Abstract
Nup159p/Rat7p is an essential FG repeat–containing nucleoporin localized at the cytoplasmic face of the nuclear pore complex (NPC) and involved in poly(A)+ RNA export and NPC distribution. A detailed structural–functional analysis of this nucleoporin previously demonstrated that Nup159p is anchored within the NPC through its essential carboxyl-terminal domain. In this study, we demonstrate that Nup159p specifically interacts through this domain with both Nsp1p and Nup82p. Further analysis of the interactions within the Nup159p/Nsp1p/Nup82p subcomplex using the nup82Δ108 mutant strain revealed that a deletion within the carboxyl-terminal domain of Nup82p prevents its interaction with Nsp1p but does not affect the interaction between Nup159p and Nsp1p. Moreover, immunofluorescence analysis demonstrated that Nup159p is delocalized from the NPC in nup82Δ108 cells grown at 37°C, a temperature at which the Nup82Δ108p mutant protein becomes degraded. This suggests that Nup82p may act as a docking site for a core complex composed of the repeat-containing nucleoporins Nup159p and Nsp1p. In vivo transport assays further revealed that nup82Δ108 and nup159-1/rat7-1 mutant strains have little if any defect in nuclear protein import and protein export. Together our data suggest that the poly(A)+ RNA export defect previously observed in nup82 mutant cells might be due to the loss from the NPCs of the repeat-containing nucleoporin Nup159p.
INTRODUCTION
Information about the mechanisms by which macromolecules are transported across the nuclear envelope has grown significantly in recent years (for reviews, see Corbett and Silver, 1997; Nigg, 1997; Ohno et al., 1998). This bidirectional transport occurs through nuclear pore complexes (NPCs),1 which are supramolecular protein assemblies spanning the nuclear envelope and estimated to be composed of ∼50 proteins, called nucleoporins (NUPs) (Rout and Blobel, 1993; Matunis and Blobel, 1996). Despite this complexity, it is now expected that all yeast nucleoporins will soon be identified, whereas progress identifying vertebrate nucleoporins is steady, albeit slower (Doye and Hurt, 1997).
On a structural level, a distinguishing feature of ∼15 nucleoporins identified in yeast or vertebrates are their repeat domains, consisting of iterative FG, FXFG, or GLFG sequences with variable spacer regions between repeats. In vitro binding studies, in conjunction with specific coimmunoprecipitations in vivo, strongly suggest that these FG-containing domains represent binding sites for a family of soluble transport receptors that belong to the importin β/karyopherin β family (reviewed in Wente et al., 1996; Corbett and Silver, 1997; Ohno et al., 1998; see also Fornerod et al., 1997; Iovine and Wente, 1997; Neville et al., 1997; Pemberton et al., 1997; Rosenblum et al., 1997; Yaseen and Blobel, 1997). In addition, other nucleoporins have been demonstrated to contain binding sites for the small GTPase Ran, an effector of nuclear transport (for reviews, see Corbett and Silver, 1997; Nigg, 1997).
Although we are beginning to understand the interactions between transport factors and individual nucleoporins, the next level of transport research will be directed at understanding these individual nucleoporins in the context of the overall architecture of the NPC. Understanding how nucleoporins assemble into functional subcomplexes and how these subcomplexes are organized within the NPC will help explain how the architectural layout of the NPC contributes to the docking and translocation processes. To that end, most of the identified vertebrate nucleoporins have been localized by immunoelectron microscopy to various NPC substructures, including the central plug, the cytoplasmic filaments, and the filamentous basket-like structure located at the nucleoplasmic face of the NPC (reviewed in Panté and Aebi, 1996; also see Hu et al., 1996; Cordes et al., 1997). In yeast, however, so far only Nup159p and Nup188p have been localized by immunoelectron microscopy to specific NPC substructures (Kraemer et al., 1995; Nehrbass et al., 1996).
Biochemical approaches have revealed that heptad repeats favoring α-helical coiled-coil structures are one of the major structural motifs through which nucleoporins bind to each other. Among the yeast nucleoporins that contain coiled-coil domains, Nsp1p, Nup49p, and Nup57p constitute a subcomplex to which Nic96p is more loosely associated (Grandi et al., 1993, 1995b). A distinct fraction of Nsp1p was also found to be associated with the carboxyl-terminal coiled-coil domain of Nup82p, an essential nucleoporin required for poly(A)+ RNA export (Grandi et al., 1995a; Hurwitz and Blobel, 1995). Another yeast NPC subcomplex identified includes Nup120p, Nup84p, Nup85p, the in vivo–cleaved carboxyl-terminal half of Nup145p, Seh1p, and a fraction of Sec13p (Siniossoglou et al., 1996; Teixeira et al., 1997). In contrast to the other well-characterized yeast and vertebrate NPC subcomplexes, none of these nucleoporins contains putative coiled-coil domains. Disruption of the genes encoding Nup84p, Nup85p, or Nup120p or deletion of the carboxyl-terminal domain of Nup145p caused a temperature-sensitive (ts) phenotype associated with defects in poly(A)+ RNA export at the restrictive temperature as well as a constitutive clustering of NPCs (Wente and Blobel, 1994; Aitchison et al., 1995a; Heath et al., 1995; Goldstein et al., 1996; Siniossoglou et al., 1996; Teixeira et al., 1997).
