait under consideration. All variables, including sex, age, body weight, and the respective phenotypes of each group were taken into consideration. We eliminated insignificant factors (P > 0.05) from our model in descending order until only significant ones remained. The remaining factors were then used to calculate the residual from the linear models, and adjusted values were obtained by adding the residuals of each group to the mean of the raw phenotype data of all the groups. Based on the results of the linear modeling, tongue weight was adjusted for body weight and tongue length (ApV and ApM). ApV was adjusted for tongue weight and ApM. Fungiform papillae number and area were not adjusted because there were no significant factors in the linear models.
Heritability
Broad-sense heritability provides an approximation of the total variance of phenotypes due to genetic factors. We measured broad-sense heritability by comparing between-strain and total differences using the method outlined by Hegmann and Possidente (1981) in which h2 = VA/(VA + 2VE), where VA = genetic variance and VE = environmental variance.
QTL mapping
QTL analysis classifies strains based on their genotypes at discrete chromosomal markers and compares these strains with a quantitative phenotypic variable, in this case, ApV (tongue length), fungiform, and filiform papillae number. If there is a strong association between the differences in phenotype and genotype, a QTL will be detected (Lu et al. 2001).
We used raw and adjusted tongue length and papillae data from BXD RI strains to map potential QTLs. QTL maps of body weight and tongue weight were also generated. We report loci with genome-wide significance and those considered suggestive based on 1000 permutation tests. (Permutation tests randomly reassign [permute] trait values across the strains; this serves to redistribute gene-to-phenotype relationships. Comparisons are then made between permutated data and original data, which are used to determine the empirical significance of the QTL.) QTL maps were generated in this study using conventional interval mapping and marker regression–based methods. Interval mapping evaluates the significance of a hypothetical QTL at regular intervals across the genome even at points where the genotype data are sparse. Trait values are compared with the probability that a specific genotype exists at a specified location. Marker regression expresses the relationship between the differences in a trait and differences in alleles at a single marker and can be computed using a regression analysis of genotype versus phenotype. The likelihood ratio statistic (LRS), a chi-square statistic, provides a measure of the linkage between variation in the phenotype and genetic differences at a specific genetic locus, and this was used to identify genome-wide significant QTLs. Logarithmic of odds (LOD) values can be obtained by dividing the LRS values by 4.6. All QTL maps were generated using WebQTL (www.genenetwork.org). Bootstrap analysis resamples the original data set and is used to evaluate the approximate confidence limits of QTL peaks.
Results
Tongue dimensions and papillae
Table 1 contains tongue dimension data (tongue lengths, width, and weight) from all 43 BXD and parental strains. Table 2 contains fungiform and filiform papillae data from 23 BXD and parental strains.
Table 1.
Age, body weight, raw, and adjusted tongue measurements
| Strain | Total cases (n = male) | Age (days) | Body weight (g) | ApM (mm) | ApV (mm) | Adjusted ApV (mm) | Tongue width (mm) | Tongue weight (g) | Adjusted tongue weight |
| BXD1 | 2 (1) | 54.00 ± 3.00 | 19.26 ± 1.90 | 6.00 ± 0.00 | 10.05 ± 0.05 | 10.08 ± 0.40 | 3.40 ± 0.60 | 0.0338 ± 0.0027 | 0.0345 ± 0.0062 |
| BXD2 | 2 (1) | 47.00 ± 1.00 | 20.30 ± 2.75 | 6.15 ± 0.85 | 9.80 ± 0.70 | 9.76 ± 0.43 | 3.95 ± 0.05 | 0.0339 ± 0.0055 | 0.0339 ± 0.0028 |
| BXD9 | 2 (1) | 44.50 ± 0.50 | 19.37 ± 1.64 | 6.65 ± 0.25 | 10.65 ± 0.45 | 10.23 ± 0.80 | 4.10 ± 0.00 | 0.0399 ± 0.0012 | 0.0372 ± 0.0022 |
| BXD12 | 4 (2) | 46.00 ± 1.15 | 18.70 ± 0.39 | 6.08 ± 0.37 | 10.2 ± 0.18 | 10.21 ± 0.52 | 3.95 ± 0.09 | 0.0332 ± 0.0001 | 0.0339 ± 0.0032 |
| BXD14 | 2 (1) | 45.50 ± 0.50 | 22.46 ± 1.39 | 6.65 ± 0.35 | 11.60 ± 0.40 | 11.13 ± 0.59 | 4.65 ± 0.15 | 0.0408 ± 0.0025 | 0.0356 ± 0.0051 |
| BXD32 | 3 (2) | 42.00 ± 1.00 | 21.03 ± 1.10 | 5.87 ± 0.14 | 10.67 ± 0.44 | 10.77 ± 0.65 | 3.73 ± 0.37 | 0.0334 ± 0.0032 | 0.0324 ± 0.0046 |
| BXD33 | 1 (0) | 46.00 ± 0 | 16.43 ± 0 | 6.20 ± 0 | 10.00 ± 0 | 9.91 ± 0 | 3.50 ± 0 | 0.0346 ± 0 | 0.0368 ± 0 |
| BXD34 | 3 (1) | 47.33 ± 4.67 | 21.87 ± 0.85 | 6.20 ± 0.46 | 10.83 ± 0.44 | 10.63 ± 0.91 | 3.6 ± 0.31 | 0.0387 ± 0.0023 | 0.0360 ± 0.0032 |
| BXD36 | 3 (1) | 42.33 ± 0.33 | 16.42 ± 1.47 | 6.03 ± 0.03 | 10.33 ± 0.03 | 10.35 ± 0.31 | 4.00 ± 0.00 | 0.0337 ± 0.0013 | 0.0358 ± 0.0033 |
| BXD39 | 2 (1) | 45.00 ± 0.00 | 20.50 ± 1.32 | 6.05 ± 0.05 | 10.95 ± 0.05 | 10.83 ± 0.30 | 3.65 ± 0.35 | 0.0382 ± 0.0018 | 0.0366 ± 0.0029 |