A few other yeast nucleoporin mutants have been shown to affect both poly(A)+ RNA export and NPC distribution (for review, see Doye and Hurt, 1997). Among them is the rat7-1/nup159-1 allele, identified in a screen for mRNA export mutants (Gorsch et al., 1995). The RAT7/NUP159 gene (hereafter referred to as NUP159) encodes an essential FG repeat-containing nucleoporin localized at the cytoplasmic face of the NPC (Gorsch et al., 1995; Kraemer et al., 1995). A specific feature of the nup159-1 mutant is that the NPC clustering phenotype is not constitutive, because shifting nup159-1 mutant cells to 37°C restores nearly wild-type NPC distribution (Gorsch et al., 1995). A detailed structural–functional analysis of Nup159p recently revealed that the two predicted coiled-coil regions located near the carboxyl-terminal domain of Nup159p are the only essential domains of the protein and suggested that this carboxyl-terminal domain is required to anchor Nup159p within the NPC (Del Priore et al., 1998).
In this study, we demonstrate that Nup159p associates with both Nup82p and Nsp1p. Within this nuclear pore subcomplex, interactions are mediated by coiled-coil domains present in the three nucleoporins. Biochemical and immunofluorescence studies performed with nup82 and nup159 mutant strains further revealed that the carboxyl-terminal domain of Nup82p anchors Nup159p at the cytoplasmic face of the NPC, whereas the carboxyl-terminal domain of Nup159p is required for the stability of the Nup159p/Nsp1p/Nup82p subcomplex. In agreement with these results, in vivo studies revealed that carboxyl-terminal truncation of Nup159p and Nup82p led to similar defects in NPC distribution and nucleocytoplasmic transport.
MATERIALS AND METHODS
Plasmid and Strain Construction
The yeast plasmids and strains used in this study are listed in Tables 1 and 2. DNA manipulations including restriction endonuclease analyses, fill-in reactions with Klenow fragment, and ligations were performed essentially as described (Maniatis et al., 1982). Microbiological techniques, including yeast growth on minimal or YPD medium, plasmid transformation, and plasmid recovery, were performed as described (Wimmer et al., 1992a), except that minimal SD medium was supplemented either with a 0.1 g/l concentration of each amino acid and nucleic acid base component (obtained from Sigma Chemical, St. Louis, MO) except those used for selection, or in the case of medium lacking uracil and/or tryptophan, by 5 g/l casamino acids (Difco, Detroit, MI), 0.1 g/l adenine, and 0.1 g/l uracil or tryptophan.
Table 1.
Plasmids used in this study
| Plasmid | Comments | Reference | |
|---|---|---|---|
| pSB32-ProtA-C-NSP1 | ARS1, CEN4, LEU2 | 4 IgG binding domains of the proteinA fused to the carboxyl-terminal domain of NSP1 and expressed under the control of the authentic NSP1 promotor. | Grandi et al., 1993 |
| pSB32-ProtA-C-nsp1-ala6 | ARS1, CEN4, LEU2 | Same construct as the pSB32-ProtA-C-NSP1 plasmid, except that within the carboxyl-terminal domain of Nsp1p, 6 positively charged amino acids were replaced by alanine residues. | Grandi et al., 1993 |
| pRS316-ProtA-C-NSP1 | ARSH4, CEN6, URA3 | The BamH1 fragment from plasmid pSB32-ProtA-NSP1 was subcloned into the BamH1 site of the pRS316 plasmid (Sikorski and Hieter, 1989). | This study |
| pSB32-GFP-C-NSP1 | ARS1, CEN4, LEU2 | A SacII fragment obtained by PCR and encoding the GFP S65T/V163A variant (Kahana and Silver, 1996) was used to replace the SacII fragment corresponding to the ProtA domain of the pSB32-ProtA-C-NSP1 plasmid. | This study |
| pSB32-GFP-C-nsp1-ala6 | ARS1, CEN4, LEU2 | A SacII fragment obtained by PCR and encoding the GFP S65T/V163A variant (Kahana and Silver, 1996) was used to replace the SacII fragment corresponding to the ProtA domain of the pSB32-ProtA-C-nsp1-ala6 plasmid. | This study |
| pUN100-ProtA-NUP49 | ARS1, CEN4, LEU2 | Containing 4 IgG binding domains of S. aureus protein A in fusion with the complete NUP49 gene. | Wimmer et al., 1992a |
| pUN100-GFP-NUP49 | ARS1, CEN4, LEU2 | Containing the GFP (S65T, V163A) fused to the amino terminus of NUP49 and with the authentic NUP49 promotor. | Belgareh and Doye, 1997 |
| pFL38-GFP-NUP49 | ARS, CEN, URA3 | The 4.26-kb SacI–BamHI fragment of GFP-NUP49 from the pUN100-GFP-NUP49 plasmid was inserted into pFL38. | Constructed by S. Camier (CEA, Saclay, France) |
| pUN100-ProtA-NUP82 | ARS1, CEN4, LEU2 | Containing the fusion gene between IgG binding domains of protein A joined to the amino-terminal end of Nup82p and expressed under the NOP1 promoter. | Grandi et al., 1995a |
| pRS316-ProtA-NUP82 | ARSH4, CEN6, URA3 | The BamHI–HindIII fragment from pUN100-ProtA-NUP82 was subcloned into the pRS316 plasmid (Sikorski and Hieter, 1989). | This study |
| pUN100-ProtA-N-nup82 | ARS1, CEN4, LEU2 | An internal PvuII–NcoI blunt-ended deletion within the pUN100-ProtA-NUP82 plasmid was used to generate a stop codon at amino acid 460 of Nup82p. | This study |
| pRS315-ProtA-C-nup82 | ARSH4, CEN6, LEU2 | The BamHI–EcoRI blunt-ended fragment from the pUN100-ProtA-NOP1 plasmid (Berges et al., 1994) containing the 2 IgG binding domains of ProtA under the control of the NOP1 promotor was fused in frame to the PvuII–HindIII fragment from the pUN100-ProtA-NUP82 plasmid that encodes the last 251 amino acids of Nup82p, and inserted in the pRS315 vector (Sikorski and Hieter, 1989) opened at its BamHI–HindIII sites. | This study |
| pUN100-GFP-NUP82 | ARS1, CEN4, LEU2 | A SphI–SacI fragment obtained by PCR and encoding the GFP S65T/V163A variant (Kahana and Silver, 1996) was used to replace the SphI–SacI fragment corresponding to the ProtA domain of the pUN100-ProtA-NUP82 plasmid. | This study |
| pRS316-NUP82 | ARSH4, CEN6, URA3 | A SacI–HindIII fragment containing the entire NUP82 gene and cloned into vector pRS316 (Sikorski and Hieter, 1989). | Grandi et al., 1995a |
| pNUP82-wt | ARSH4, CEN6, LEU2 | A PCR product of NUP82, fused in frame with two epitopes of the influenza HA protein, and inserted into pRS315. | Hurwitz and Blobel, 1995 |
| pRS314-NUP82-HA | ARSH4, CEN6, TRP1 | A HindIII blunt-ended fragment from the pNUP82-wt plasmid (Hurwitz and Blobel, 1995) encoding the HA-epitope-tagged Nup82p was inserted into the SmaI site of the pRS314 plasmid (Sikorski and Hieter, 1989). | This study |
| pNUP82-Δ108 | 2μ, LEU2 | A PCR product missing the last 324 nucleotides of NUP82, fused in frame with two epitopes of the influenza hemagglutinin protein and inserted into a modified Yeplac181 plasmid | Hurwitz and Blobel, 1995 |
| pBM3405 | 2μ, URA3 | Mig1-GFP-β-galactosidase fusion in a 2μ, URA3 vector | De Vit et al., 1997 |
| pKW430 | 2μ, URA3 | Contains the NLS-GFP2-NES fusion expressed under the control of the ADH1 promotor in the pRS424 plasmid | Stade et al., 1997 |
Table 2.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| BJ2168 | Mata leu2 trp1 ura3-52 pep4-3 pre1-407 prb1-1122 | Aris and Blobel, 1988 |
| FY86 | Mata his3Δ200 ura3-52 leu2Δ1 | Gorsch et al., 1995 |
| VDPY121 (nup159-ΔN) | Mata his3Δ200 ura3-52 leu2Δ1 rat7::HIS3 (pVDP16-LEU2–rat7-ΔN) | Del Priore et al., 1998 |
| VDPY122 (nup159-C) | Mata his3Δ200 ura3-52 leu2Δ1 rat7::HIS3 (pVDP17-LEU2-rat7-C) | Del Priore et al., 1998 |
| LGY101 (nup159-1) | Mata his3Δ200 ura3-52 leu2Δ1 rat7-1ts | Gorsch et al., 1995 |
| NUP82-WT | Mata his3Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 nup82::HIS3 (pNUP82-WT-LEU2) | Hurwitz and Blobel, 1995 |
| nup82-Δ87 | Mata his3Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 nup82::HIS3 (pNUP82-Δ87-LEU2) | Hurwitz and Blobel, 1995 |
| nup82-Δ108 | Mata his3Δ200 leu2-3,112 lys2-801 trp1-1 ura3-52 nup82::HIS3 (pNUP82-Δ108-LEU2) | Hurwitz and Blobel, 1995 |
| NUP82 shuffle | Mata ade2 trp1 leu2 ura3 his3 nup82::HIS3 (pRS316-URA3-NUP82) | Grandi et al., 1995a |
| GFP-NUP82 (YV201) | Mata ade2 trp1 leu2 ura3 his3 nup82::HIS3 (pUN100-LEU2-GFP-NUP82) | This study |
| GFP-NUP82/nup159-1 (YV384) | Mat? trp? leu2 ura3 his3 nup82::HIS3 rat7-1ts (pUN100-LEU2-GFP-NUP82) | This study |
| xpo1-1 | Mat? ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 xpo1::LEU2 (pKW457=pRS313-HIS3-xpo1-1) | Stade et al., 1997 |
| nsp1-ala6 | Mata ade2 trp1 leu2 ura3 nsp1-ala6::HIS3 | Wimmer et al., 1992b |
| R24 | Mata ade2 ade3 leu2 ura3 trp1 HIS3::nsp1 (pCH1122-URA3-ADE3-NSP1) | Wimmer et al., 1992a |
| ProtA-C-NSP1 | Mata ade2 ade3 leu2 ura3 trp1 HIS3::nsp1 (pSB32-ProtA-C-NSP1) | This study |
| ProtA-C-nsp1-ala6 | Mata ade2 ade3 leu2 ura3 trp1 HIS3::nsp1 (pSB32-ProtA-C-nsp1-ala6) | This study |
| GFP-C-NSP1 | Mata ade2 ade3 leu2 ura3 trp1 HIS3::nsp1 (pSB32-GFP-C-NSP1) | This study |
| GFP-C-nsp1-ala6 | Mata ade2 ade3 leu2 ura3 trp1 HIS3::nsp1 (pSB32-GFP-C-nsp1-ala6) | This study |
To construct the ProtA-C-NSP1, ProtA-C-nsp1-ala6, GFP-C-NSP1, and GFP-C-nsp1-ala6 strains, the pSB32-ProtA-C-NSP1, pSB32-ProtA-C-nsp1-ala6, pSB32-GFP-C-NSP1, and pSB32-GFP-C-nsp1-ala6 plasmids, respectively, were introduced into the R24 strain that carries a disrupted chromosomal NSP1 gene and is complemented by the wild-type NSP1 allele on an ADE3/URA3-containing plasmid (Wimmer et al., 1992a). Colonies were selected on SD-Leu plates, and the transformants were plated on 5-fluoro orotic acid–containing medium to select for cells that have lost the pCH1122-URA3-ADE3-NSP1 plasmid. As expected, the ProtA-C-nsp1-ala6 and GFP-C-nsp1-ala6 strains displayed a ts phenotype.