| BXD40 | 2 (1) | 47.00 ± 0.00 | 18.01 ± 1.22 | 5.90 ± 0.02 | 10.05 ± 0.05 | 10.13 ± 0.33 | 4.00 ± 0.00 | 0.0338 ± 0.0005 | 0.0355 ± 0.0030 |
| BXD42 | 3 (2) | 42.33 ± 1.33 | 14.63 ± 0.93 | 5.97 ± 0.03 | 10.43 ± 0.19 | 10.51 ± 0.43 | 3.97 ± 0.03 | 0.0352 ± 0.0010 | 0.0358 ± 0.0031 |
| BXD43 | 2 (1) | 43.50 ± 0.50 | 18.05 ± 2.17 | 5.50 ± 0.00 | 9.25 ± 0.25 | 9.61 ± 0.44 | 4.20 ± 0.30 | 0.0301 ± 0.0030 | 0.0340 ± 0.0034 |
| BXD44 | 3 (1) | 46.67 ± 1.67 | 16.26 ± 0.93 | 5.77 ± 0.03 | 9.77 ± 0.09 | 10.10 ± 0.43 | 3.80 ± 0.15 | 0.0270 ± 0.0026 | 0.0307 ± 0.0044 |
| BXD45 | 2 (1) | 44.00 ± 0.00 | 17.24 ± 0.97 | 5.00 ± 0.00 | 10.30 ± 0.00 | 10.80 ± 0.00 | 4.20 ± 0.10 | 0.0345 ± 0.0007 | 0.0383 ± 0.0000 |
| BXD50 | 2 (1) | 46.50 ± 1.50 | 16.40 ± 2.55 | 6.00 ± 0.00 | 10.85 ± 0.15 | 10.91 ± 0.35 | 4.25 ± 0.25 | 0.0327 ± 0.0026 | 0.0341 ± 0.0029 |
| BXD51 | 6 (2) | 49.33 ± 0.21 | 18.37 ± 0.44 | 6.20 ± 0.18 | 10.97 ± 0.24 | 10.80 ± 0.49 | 4.08 ± 0.13 | 0.0371 ± 0.0011 | 0.0366 ± 0.0037 |
| BXD55 | 2 (1) | 49.00 ± 2.00 | 19.81 ± 3.96 | 6.05 ± 0.05 | 10.50 ± 0.30 | 10.61 ± 0.38 | 3.70 ± 0.40 | 0.0300 ± 0.0058 | 0.0295 ± 0.0047 |
| BXD56 | 2 (1) | 48.00 ± 0.00 | 20.99 ± 1.35 | 5.85 ± 0.05 | 10.15 ± 0.05 | 10.38 ± 0.36 | 3.85 ± 0.05 | 0.0293 ± 0.002 | 0.0291 ± 0.0052 |
| BXD60 | 2 (1) | 45.00 ± 0.00 | 21.64 ± 2.39 | 7.00 ± 0.00 | 11.30 ± 0.40 | 10.63 ± 0.73 | 4.15 ± 0.05 | 0.0423 ± 0.002 | 0.0373 ± 0.0032 |
| BXD61 | 3 (2) | 45.67 ± 0.67 | 21.28 ± 1.78 | 5.93 ± 0.28 | 10.03 ± 0.13 | 10.08 ± 0.34 | 3.57 ± 0.38 | 0.0341 ± 0.0025 | 0.0337 ± 0.0034 |
| BXD62 | 3 (2) | 47.67 ± 3.18 | 19.85 ± 0.42 | 6.3 ± 0.3 | 10.47 ± 0.27 | 10.39 ± 0.44 | 4.63 ± 0.32 | 0.0326 ± 0.0021 | 0.0317 ± 0.0052 |
| BXD63 | 1 (0) | 44.00 ± 0 | 16.05 ± 0 | 5.90 ± 0 | 10.00 ± 0 | 10.13 ± 0 | 3.7 ± 0 | 0.0319 ± 0 | 0.0350 ± 0.0 |
| BXD65 | 3 (1) | 46.33 ± 0.67 | 18.77 ± 1.20 | 5.93 ± 0.07 | 10.53 ± 0.48 | 10.46 ± 0.79 | 4.20 ± 0.12 | 0.0381 ± 0.0003 | 0.0386 ± 0.0033 |
| BXD66 | 2 (1) | 46.50 ± 3.50 | 16.34 ± 0.88 | 6.10 ± 0.10 | 9.95 ± 0.05 | 9.85 ± 0.32 | 4.10 ± 0.10 | 0.0364 ± 0.0015 | 0.0390 ± 0.0032 |
| BXD68 | 2 (2) | 45.50 ± 2.50 | 18.66 ± 2.05 | 6.35 ± 0.15 | 10.5 ± 0.00 | 10.56 ± 0.28 | 3.50 ± 0.30 | 0.0272 ± 0.0029 | 0.0268 ± 0.0041 |
| BXD69 | 4 (2) | 43.00 ± 0.58 | 19.98 ± 0.65 | 6.10 ± 0.15 | 10.35 ± 0.23 | 10.26 ± 0.49 | 4.15 ± 0.22 | 0.0362 ± 0.0016 | 0.0357 ± 0.0039 |
| BXD70 | 3 (2) | 40.33 ± 0.33 | 17.94 ± 1.35 | 6.00 ± 0.06 | 10.73 ± 0.38 | 10.74 ± 0.68 | 4.37 ± 0.09 | 0.0345 ± 0.0001 | 0.0351 ± 0.0027 |
| BXD71 | 3 (2) | 43.33 ± 3.28 | 18.73 ± 2.41 | 6.23 ± 0.12 | 11.00 ± 0.15 | 10.84 ± 0.50 | 4.2 ± 0.32 | 0.0365 ± 0.0027 | 0.0356 ± 0.0032 |
| BXD73 | 2 (1) | 46.50 ± 1.50 | 17.59 ± 1.37 | 6.20 ± 0.00 | 10.40 ± 0.30 | 10.37 ± 0.52 | 3.80 ± 0.10 | 0.0327 ± 0.0020 | 0.0335 ± 0.0028 |
| BXD75 | 2 (1) | 45.50 ± 2.50 | 18.23 ± 0.57 | 5.95 ± 0.85 | 10.10 ± 0.70 | 10.18 ± 0.49 | 3.65 ± 0.05 | 0.0329 ± 0.0035 | 0.0343 ± 0.0023 |
| BXD77 | 4 (2) | 42.5 ± 1.32 | 20.92 ± 1.92 | 6.03 ± 0.33 | 10.925 ± 0.30 | 10.90 ± 0.49 | 4.25 ± 0.22 | 0.0352 ± 0.0020 | 0.0335 ± 0.0039 |
| BXD80 | 2 (1) | 45.50 ± 0.50 | 21.24 ± 2.15 | 6.90 ± 0.00 | 10.60 ± 0.40 | 10.1 ± 0.56 | 4.05 ± 0.25 | 0.0372 ± 0.0038 | 0.0337 ± 0.0039 |
| BXD83 | 2 (1) | 45.00 ± 1.00 | 19.33 ± 0.97 | 6.55 ± 0.25 | 10.10 ± 0.10 | 9.77 ± 0.57 | 4.15 ± 0.05 | 0.0375 ± 0.0029 | 0.0368 ± 0.0040 |
| BXD84 | 3 (1) | 45.00 ± 2.52 | 17.39 ± 1.08 | 6.33 ± 0.33 | 11.13 ± 0.13 | 11.04 ± 0.53 | 3.97 ± 0.15 | 0.0327 ± 0.0007 | 0.0322 ± 0.0026 |
| BXD85 | 2 (1) | 43.50 ± 2.50 | 18.08 ± 0.07 | 6.00 ± 0.00 | 10.90 ± 0.90 | 10.86 ± 1.20 | 4.15 ± 0.05 | 0.0361 ± 0.0008 | 0.0363 ± 0.0044 |
| BXD86 | 3 (1) | 44.67 ± 1.33 | 15.90 ± 1.52 | 5.77 ± 0.23 | 10.43 ± 0.19 | 10.66 ± 0.51 | 3.90 ± 0.10 | 0.0304 ± 0.0026 | 0.0331 ± 0.0036 |
| BXD87 | 3 (1) | 39.67 ± 0.33 | 18.04 ± 1.25 | 5.77 ± 0.28 | 10.43 ± 0.35 | 10.65 ± 0.51 | 3.93 ± 0.38 | 0.0309 ± 0.0003 | 0.0337 ± 0.0042 |
| BXD89 | 3 (1) | 43.00 ± 0.58 | 18.58 ± 1.82 | 6.30 ± 0.15 | 11.13 ± 0.12 | 11.00 ± 0.48 | 4.10 ± 0.06 | 0.0345 ± 0.0019 | 0.0333 ± 0.0031 |
| BXD90 | 2 (1) | 48.00 ± 5.00 | 22.66 ± 3.47 | 6.30 ± 0.30 | 10.50 ± 0.40 | 10.20 ± 0.49 | 4.25 ± 0.25 | 0.0402 ± 0.0017 | 0.0373 ± 0.0040 |
| BXD92 | 2 (1) | 45.00 ± 0.00 | 21.86 ± 5.98 | 5.45 ± 0.25 | 9.90 ± 0.20 | 10.28 ± 0.48 | 3.95 ± 0.15 | 0.0300 ± 0.0041 | 0.0305 ± 0.0032 |
| BXD95 | 2 (1) | 47.00 ± 4.00 | 21.48 ± 2.59 | 6.10 ± 0.10 | 10.65 ± 0.35 | 10.39 ± 0.66 | 4.15 ± 0.15 | 0.0418 ± 0.0280 | 0.0399 ± 0.0040 |
| BXD96 | 5 (3) | 43.60 ± 0.24 | 20.56 ± 0.98 | 6.18 ± 0.14 | 10.42 ± 0.21 | 10.43 ± 0.50 | 3.56 ± 0.22 | 0.0314 ± 0.0033 | 0.0303 ± 0.0054 |
| C57BL/6J | 3 (3) | 44.33 ± 0.88 | 19.283 ± 1.41 | 6.53 ± 0.42 | 11.07 ± 0.48 | 10.71 ± 0.51 | 4.07 ± 0.07 | 0.0386 ± 0.0031 | 0.0365 ± 0.0042 |
| DBA/2J | 4 (2) | 47.75 ± 0.25 | 13.124 ± 0.81 | 6.23 ± 0.23 | 10.45 ± 0.20 | 10.43 ± 0.55 | 4.08 ± 0.05 | 0.0316 ± 0.0014 | 0.0353 ± 0.0035 |
All data are presented as mean ± standard error of the mean.