Similarly, to construct the GFP-NUP82 strain, the pUN100-GFP-NUP82 plasmid was introduced into the “NUP82 shuffle” strain (Grandi et al., 1995a). Colonies were selected on SD-Leu plates, and the transformants were plated on 5-fluoro orotic acid–containing medium to select for cells that have lost the URA3-containing pRS316-NUP82 plasmid.
To combine the nup159-1 allele and the GFP-NUP82 construct, the GFP-NUP82 strain was mated with the LGY101 strain that carries an integrated nup159-1 allele. Diploids growing on SD-leu-trp medium were sporulated. Haploid progeny was selected that were his+, leu+, and temperature sensitive.
Preparation of Whole-Cell Extract
Whole-cell extracts were prepared by resuspending freshly harvested cells from a 20-ml culture (OD600, 1.0) in 0.5 ml of lysis buffer (PBS, 2 mM EDTA, 2.5 mM PMSF, 2.5 mg/ml protease inhibitors). Glass beads (0.4 g) were added, and the samples were incubated during 0.5 h at 4°C with continuous vortexing. The extracts were centrifuged, and the supernatants were diluted with 2× Laemmli buffer (1× Laemli buffer: 62.5 mM Tris, pH 6.8, 10% glycerol, 3% SDS, 5% β-mercaptoethanol) and boiled for 3 min. Aliquots corresponding to 20 μg proteins (as measured by the dye-binding assay of Bradford, 1976) were analyzed on 8% SDS-polyacrylamide gels.
Purification of Protein A Fusion Proteins by IgG-Sepharose Chromatography
Affinity purification of protein A (ProtA)–tagged nucleoporins by IgG-Sepharose chromatography from whole-cell lysates under nondenaturing conditions was performed essentially as described (Grandi et al., 1993; Siniossoglou et al., 1996). Briefly, yeast cells expressing the various protein A–tagged nucleoporins were grown to an OD600 of 1.0 and converted into spheroplasts by treatment with Zymolyase (Seikagaku America, Rockville, MD). Spheroplasts were lysed in 30 ml of lysis buffer (0.5% Triton X-100, 20 mM Tris, pH 8.0, 150 mM KCl, 5 mM MgCl2, and a mixture of protease inhibitors) by performing 10 strokes with a Dounce homogenizer. The lysed spheroplasts were centrifuged for 10 min at 40,000 × g, and the supernatant was loaded onto a 3-mm-diameter column packed with IgG-Sepharose (Pharmacia, Uppsala, Sweden) and equilibrated with lysis buffer. The column was then successively washed with 10–15 ml lysis buffer and 5 ml 5 mM NH4OAc (pH 5.0). The bound proteins were eluted with 1.5 ml 0.5 M acetic acid/NH4OAc (pH 3.4), lyophilized, and resuspended in Laemmli buffer. For the experiments presented in Figure 2B, the originally published lysis buffer (2% Triton X-100, 20 mM NaCl, 0.2 mM MgCl2, 20 mM Tris-HCl, pH 8.0) and washing conditions (Grandi et al., 1993) were used and gave similar results similar to those obtained with the above-mentioned protocol.
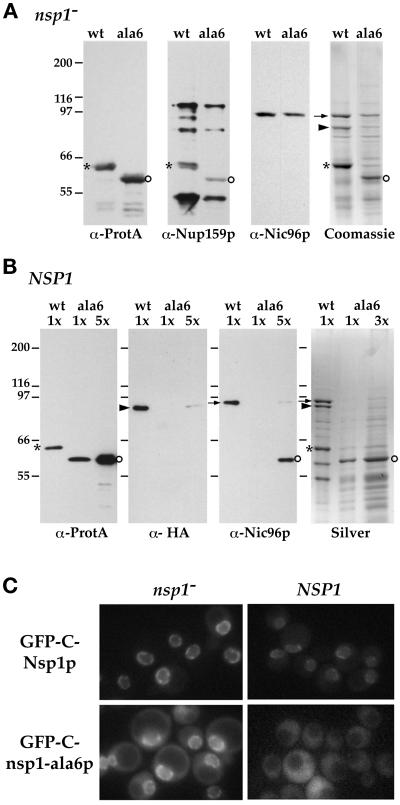
Figure 2.