Table 2.
Age, body weight, raw, and adjusted papillae number and area
| Strain | Total cases (n = male) | Age (days) | Body weight (g) | Fungiform papillae number | Total filiform papillae number | Adjusted total filiform papillae number | Fungiform papillae area (mm2) |
| BXD1 | 1 (0) | 57.00 ± 0 | 17.36 ± 0 | 59.00 ± 0 | 418.00 ± 0 | 344.30 ± 0 | 0.057 ± 0 |
| BXD12 | 2 (0) | 44.00 ± 0.00 | 18.39 ± 0.33 | 98.50 ± 3.50 | 326.50 ± 16.50 | 328.78 ± 49.47 | 0.124 ± 0.049 |
| BXD14 | 2 (1) | 45.50 ± 0.50 | 22.46 ± 1.39 | 107.00 ± 14.00 | 264.5 ± 9.50 | 258.01 ± 45.39 | 0.181 ± 0.033 |
| BXD32 | 2 (2) | 43.00 ± 0.00 | 21.57 ± 1.66 | 81.00 ± 2.00 | 377.00 ± 33.00 | 385.13 ± 65.97 | 0.122 ± 0.029 |
| BXD34 | 3 (1) | 47.33 ± 4.67 | 21.87 ± 0.85 | 104.33 ± 5.93 | 334.33 ± 6.96 | 317.13 ± 66.70 | 0.069 ± 0.005 |
| BXD36 | 3 (1) | 42.33 ± 0.33 | 16.42 ± 1.47 | 83.00 ± 12.86 | 261.33 ± 60.70 | 273.36 ± 93.13 | 0.105 ± 0.021 |
| BXD42 | 2 (1) | 43.00 ± 2.00 | 15.02 ± 1.48 | 99.5 ± 2.5 | 258.98 ± 50.85 | 267.11 ± 95.51 | 0.081 ± 0.003 |
| BXD45 | 2 (1) | 44.00 ± 0.00 | 17.24 ± 0.97 | 91.50 ± 0.50 | 297.50 ± 26.50 | 299.78 ± 59.47 | 0.134 ± 0.001 |
| BXD50 | 2 (1) | 46.50 ± 1.50 | 16.40 ± 2.55 | 103.50 ± 12.50 | 301.00 ± 42.00 | 288.67 ± 66.21 | 0.231 ± 0.060 |
| BXD51 | 5 (2) | 49.2 ± 0.20 | 18.34 ± 0.53 | 87.4 ± 7.21 | 422.80 ± 27.25 | 394.69 ± 60.30 | 0.181 ± 0.030 |
| BXD62 | 2 (1) | 49.00 ± 5.00 | 20.12 ± 0.55 | 112.00 ± 7.00 | 373.53 ± 8.53 | 346.58 ± 60.30 | 0.159 ± 0.044 |
| BXD65 | 3 (1) | 46.33 ± 0.67 | 18.77 ± 1.20 | 107.67 ± 12.35 | 299.00 ± 22.61 | 287.64 ± 59.39 | 0.119 ± 0.023 |
| BXD66 | 2 (1) | 46.50 ± 3.50 | 16.34 ± 0.88 | 96.00 ± 13.00 | 326.50 ± 7.50 | 314.17 ± 60.93 | 0.150 ± 0.014 |
| BXD69 | 2 (1) | 42.00 ± 0.00 | 20.47 ± 0.75 | 104.00 ± 15.00 | 319.00 ± 27.00 | 332.97 ± 59.97 | 0.107 ± 0.000 |
| BXD70 | 2 (1) | 40.50 ± 0.50 | 18.29 ± 2.25 | 85.50 ± 8.50 | 365.00 ± 47.00 | 387.74 ± 82.89 | 0.143 ± 0.011 |
| BXD71 | 2 (1) | 42.50 ± 5.5 | 16.60 ± 1.94 | 124.00 ± 3.00 | 216.90 ± 40.99 | 227.94 ± 41.81 | 0.221 ± 0.073 |
| BXD77 | 3 (1) | 43.00 ± 1.73 | 19.27 ± 1.37 | 97.33 ± 11.32 | 313.72 ± 58.12 | 321.85 ± 100.81 | 0.150 ± 0.044 |
| BXD84 | 3 (1) | 45.00 ± 2.52 | 17.38 ± 1.08 | 106.33 ± 6.67 | 365.00 ± 44.84 | 361.44 ± 63.42 | 0.228 ± 0.014 |
| BXD85 | 2 (1) | 43.50 ± 2.50 | 18.08 ± 0.07 | 119.50 ± 5.50 | 321.50 ± 30.50 | 326.70 ± 48.86 | 0.174 ± 0.017 |
| BXD86 | 3 (1) | 44.67 ± 1.33 | 15.90 ± 1.53 | 114.00 ± 2.52 | 356.67 ± 53.65 | 355.05 ± 79.65 | 0.165 ± 0.004 |
| BXD87 | 3 (1) | 39.67 ± 0.33 | 18.04 ± 1.25 | 112.00 ± 2.645 | 313.67 ± 24.52 | 341.27 ± 55.71 | 0.136 ± 0.027 |
| BXD89 | 2 (1) | 43.50 ± 0.5 | 18.79 ± 3.14 | 99.50 ± 11.50 | 218.19 ± 18.82 | 223.39 ± 54.71 | 0.117 ± 0.011 |
| BXD95 | 2 (1) | 47.00 ± 4.0 | 21.48 ± 2.59 | 104.00 ± 4.00 | 280.10 ± 66.90 | 264.85 ± 123.25 | 0.071 ± 0.003 |
| C57BL/6J | 3 (3) | 44.33 ± 0.88 | 19.28 ± 1.41 | 115.67 ± 7.75 | 224.28 ± 39.73 | 242.61 ± 67.59 | 0.092 ± 0.028 |
| DBA/2J | 4 (2) | 47.75 ± 0.25 | 13.12 ± 0.81 | 96.25 ± 2.75 | 359.51 ± 27.39 | 336.87 ± 61.42 | 0.089 ± 0.012 |
All data are presented as mean ± standard error of the mean.