In the presence of wild-type Nsp1p, Nsp1-ala6p no longer interacts with Nup82p and Nic96p and is not targeted to the NPC. (A) Yeast cells with a disrupted chromosomal copy of NSP1 and complemented by ProtA-C-Nsp1p (wt) or ProtA-C-nsp1-ala6p (ala6) were grown at 24°C. Whole-cell lysates from these two strains were affinity purified by IgG-Sepharose chromatography. The affinity-purified fractions were analyzed by Western blotting for the presence of ProtA-C-Nsp1p (asterisks) or ProtA-C-nsp1-ala6p (open circles), Nup159p (using the rat7#5 antibody), and Nic96p, or by Coomassie blue staining. In this nsp1− background, both ProtA-C-Nsp1p and ProtA-C-nsp1-ala6p were able to interact with Nup159p and Nic96p. Nic96p (arrow) and Nup82p (arrowhead) could also be detected by Coomassie blue staining of the two eluates. Note that in this strain background, Nup159p was degraded during the purification procedure and was therefore hardly detectable by Coomassie blue staining. (B) Whole-cell lysates from NUP82-HA cells carrying a wild-type chromosomal copy of NSP1 and expressing ProtA-C-Nsp1p (wt) or ProtA-C-nsp1-ala6p (ala6) were affinity purified byIgG-Sepharose chromatography as described in MATERIALS AND METHODS. The eluates were analyzed by silver staining (Silver) or by Western blot using an anti-IgG coupled to HRP to detect ProtA-C-Nsp1p (asterisks) or ProtA-C-nsp1-ala6p (open circles), an anti-HA antibody to detect Nup82-HAp (arrrowhead), or an antiNic96p (arrow). In the presence of wild-type Nsp1p, the interaction between ProtA-C-nsp1-ala6p and both Nup82-HAp and Nic96p was inhibited (a very faint band was detectable only when a fivefold excess of the ProtA-C-nsp1-ala6 eluate was loaded). (C) The GFP-C-NSP1 and GFP-C-nsp1-ala6 fusion genes were expressed in an NSP1-disrupted strain (nsp1−) and in a wild-type strain (NSP1). The GFP-C-Nsp1p and GFP-C-nsp1-ala6p signals were observed by fluorescence microscopy in living cells grown at 24°C as described in MATERIALS AND METHODS.
Western Blot Analysis
Proteins were separated on 8% SDS-polyacrylamide gels and transferred to nitrocellulose (Schleicher & Schuell, Keene, NH). Membranes were blocked in Tris-buffered saline plus 0.1% Tween 20 and 5% nonfat milk powder. To decrease nonspecific cross-reactivity of the antibodies with the protein A moiety of the fusion proteins, 10% human serum was added during the various incubation steps. Antibodies were diluted in Tris-buffered saline plus 0.1% Tween 20 and 5% nonfat milk powder at the following dilutions: IgGs coupled to horseradish peroxidase (HRP; Dakopatts, Glostrup, Denmark), 1:5000; rat7#4, raised in guinea pig against the repeat region of Nup159p (Gorsch et al., 1995), 1:10,000; rat7#5, raised in rabbit against the carboxyl domain of Nup159p (Del Priore et al., 1998), 1:5000; anti-Nsp1p, directed at the repeated domains of Nsp1p (Nehrbass et al., 1990), 1:2000; anti-hemagglutinin (HA), 12CA5 mAb (Babco, Richmond, CA), 1:2500; anti-Nic96p, raised in rabbit against the amino-terminal domain of Nic96p (Grandi et al., 1995b), 1:500; and anti-Nup133p directed at the amino-terminal domain of Nup133p (Belgareh and Doye, 1997), 1:250. The secondary antibodies were goat anti-mouse or anti-rabbit IgGs coupled to HRP (Jackson ImmunoResearch), 1:5000, and goat anti-guinea pig IgG coupled to alkaline phosphatase (Sigma), 1:1000.
Fluorescence and Immunofluorescence Experiments
Indirect immunofluorescence was performed as described previously (Gorsch et al., 1995). Briefly, cells were grown overnight to early log phase, shifted for 3 h to 37°C, fixed with 3.7% formaldehyde (Fisher Scientific, Pittsburgh, PA), washed, and converted to spheroplasts by using 300 mg/ml Zymolyase 100T. Cells were incubated overnight at 4°C with the rat7#4 antibody (Gorsch et al., 1995) diluted 1:3000. Cells were washed and then incubated with an FITC-conjugated anti-guinea pig IgG secondary antibody (Vector Laboratories, Burlingame, CA) at a dilution of 1:250 and viewed and photographed with a Zeiss (Thornwood, NY) Axiphot microscope equipped with a charge-coupled device (CCD) camera.
Protein export assays were performed as described (Stade et al., 1997) using the NLS-GFP2-NES (nuclear localization signal-2 green fluorescent protein molecules-nuclear export signal) reporter protein. Cells were grown to early log phase, shifted to 37°C for 2 h, collected by brief centrifugation, and resuspend in 1 ml medium. Four microliters of cells were spotted onto coverslips coated with 1% polyethylenimine and viewed as above.
To visualize NPC distribution in living cells, the strains were transformed with the pFL38-GFP-NUP49 plasmid which encodes a form of Nup49p tagged with GFP (Belgareh and Doye, 1997). To visualize GFP-Nup82p, cells were grown in SD medium lacking uracil to midlog phase at the indicated temperature and shifted for 2 h to 37°C. To inhibit protein synthesis, cultures were treated with either 10 or 30 μg cycloheximide per ml, added 5 min before shifting to 37°C.
For the in vivo analysis of nuclear protein import, wild-type or nucleoporin mutant cells expressing the Mig1p-GFP-LacZ reporter (De Vit et al., 1997) were grown overnight at 25°C in SD-uracil medium. This primary culture was used to inoculate secondary cultures of SGly-uracil medium (5% glycerol, 6.7 g/l yeast nitrogen base without amino acids, 5% casamino acids, 0.1 mg/l adenine and tryptophan). Cells were then grown at 25°C to an OD600 of ∼0.5 and either kept at 25°C or shifted for 2 h to 37°C. One milliliter of cell suspension was transferred to a prewarmed Eppendorf (Madison, WI) tube, briefly centrifuged, resuspended in 100 μl SGly-uracil medium, and kept at the appropriate temperature. At this stage, 10 μl of a 20% glucose solution was added to induce the nuclear import of the Mig1p-GFP-LacZ reporter (defining this time as t = 0). Five microliters of cells were rapidly placed between a slide and a coverslip and observed by fluorescence microscopy using the FITC channel. A given field was recorded at various times after the addition of glucose (routinely t = 1, 3, 5, and 8 min) using a CCD camera. Alternatively, cells were kept at 25 or 37°C for 5 or 10 min after glucose addition, and independent fields were recorded.