Heritability of phenotypes
Broad-sense heritability (h2) was calculated using raw data for ApV, fungiform papillae number, filiform papillae number, and fungiform papillae area, as well as body weight and tongue weight. Heritability for ApV was 28% but decreased to 12% when adjusted for ApM and tongue weight. Fungiform number and area heritability were 36% each. Filiform heritability was 31% but decreased to 14% when adjusted for age. Heritability for body weight was 23% while tongue weight was 31% and decreased to 14% when adjusted for body weight and tongue length (ApM and ApV).
Sex and body weight
Age, sex, body weight, ApM, ApV, tongue width, and tongue weight were examined using a Pearson product-moment correlation, and these results are shown in Table 3. Sex had a significant negative correlation with body weight (r = −0.49) and accounted for 24% of the variance in body weight (F1,116 = 36.3, P < 0.0001).
Table 3.
Age, sex, body weight, and tongue correlations
| Age | Sex | Body weight | ApM | ApV | Tongue width | Tongue weight | |
| Age | 1 | ||||||
| Sex | 0.05 | 1 | |||||
| Body weight | 0.07 | −0.49* | 1 | ||||
| ApM | 0.15 | −0.05 | 0.23* | 1 | |||
| ApV | −0.04 | −0.04 | 0.18 | 0.44* | 1 | ||
| Tongue width | −0.1 | 0.03 | −0.003 | 0.06 | 0.22 | 1 | |
| Tongue weight | 0.06 | −0.20* | 0.52* | 0.40* | 0.37* | 0.17 | 1 |
P value <0.05.
Body weight and tongue weight
Body weight correlated significantly with tongue weight (r = 0.52). Using regression analysis, body weight accounted for 27% of the total variation in tongue weight (F1,116 = 42.3, P < 0.0001).
Tongue length and tongue weight
Measures of tongue length (ApM and ApV) correlated with each other (r = 0.44, P < 0.0001) and with tongue weight. ApV correlated with tongue weight (r = 0.37) and accounted for 14% of the variance in tongue weight (F1,115 = 18.5, P < 0.0001). ApM also correlated significantly with tongue weight (r = 0.40). ApM accounted for 16% of the variance in tongue weight (F1,116 = 22, P < 0.0001).
Parental strain differences
Body weights of the parental strains differed significantly (P = 0.0098); C57BL/6J had an average body weight of 19.28 ± 1.41 g, and DBA/2J had an average body weight of 13.12 ± 1.15 g. Averaged tongue weights did not differ significantly between the 2 parental strains (P = 0.07); C57BL/6J had a tongue weight of 0.039 ± 0.003 g, and DBA/2J had a tongue weight of 0.032 ± 0.002 g. C57BL/6J had a slightly greater ApV (11.07 ± 0.48 mm) compared with DBA/2J (10.45 ± 0.29 mm). The parental strains differed significantly in number of total fungiform papillae (P = 0.04) with C57BL/6J containing an average of 115.67 ± 7.75 fungiform papillae, whereas DBA/2J had an average of 96.25 ± 3.89 fungiform papillae. DBA/2J had a significantly greater number of total filiform papillae (P = 0.03). The parental strains did not differ significantly in fungiform papillae area (P = 0.90); C57BL/6J had an average fungiform papillae area of 0.0922 ± 0.028 mm2, whereas DBA/2J had an average of 0.089 ± 0.017 mm2.
Age and total filiform papillae number
Age, sex, body weight, ApM, ApV, tongue width, tongue weight, total fungiform number, total filiform number, and fungiform area were examined using a Pearson's Product-Moment correlation, and these results are shown in Table 4. Total filiform papillae number was correlated with age (r = 0.31). Approximately 10% of the variance in total filiform papillae number was accounted for by age (F1,60 = 6.33, P = 0.0146).
Table 4.
Age, sex, body weight, and papillae correlations
| Age | Sex | Body weight | ApM | ApV | Tongue width | Tongue weight | Total fungiform | Total filiform | Fungiform area | |
| Age | 1 | |||||||||
| Sex | 0.16 | 1 | ||||||||
| Body weight | −0.01 | −0.38* | 1 | |||||||
| ApM | 0.26* | 0.04 | 0.24 | 1 | ||||||
| ApV | −0.06 | −0.06 | 0.33* | 0.38 | 1 | |||||
| Tongue width | −0.06 | −0.02 | −0.001 | 0.05 | 0.13 | 1 | ||||
| Tongue weight | 0.08 | −0.18 | 0.72* | 0.28* | 0.123 | −0.003 | 1 | |||
| Total fungiform | −0.10 | −0.02 | 0.06 | 0.21 | −0.02 | 0.20 | 0.09 | 1 | ||
| Total filiform | 0.31* | 0.1 | −0.05 | 0.08 | −0.04 | −0.11 | −0.14 | −0.21 | 1 | |
| Fungiform area | −0.20 | −0.06 | 0.01 | 0.11 | 0.23 | 0.19 | −0.07 | 0.14 | 0.16 | 1 |
P value <0.05.
Tongue size and papillae area and number
Tongue size correlated with fungiform papillae area and number. ApM correlated with fungiform papillae number (r = 0.21) and accounted for 4% of the variance in fungiform papillae number (F1,60 = 2.65, P = 0.1086). ApV also correlated with fungiform papillae area (r = 0.23) and accounted for 5% of the variance in fungiform papillae area (F1,59 = 3.18, P = 0.0799). Tongue width correlated negatively with total fungiform number (r = −0.20) and accounted for 4% of the variance in total fungiform papillae number (F1,60 = 2.39, P = 0.1275). Tongue width also correlated with fungiform papillae area (r = −0.19) and accounted for 4% of the variance in fungiform papillae area (F1,60 = 2.26, P = 0.1381). Tongue weight correlated with total filiform papillae number (r = −0.14, P = 0.29).
Total fungiform papillae number and area and total filiform papillae number
Total fungiform papillae number was negatively correlated with total filiform papillae number (r = −0.21, P = 0.0962). Fungiform papillae area was correlated with total fungiform papillae number (r = 0.14, P = 0.2706).
QTL modulating body weight
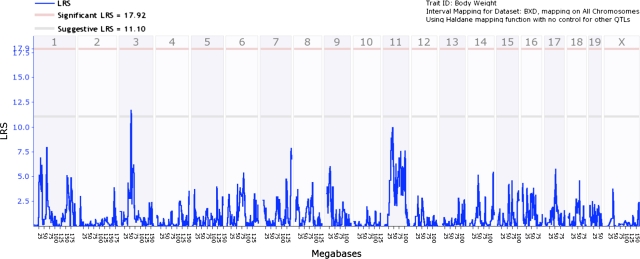
To confirm that our detected QTLs for tongue size and papillae number are largely independent of body weight variation, we examined raw body weight using only BXD RI strains (n = 43). Figure 2 shows an interval genome-wide QTL map of body weight using raw data. Clearly, there were no significant QTLs but only a suggestive signal on chromosome 3. However, this suggestive signal did not overlap with any of the significant or suggestive QTLs for tongue size and papillae number or area.
Figure 2.
Genome-wide linkage map of body weight. Blue trace shows the LRS for body weight. Note that a suggestive QTL was detected on chromosome 3. Lower gray horizontal line: suggestive LRS genome-wide threshold at P ≤ 0.63. Upper red horizontal line: significant LRS genome-wide threshold at P ≤ 0.05.
QTL modulating tongue weight
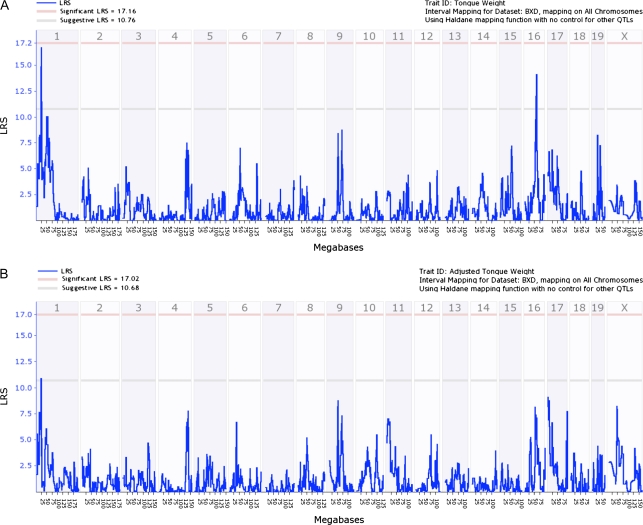
QTL mapping of tongue weight was completed using 43 BXD RI strains. Figure 3A shows an interval genome-wide QTL map of tongue weight using raw data where 2 suggestive QTLs were found on chromosomes 1 and 16. However, after adjusting for body weight and tongue length (ApM and ApV), the suggestive QTL on chromosome 1 diminished in strength (Figure 3B). The suggestive QTL on chromosome 16 was not observed in the second map.
Figure 3.
Genome-wide linkage QTL maps of tongue weight. (A) Interval genome-wide map QTL of tongue weight using raw data. Suggestive QTLs can be seen on chromosomes 1 and 16. (B) Tongue weight QTL using adjusted data.
QTL modulating tongue length (ApV)
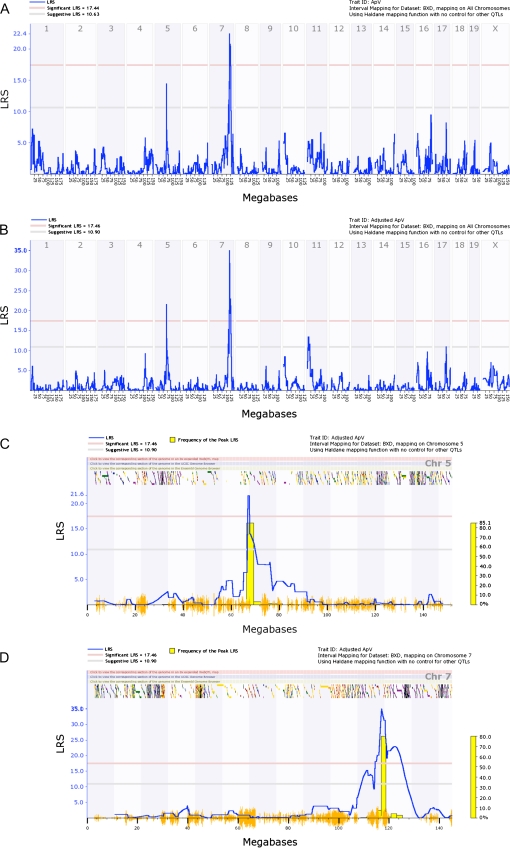
Figure 4A shows a simple interval QTL map encompassing all the genome for raw ApV. There was a highly significant QTL on chromosome 7 and a suggestive QTL on chromosome 5. After adjusting for ApM and tongue weight, the QTL (Figure 4B) on chromosome 5 increased in significance, whereas the QTL on chromosome 7 remained relatively unchanged. Suggestive QTLs were also observed on chromosomes 11 and 17. We used the adjusted value for ApV for all analyses.
Figure 4.
Genome-wide linkage maps of tongue length (ApV). (A) Interval genome-wide QTL map using raw data showing a significant QTL on chromosome 7 and a suggestive signal on chromosome 5. When using adjusted values (B), the QTL on chromosome 5 crosses the significance threshold, whereas the QTL on chromosome 7 remains at relatively the same significance level. (C and D) interval QTL maps with bootstrap analysis of chromosomes 5 and 7 using adjusted ApV values, respectively. Lower gray horizontal line: suggestive LRS genome-wide threshold at P ≤ 0.63. Upper red horizontal line: significant LRS genome-wide threshold at P ≤ 0.05. Yellow histogram: frequency of peak LRS (bootstrap analysis). Orange seismograph marks indicate SNP density.
Marker regression analysis using WebQTL revealed 22 loci on chromosomes 5 and 7 with LRS values above the 17.32 significance threshold (Table 5). Interval maps of chromosome 5 (Figure 4C) and chromosome 7 (Figure 4D) are shown. We utilized a 1.5 LOD support interval around the peak significant value to further delineate the QTL region. The QTL on chromosome 5 spanned a relatively small region, beginning at 66.0 Mb and ending at 68.0 Mb. On the other hand, the QTL on chromosome 7 spanned a much larger region, beginning at 114.5 Mb and ending at 126.0 Mb. A pair-scan analysis revealed no interaction between the 2 QTLs on chromosomes 5 and 7 (data not shown). This suggests that the 2 QTLs operate independently of each other.
Table 5.
Chromosomes 5 and 7 loci
| Adjusted ApV* | |
| Locus | LRS |
| Chromosome 5 | |
| rs4225252 | 20.608 |
| rs3719870 | 20.608 |
| rs3711269 | 20.608 |
| rs13478309 | 20.608 |
| Chromosome 7 | |
| rs13479471 | 18.123 |
| rs6160824 | 18.123 |
| rs13479470 | 18.202 |
| rs6241342 | 19.285 |
| rs6271956 | 19.285 |
| mCV24779699 | 20.193 |
| rs3656074 | 20.193 |
| D7Mit330 | 21.565 |
| rs3709679 | 21.565 |
| rs6322316 | 21.565 |
| rs6366212 | 23.180 |
| rs3722112 | 23.180 |
| rs13479483 | 27.801 |
| rs4226870 | 31.446 |
| rs6305308 | 31.446 |
| gnf07.114.758 | 32.023 |
| rs6340473 | 32.366 |
| rs13479476 | 32.366 |
| rs13479477 | 35.134 |
| Total fungiform | |
| rs13476330 | 15.474 |
| gnf02.003.251 | 15.474 |
| Fungiform area* | |
| rs13483677 | 18.224 |
| rs13483679 | 18.224 |
P value <0.05.