For in vivo fluorescence analysis, 5–10 μl of cells expressing the appropriate GFP-fusion protein were placed on a microscope slide and covered with a coverslip. Images were obtained by using a cooled CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) on a microscope (Leica, Deerfield, IL) equipped with the following filter set: excitation, 450–490 nm; dichroic, 510 nm; emission, 515–560 nm (filter L4, Leica), a 100-W mercury arc lamp, and a 100×, numerical aperture 1.4 objective lens. Identical exposure conditions were used for all comparable images, and composites were prepared using Adobe (Mountain View, CA) Photoshop with identical brightness and contrast optimization for all comparable images within a figure.
Electron Microscopy Experiments
Cells were examined by electron microscopy as previously described (Byers and Goetsch, 1975; Wright and Rine, 1989; Goldstein et al., 1996), except that cells were incubated for 4 h at 16°C. Thin sections of 90 nm were cut on a MT5000 ultramicrotome (Dupont Sorvall, Norwalk, CT), poststained in 2% uranyl acetate and Reynold’s lead citrate, and then examined and photographed using a Jeol (Tokyo, Japan) 100CX electron microscope at 80 kV.
RESULTS
Nup159p Is Part of the Nsp1p- and Nup82p-containing NPC Subcomplex
Most of the yeast nucleoporins that contain heptad repeats have been shown to be part of one of two distinct nuclear pore subcomplexes: the Nsp1p/Nup49p/Nup57p/Nic96p subcomplex and the Nsp1p/Nup82p subcomplex (Grandi et al., 1993, 1995a,b). Because the identification of yeast nucleoporins is close to completion (Doye and Hurt, 1997), we were interested in determining with which nucleoporins the heptad repeat containing nucleoporin Nup159p interacted. We began by determining whether Nup159p might associate with either of these two subcomplexes. Because Nup159p is highly sensitive to proteolysis (Gorsch et al., 1995; Kraemer et al., 1995), affinity chromatography under nondenaturing conditions was carried out using whole-cell lysates obtained from protease-deficient BJ2168 strains (Figure 1, A and B) expressing ProtA-Nup49p, ProtA-C-Nsp1p, or ProtA-Nup82p (the ProtA tag refers to two or four IgG binding sequences from the Staphylococcus aureus protein A; ProtA-C-Nsp1p only contains the essential carboxyl-terminal domain of Nsp1p; see Table 1). The purified fractions, isolated by IgG-Sepharose chromatography, were probed with IgG coupled to HRP to detect the fusion proteins (α-ProtA) or with a polyclonal antibody directed against the central FG repeat–containing domain of Nup159p (Figure 1A). Full-length Nup159p migrating with an apparent molecular mass of 205 kDa (Gorsch et al., 1995; Kraemer et al., 1995), as well as faster-migrating bands corresponding to Nup159p breakdown products, were detected with the anti-Nup159p antibody in both the ProtA-C-Nsp1p and ProtA-Nup82p eluates but not in the ProtA-Nup49p fraction. These results thus demonstrate that Nup159p interacts, directly or indirectly, with both Nup82p and the carboxyl-terminal domain of Nsp1p.
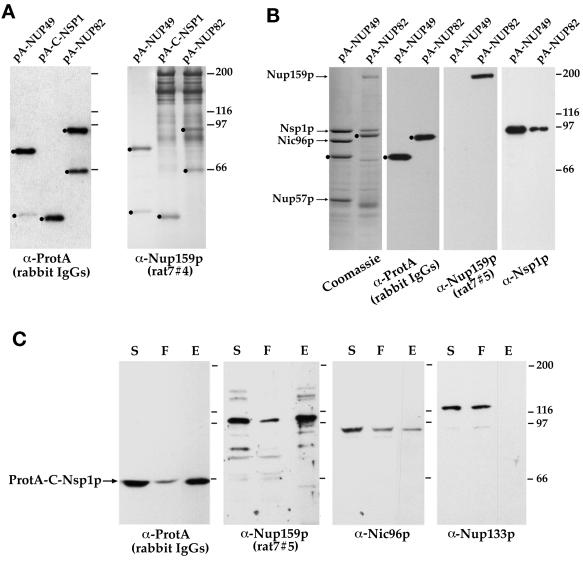
Figure 1.
Nup159p interacts with Nup82p and Nsp1p. (A and B) ProtA-Nup49p, ProtA-Nup82p, and ProtA-C-Nsp1p expressed in the protease-deficient BJ2168 strain were affinity purified by chromatography on IgG-Sepharose as described in MATERIALS AND METHODS. (A) Purified fractions from the ProtA-Nup49p, ProtA-C-Nsp1p, and ProtA-Nup82p affinity columns were analyzed by immunoblotting after SDS-PAGE, using IgG coupled to HRP to visualize the ProtA-fusion proteins (α-ProtA) and an anti-Nup159p antibody directed against the repeat motifs of Nup159p (rat7#4, see MATERIALS AND METHODS). The positions of the ProtA fusion proteins (dots) are indicated. Note that the rat7#4 antibody also cross-reacts with ProtA chimeras. (B) Eluted fractions from the ProtA-Nup49p and ProtA-Nup82p affinity columns were analyzed by SDS-PAGE followed by Coomassie blue staining or by Western blotting using IgG coupled to HRP to visualize the ProtA constructs, an antibody directed against the carboxyl-terminal domain of Nup159p (rat7#5), and an anti-Nsp1p antibody directed against the repeat domain of Nsp1p. Coomassie blue staining reveals Nsp1p, Nic96p, and Nup57p in the ProtA-Nup49p–purified fraction, whereas Nsp1p and Nup159p copurify with the ProtA-Nup82p fusion protein. The positions of Nup159p, Nsp1p, Nic96p, Nup57p (arrows), and the ProtA fusion proteins (dots) are indicated. (C) Affinity purification of ProtA-C-Nsp1p expressed in an NSP1-disrupted strain. Equivalent amounts of the load (soluble fraction from the whole-cell lysates [S]), of the unbound (flow-through [F]), and of the bound and subsequently eluted (E) fractions were analyzed by Western blotting. IgGs coupled to HRP were used to detect ProtA-C-Nsp1p, the rat7#5 antibody to visualize Nup159p, and antibodies directed against Nic96p and Nup133p were used as controls. Note that the NSP1-disrupted strain is not protease deficient, leading to a substantial degradation of Nup159p with a major degradation product migrating at ∼100 kDa.