QTL modulating fungiform and filiform papillae number
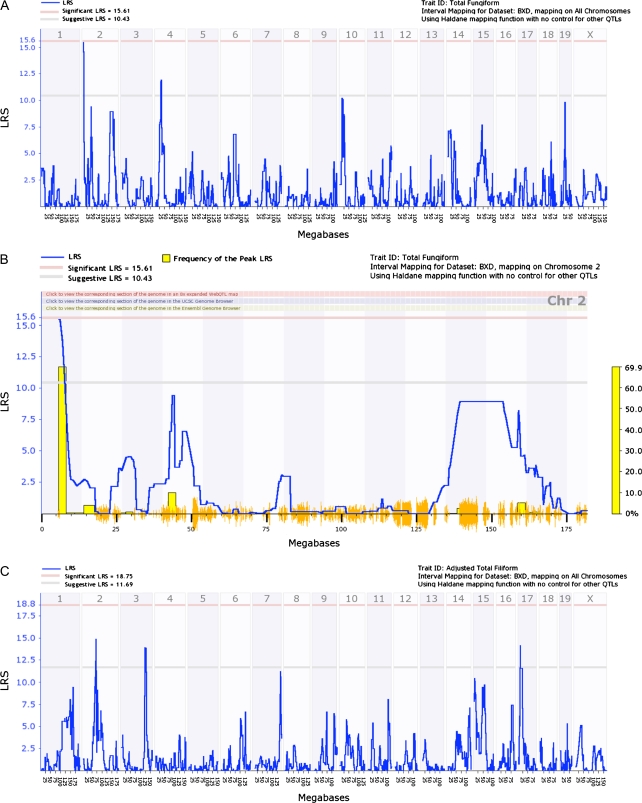
Figure 5A shows an interval genome-wide QTL map for fungiform papillae number. There were 2 suggestive QTLs located on chromosomes 2 and 4. Marker regression revealed 2 loci on chromosome 2 above the suggestive LRS value of 10.52 (Table 5). An interval QTL map of chromosome 4 is shown in Figure 5B. The nearly significant QTL spanned a region between 5.8 and 8.0 Mb. For filiform papillae number, suggestive QTLs were detected on chromosomes 14 and 17. After adjusting for age, suggestive QTLs on chromosomes 2, 3, and 17 were observed (Figure 5C).
Figure 5.
Genome-wide linkage maps of papillae numbers. (A) Interval genome-wide QTL map using raw data. A suggestive QTL was found on chromosomes 2 and 4. (B) Interval QTL map with bootstrap analysis for chromosome 2 using raw fungiform papillae number values. (C) Interval genome-wide QTL map for adjusted filiform papillae number. Suggestive QTLs were found on chromosomes 2, 3, and 17.
QTL modulating fungiform papillae area
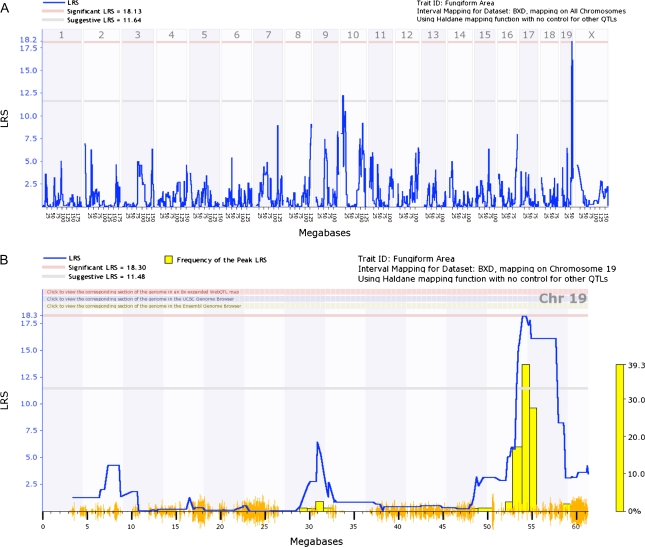
Figure 6A shows an interval genome-wide QTL for fungiform area. There was a suggestive QTL on chromosome 10 and a significant QTL on chromosome 19. Figure 6B showed an interval QTL map for chromosome 19. Marker regression analysis revealed 2 loci above the 18.22 LRS significance threshold (Table 5). The QTL on chromosome 19 spanned a region from 53.0 to 57.8 Mb.
Figure 6.
Genome-wide linkage maps of fungiform papillae area. (A) Interval genome-wide QTL map using raw data. A significant QTL was found on chromosome 19, and a suggestive QTL was found on chromosome 10. (B) Interval QTL map with bootstrap analysis of chromosome 19.
Candidate genes
For QTLs associated with tongue length, we detected 14 genes that lie under the chromosome 5 interval of 66.0–68.0 Mb and 84 genes under the chromosome 7 interval of 114.5–126.0 Mb. We detected 12 genes under the chromosome 1 QTL interval of 22.0–26.0 Mb for tongue weight. We also detected 8 genes under the QTL interval of 5.8–8.0 Mb on chromosome 2 for fungiform papillae and 37 genes under the QTL interval of 53.0–57.8 Mb on chromosome 19 for fungiform papillae area.
Tongue length—For QTLs associated with tongue length, we detected 14 genes that lie under the chromosome 5 interval of 66.0–68.0 Mb and 84 genes under the chromosome 7 interval of 114.5–126 Mb. Three genes within this former region have a relatively high SNP number: amyloid beta (A4) precursor protein-binding, family B, member 2 (Appb2) with 155 SNPs; solute carrier family 30 (zinc transporter), member 9 (Slc30a9) with 118 SNPs; and Bcl3-binding protein (B3bp) with 111 SNPs. The QTL on chromosome 7 contains 13 genes with SNP numbers greater than 100. These include SRY-box containing gene (Sox6) with 3174 SNPs, transmembrane channel-like gene family 5 (Tmc5) with 347 SNPs, pleckstrin homology domain containing family A member 7 (Plekha7) with 268 SNPs, synaptotagmin XVII (Syt17) with 267 SNPs, ATP-binding cassette, subfamily A (ABC1), member 14 (Abca14) with 261 SNPs, transmembrane channel-like gene family 7 (Tmc7) with 212 SNPs, demethyl-Q 7 (Coq7) with 163 SNPs, and G protein–coupled receptor, family C, group 5, member B (Gprc5b) with 134 SNPs.
Tongue weight—We detected 12 genes under the chromosome 1 suggestive QTL interval of 22.0–26.0 Mb for tongue weight. Two important candidate genes within this interval are noteworthy: procollagen, type IX, alpha 1 (Col9a1) with 17 SNPs and procollagen, type XIX, alpha 1 (Col19a1) with 707 SNPs. Col19a1 is expressed in differentiating muscle cells and is reportedly involved in esophageal muscle development (Sumiyoshi et al. 2004).
Total fungiform papillae number—We detected 8 genes under the QTL interval of 5.8–8.0 Mb on chromosome 2 for fungiform papillae number, none of which had a SNP number greater than 100. The QTL on chromosome 2 also covers a small interval of 5.8–8.0 Mb and includes 8 genes, none of which have a SNP number over 100.
Fungiform papillae area—Thirty-seven genes were detected under the QTL interval of 53.0–57.8 Mb on chromosome 19 for fungiform papillae area. Of these genes, the following 6 genes had a SNP number greater than 100: attractin-like 1 (Atrnl1) with 1159 SNPs, actin-binding LIM protein 1 (Ablim1) with 896 SNPs, vesicle transport through interaction with t-SNAREs homolog 1A (yeast) (Vti1a) with 473 SNPs, actin filament–associated protein 1-like 2 (Afap1l2) with 233 SNPs, X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (Xpnpep1) with 205 SNPs, and nebulin-related anchoring protein (Nrap) with 124 SNPs.
Discussion
Synopsis
We measured a number of key lingual phenotypes related directly and indirectly to ingestion and gustation across a large number of genetically well-characterized BXD strains generated from a cross between 2 fully sequenced strains of mice—C57BL/6J and DBA/2J. We measured tongue length, width, and weight, as well as filiform and fungiform papillae number and area. Variation in lingual traits is substantial (1.2- to 2-fold differences), and a significant fraction of this variation is due to gene variants that we have been able to map to chromosomes 5 and 7 (tongue length) and chromosome 19 (fungiform papillae area).
Effects of tongue and body weights
Variations in tongue weight and body weight correlate significantly. Tongue weight was also modestly correlated with tongue length (ApM and ApV). In order to examine these seemingly overlaying phenotypes, we used a linear model to generate adjusted ApV values that removed the effects of ApM and tongue weight, the most significant predictors. To confirm our QTLs, we performed a simple regression of the raw data from both body and tongue weights. As evident from body weight and tongue weight QTL maps (Figures 2 and 3), there were no signals that overlapped with our proposed QTLs for ApV, fungiform number, and fungiform papillae area. Although tongue weight and tongue length were correlated, the QTL data showed that these factors have little effect on one another.
Differences between parental strains
In the present study, C57BL/6J mice have a slightly longer tongue than DBA/2J mice by approximately 5–6%, although these differences are not significant; this trend matches that of another report (Boughter et al. 2007). C57BL/6J also possess 15–20% more fungiform papillae than DBA/2J mice. However, DBA/2J mice have significantly more total filiform papillae by approximately 35–40%. This finding may be relevant to the significant differences in taste sensitivity of C57BL/6J mice compared with DBA/2J mice for sweet- and bitter-tasting compounds (Kotlus and Blizard 1998; Blizard et al. 1999; Boughter et al. 2005; Blizard 2007), and this difference is highly likely to account for much of the marked strain difference in ethanol acceptance and drinking of these 2 strains.
Tongue length (ApV) QTL
Significant QTLs were observed on chromosomes 5 and 7 that are associated with tongue length (ApV). QTL mapping was conducted using ApV values adjusted for ApM and tongue weight, but as a comparison, raw ApV values were also mapped. When adjusted values are used, the significant QTL on chromosome 7 increased and the suggestive QTL on chromosome 5 increased to the significant threshold. Furthermore, a suggestive QTL was found on chromosome 17 using the adjusted values, whereas with the raw values, it was not observed. Using marker regression, we report 4 loci on chromosome 5 and 19 loci on chromosome 7 that are linked to the adjusted ApV. This finding demonstrates the presence of several genetic loci that contribute to variation in this trait. Such variation could underlie important motor behaviors such as fluid licking (Weijnen 1998). For example, it has been recently reported that DBA/2J mice lick at a higher rate than do C57BL/6J mice (Boughter et al. 2007).
Total fungiform papillae number QTL
We reported a genome-wide suggestive QTL on chromosome 2 that was associated with total fungiform papillae number. Mapping was conducted using raw fungiform papillae values. However, when adjusted values were used, the suggestive QTL on chromosome 2 reached significance. QTLs on chromosomes 4 and 19 were also suggestive, and another locus on chromosome 2 was suggestive when adjusted values were used. A relationship between fungiform papillae number and taste sensitivity has been reported for both mice and humans (Miller and Whitney 1989; Bartoshuk et al. 1994).
Fungiform papillae area QTL
We report a genome-wide significant QTL on chromosome 19 that is associated with fungiform papillae area as well as a suggestive QTL on chromosome 10. Using marker regression analysis, we report 2 loci on chromosome 19 that are linked to fungiform papillae area.
Candidate genes
For QTLs that were significant or highly suggestive, we searched for genes within QTL intervals. Using WebQTL, we identified a number of candidate genes (8–84 genes, depending on the QTL). For each QTL, the list was first narrowed by examining the number of SNPs present within each gene; with one exception (see below), we reported only those genes with 100 or more SNPs (Peirce et al. 2004). In general, a higher SNP density region of the genome is more likely to contain a candidate gene. The 100 SNPs threshold is therefore an arbitrary number chosen for narrowing the number of genes to a workable number. More importantly, we also considered the biological relevance of each candidate gene. Analyses that take advantage of microarray data, which use RNA expression profiles to detect genes that are correlated with a particular phenotype of interest, were unavailable for the tongue from WebQTL.
Several of these genes possess biological relevance. For example, for tongue length, Sox6 has been shown to be involved in muscle development and Sox6 knockout mice express skeletal and cardiac muscle degeneration (Hagiwara et al. 2000), but it is unknown whether this gene is related to tongue size or tongue development. Additionally, Gprc5d, a member of the Gprc family, is expressed in the center region of filiform papillae (Inoue, Nambu, and Shimomura 2004), but little is known about its function. Furthermore, 2 interesting candidate genes were detected for fungiform papillae area. Nrap is expressed in skeletal and cardiac muscle (Mohiddin et al. 2003). Furthermore, the expression of Nrap has also been shown to be important in musculature development in pigs (Murani et al. 2007). Ablim1 mediates interactions between actin filaments and cytoplasmic targets, a process critical to cellular morphogenesis and differentiation (Roof et al. 1997).
Conclusion
We have identified significant QTLs on chromosomes 5 and 7 associated with tongue length (ApV), a suggestive QTL on chromosome 2 associated with fungiform papillae number, a significant QTL on chromosome 19 associated with fungiform papillae area, and a suggestive QTL associated with tongue weight on the proximal arm of chromosome 1. These findings are a step toward identifying genes involved in controlling tongue size and papillae number and development. Further studies are needed to more accurately identify candidate genes.
Funding
National Institutes of Health (5R01DC000066 to R.S.W., NS052366 to J.D.B.); Funding Agency (PA96-035).
Acknowledgments
The authors thank Qiuhong Yang for editing.