To determine the relative abundance of Nsp1p and Nup159p in the ProtA-Nup82p complex, affinity-purified fractions obtained from the protease-deficient BJ2168 strain expressing ProtA-Nup82p (or ProtA-Nup49p used as a control) were stained with Coomassie blue. In this experiment, Nup159p and Nsp1p were especially well preserved from proteolysis, which enabled us to detect these two proteins by Coomassie blue staining (Figure 1B). Under those conditions, no other major purifying proteins could be detected in the ProtA-Nup82p eluate.
To quantify the fraction of Nup159p that is associated to the Nsp1p/Nup82p subcomplex, affinity purification of ProtA-C-Nsp1p expressed in an NSP1-disrupted strain was performed, and aliquots of the load (soluble fraction from the whole-cell lysates), of the unbound (flow-through) and of the bound and subsequently eluted fractions were coincidentally analyzed. As shown in Figure 1C, this revealed that a major fraction of Nup159p (equivalent to the fraction of ProtA-C-Nsp1p that is bound and subsequently eluted from the IgG-Sepharose column) can be copurified with ProtA-C-Nsp1p under our lysis conditions (the NSP1-disrupted strain is not protease deficient, leading to a substantial degradation of Nup159p). In contrast, <50% of Nic96p copurified with ProtA-C-Nsp1p under these conditions. Nic96p being one of the most abundant nucleoporins (Aitchison et al., 1995b), this result might reflect the fact that, unlike Nup159p, which mainly interacts with the Nsp1p-Nup82p complex, Nic96p might be associated with several NPC subcomplexes. These results, together with the presence of equivalent amounts of Nup159p in both ProtA-Nup82p and ProtA-C-Nsp1p eluates (Figure 1A) and the previously reported interaction between Nsp1p and Nup82p (Grandi et al., 1995a), therefore demonstrate that these three proteins are the major components of a distinct nuclear pore subcomplex.
Mutations within the Carboxyl-terminal Domain of Nsp1p Affect Its Interaction with Both the Nup82p/Nup159p- and the Nic96p-containing Subcomplexes
Previous studies, based on the affinity purification of ProtA-C-nsp1-ala6p expressed in a wild-type NSP1 (wtNSP1) background (Grandi et al., 1993) indicated that the ProtA-C-nsp1-ala6 mutant allele was not able to interact with Nic96p but was still able to interact with an 80-kDa species comigrating with the 80-kDa band seen with intact ProtA-C-Nsp1p (and subsequently identified as Nup82p; Grandi et al., 1995a). Unexpectedly, we found that when ProtA-C-nsp1-ala6p was expressed in an nsp1-disrupted strain, both Nup159p and Nic96p copurified as efficiently with ProtA-C-nsp1-ala6p as with ProtA-C-Nsp1p (Figure 2A). When similar studies were performed in a wtNsp1p background (as originally published), the use of HA-tagged Nup82p revealed that the interaction between ProtA-C-nsp1-ala6p and both Nic96p and Nup82p was impaired (Figure 2B). Accordingly, the 80-kDa species comigrating with the 80-kDa band seen with ProtA-C-Nsp1p (Grandi et al., 1995a) is not Nup82p but most likely corresponds to a nonspecific interacting protein. In agreement with these biochemical data, we observed that unlike GFP-C-Nsp1p, GFP-nsp1-ala6p is not efficiently targeted to the NPC in wild-type cells (i.e., when wtNsp1p is present) and only becomes targeted in the absence of Nsp1p (Figure 2C). These results therefore demonstrate that the interaction between nsp1-ala6 and both Nic96p and Nup82p is compromised in the presence of competing Nsp1p. Accordingly, identical or overlapping domain(s) within the heptad repeat sequence of Nsp1p might be involved in the interaction in the context of both the Nsp1p/Nup159p/Nup82p and the Nsp1p/Nup49p/Nup57p/Nic96p NPC subcomplexes.