References
- Bachmanov AA, Reed DR, Ninomiya Y, Inoue M, Tordoff MG, Price RA, Beauchamp GK. Sucrose consumption in mice: major influence of two genetic loci affecting peripheral sensory responses. Mamm Genome. 1997;8:545–548. doi: 10.1007/s003359900500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow L. Gustatory system development. New York: Wiley; 2000. [Google Scholar]
- Bartoshuk LM, Duffy VB, Miller IJ. PTC/PROP tasting: anatomy, psychophysics, and sex effects. Physiol Behav. 1994;56:1165–1171. doi: 10.1016/0031-9384(94)90361-1. [DOI] [PubMed] [Google Scholar]
- Blizard DA. Sweet and bitter taste of ethanol in C57BL/6J and DBA2/J mouse strains. Behav Genet. 2007;37:146–159. doi: 10.1007/s10519-006-9121-4. [DOI] [PubMed] [Google Scholar]
- Blizard DA, Kotlus B, Frank ME. Quantitative trait loci associated with short-term intake of sucrose, saccharin and quinine solutions in laboratory mice. Chem Senses. 1999;24:373–385. doi: 10.1093/chemse/24.4.373. [DOI] [PubMed] [Google Scholar]
- Boughter JD, Jr, Baird JP, Bryant J, St John SJ, Heck D. C57BL/6J and DBA/2J mice vary in lick rate and ingestive microstructure. Genes Brain Behav. 2007;6:619–627. doi: 10.1111/j.1601-183X.2006.00293.x. [DOI] [PubMed] [Google Scholar]
- Boughter JD, Jr, Raghow S, Nelson TM, Munger SD. Inbred mouse strains C57BL/6J and DBA/2J vary in sensitivity to a subset of bitter stimuli. BMC Genet. 2005;6:36. doi: 10.1186/1471-2156-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RM. Tongue topography. New York: Springer; 1971. [Google Scholar]
- Finger TE, Simon SA. Cell biology of taste epithelium. New York: Wiley; 2000. [Google Scholar]
- Hagiwara N, Klewer SE, Samson RA, Erickson DT, Lyon MF, Brilliant MH. Sox6 is a candidate gene for p100H myopathy, heart block, and sudden neonatal death. Proc Natl Acad Sci USA. 2000;97:4180–4185. doi: 10.1073/pnas.97.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–114. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- Inoue M, Beauchamp GK, Bachmanov AA. Gustatory neural responses to umami taste stimuli in C57BL/6ByJ and 129P3/J mice. Chem Senses. 2004;29:789–795. doi: 10.1093/chemse/bjh083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Nambu T, Shimomura T. The RAIG family member, GPRC5D, is associated with hard-keratinized structures. J Invest Dermatol. 2004;122:565–573. doi: 10.1046/j.0022-202X.2004.12628.x. [DOI] [PubMed] [Google Scholar]
- Kotlus BS, Blizard DA. Measuring gustatory variation in mice: a short-term fluid-intake test. Physiol Behav. 1998;64:37–47. doi: 10.1016/s0031-9384(98)00016-x. [DOI] [PubMed] [Google Scholar]
- Krimm RF. Factors that regulate embryonic gustatory development. BMC Neurosci. 2007;8:54. doi: 10.1186/1471-2202-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CX, Wei X, Lu L, Peirce JL, Williams RW, Waters RS. Genetic analysis of barrel field size in the first somatosensory area (SI) in inbred and recombinant inbred strains of mice. Somatosens Mot Res. 2005;22:141–150. doi: 10.1080/08990220500262182. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Blizard DA, Gerhard GS, Vandenbergh DJ, Stout JT, Vogler GP, McClearn GE, Larsson L. Genetic determinants of weight of fast- and slow-twitch skeletal muscle in 500-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genomics. 2005;21:184–192. doi: 10.1152/physiolgenomics.00209.2004. [DOI] [PubMed] [Google Scholar]
- Lu L, Airey DC, Williams RW. Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci. 2001;21:3503–3514. doi: 10.1523/JNEUROSCI.21-10-03503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Jr, Smith DV. Quantitative taste bud distribution in the hamster. Physiol Behav. 1984;32:275–285. doi: 10.1016/0031-9384(84)90142-2. [DOI] [PubMed] [Google Scholar]
- Miller IJ, Jr, Whitney G. Sucrose octaacetate-taster mice have more vallate taste buds than non-tasters. Neurosci Lett. 1989;100:271–275. doi: 10.1016/0304-3940(89)90697-6. [DOI] [PubMed] [Google Scholar]
- Mistretta CM, Liu HX. Development of fungiform papillae: patterned lingual gustatory organs. Arch Histol Cytol. 2006;69:199–208. doi: 10.1679/aohc.69.199. [DOI] [PubMed] [Google Scholar]
- Mohiddin SA, Lu S, Cardoso JP, Carroll S, Jha S, Horowits R, Fananapazir L. Genomic organization, alternative splicing, and expression of human and mouse N-RAP, a nebulin-related LIM protein of striated muscle. Cell Motil Cytoskeleton. 2003;55:200–212. doi: 10.1002/cm.10123. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Hamre KM, Holmes A, Lu L, Williams RW. Genetic and structural analysis of the basolateral amygdala complex in BXD recombinant inbred mice. Behav Genet. 2007;37:223–243. doi: 10.1007/s10519-006-9122-3. [DOI] [PubMed] [Google Scholar]
- Murani E, Muraniova M, Ponsuksili S, Schellander K, Wimmers K. Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol. 2007;7:109. doi: 10.1186/1471-213X-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson TM, Munger SD, Boughter JD., Jr Haplotypes at the Tas2r locus on distal chromosome 6 vary with quinine taste sensitivity in inbred mice. BMC Genet. 2005;6:32. doi: 10.1186/1471-2156-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschl C, Brockmann GA, Knott SA. Multiple-trait QTL mapping for body and organ weights in a cross between NMRI8 and DBA/2 mice. Genet Res. 2007;89:47–59. doi: 10.1017/S001667230700852X. [DOI] [PubMed] [Google Scholar]
- Nosrat IV, Agerman K, Marinescu A, Ernfors P, Nosrat CA. Lingual deficits in neurotrophin double knockout mice. J Neurocytol. 2004;33:607–615. doi: 10.1007/s11068-005-3330-2. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Chesler EJ, Williams RW, Lu L. Genetic architecture of the mouse hippocampus: identification of gene loci with selective regional effects. Genes Brain Behav. 2003;2:238–252. doi: 10.1034/j.1601-183x.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997;138:575–588. doi: 10.1083/jcb.138.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi H, Mor N, Lee SY, Doty S, Henderson S, Tanaka S, Yoshioka H, Rattan S, Ramirez F. Esophageal muscle physiology and morphogenesis require assembly of a collagen XIX-rich basement membrane zone. J Cell Biol. 2004;166:591–600. doi: 10.1083/jcb.200402054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BA. Recombinant inbred strains. 2nd ed. Oxford: Oxford University Press; 1989. [Google Scholar]
- Weijnen JA. Licking behavior in the rat: measurement and situational control of licking frequency. Neurosci Biobehav Rev. 1998;22:751–760. doi: 10.1016/s0149-7634(98)00003-7. [DOI] [PubMed] [Google Scholar]
- Williams RW. Neuroscience meets quantitative genetics: using morphometric data to map genes that modulate CNS architecture. Washington (DC): Society for Neuroscience; 1998. [Google Scholar]
- Zhang J, Xiong Y, Zuo B, Lei M, Jiang S, Li F, Zheng R, Li J, Xu D. Detection of quantitative trait loci associated with several internal organ traits and teat number trait in a pig population. J Genet Genomics. 2007;34:307–314. doi: 10.1016/S1673-8527(07)60032-0. [DOI] [PubMed] [Google Scholar]