A Deletion within the Heptad Repeat–containing Carboxyl-terminal Domain of Nup82p Destabilizes Its Interaction with the Nsp1p/Nup159p Complex
The heptad repeat-containing carboxyl-terminal domain of Nup82p has been shown previously to interact with the essential carboxyl-terminal domain of Nsp1p that also contains heptad repeats (Grandi et al., 1995a). To examine whether this domain of Nup82p is also involved in the interaction with Nup159p, the ProtA-N-nup82p and ProtA-C-nup82p constructs, containing the amino- and carboxyl-terminal domains of Nup82p, respectively (see Table 1), and the ProtA-Nup82p construct were introduced into the BJ2168 protease-deficient strain, and affinity chromatography was performed on whole-cell extracts. Equal amounts of eluates were tested for the presence of Nup159p, and, as control, for the presence of Nsp1p (Figure 3A). Nup159p was found to interact with the carboxyl-terminal domain of Nup82p (although to a lower extent compared with Nsp1p), whereas no interaction was detected with the amino-terminal construct. This result therefore suggests that the heptad repeat–containing carboxyl-terminal domain of Nup82p is involved in the interaction with both Nsp1p and Nup159p. However, as previously reported (Grandi et al., 1995a), neither ProtA-N-nup82p nor ProtA-C-nup82p is able to complement the lethal phenotype of a nup82-disrupted strain. Furthermore, analysis of the localization of GFP-tagged N-nup82p or C-nup82p did not reveal a specific targeting of either of these two constructs in a wild-type strain (Belgareh and Doye, unpublished results). Accordingly, the experiments presented in Figure 3A could only be conducted in the presence of wtNup82p, which might compete with ProtA-N-nup82p or ProtA-C-nup82p (as observed above in the case of ProtA-C-nsp1-ala6p).
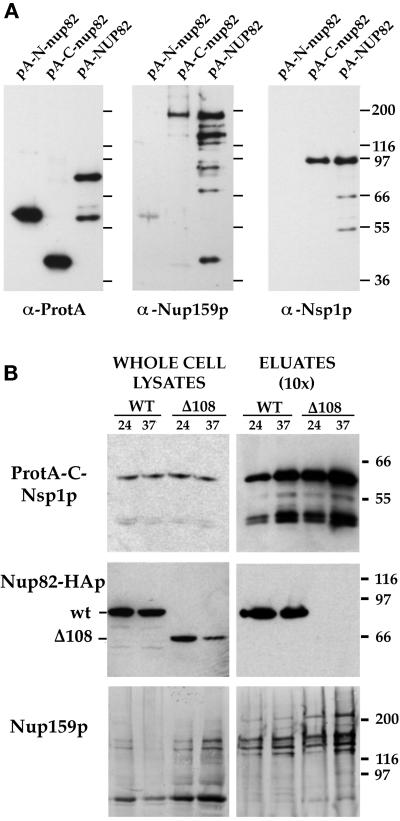
Figure 3.
Nsp1p and Nup159p form a core complex independent of interaction with the carboxyl-terminal domain of Nup82p. (A) Affinity purification by IgG-Sepharose chromatography of ProtA-N-nup82p, ProtA-C-nup82p, and ProtA-Nup82p expressed in strains derived from BJ2168. The purified fractions were analyzed by Western blotting using IgG coupled to HRP to detect the ProtA fusions, an anti-Nup159p antibody directed against the carboxyl-terminal domain of Nup159p (rat7#5), and an antibody directed against the repeat domain of Nsp1p. (B) Whole-cell lysates from NUP82 (WT) and nup82-Δ108 (Δ108) strains transformed with the pRS316-ProtA-C-Nsp1p plasmid and maintained at 24°C or shifted to 37°C for 3 h were affinity purified by IgG-Sepharose chromatography. Whole-cell lysates and affinity-purified fractions (eluates, 10-fold equivalent) were analyzed by Western blotting for the presence of ProtA-C-Nsp1p using an anti-IgG coupled to HRP, for the presence of Nup82-HAp and Nup82-Δ108-HAp with a monoclonal antibody directed against the HA epitope, and for the presence ofNup159p with the rat7#4 antibody. Although Nup82-Δ108-HAp does not interact with ProtA-C-Nsp1p at either 24 or 37°C, Nup159p still copurifies with ProtA-C-Nsp1p in the nup82Δ108 strain. Note that the NUP82-wt and nup82Δ108 strains are not protease deficient, leading to a substantial degradation of Nup159p. The shorter degradation product of Nup159p did not copurify with ProtA-C-Nsp1p.
To further characterize the role of the coiled-coil domain of Nup82p in the assembly of the Nsp1p/Nup159p/Nup82p complex, we therefore introduced the ProtA-C-Nsp1p construct into the NUP82-wt and the nup82Δ108 strains, producing either full-length HA epitope-tagged Nup82p or a mutant form of Nup82p deleted for its last 108 amino acids and thus lacking the second half of its putative coiled-coil domain (Hurwitz and Blobel, 1995). Cells expressing Nup82Δ108p on a high-copy plasmid as the only form of Nup82p were temperature sensitive for growth (Hurwitz and Blobel, 1995), whereas cells in which the mutant allele was present on a centromeric plasmid grew very poorly, even at permissive temperature (our unpublished results). As previously reported (Hurwitz and Blobel, 1995), immunoblotting using the anti-HA monoclonal antibody revealed a decrease in the total amount of Nup82Δ108p when nup82Δ108 cells were shifted for 3 h to 37°C (Figure 3B), whereas wild-type Nup82p remains stable. Affinity purification on IgG-Sepharose was carried out using whole-cell lysates from NUP82-wt or nup82Δ108 strains expressing ProtA-C-Nsp1p and grown at permissive temperature (24°C) or shifted to 37°C for 3 h. Western blot analysis revealed that, unlike wild-type Nup82p, the Nup82Δ108p protein did not copurify with ProtA-C-Nsp1p, even when cells were grown at permissive temperature (Figure 3B). Similar results were obtained with the nup82Δ87 strain (our unpublished results), even though this strain does not display a ts phenotype (Hurwitz and Blobel, 1995). This indicates that a deletion within the carboxyl-terminal domain of Nup82p destabilizes its interaction with Nsp1p (i.e., this interaction is no longer stable under our lysis conditions). Interestingly, however, the anti-Nup159p antibody revealed the presence of equivalent amount of Nup159p in the ProtA-C-Nsp1p eluates from both NUP82-wt, and nup82Δ108 strains (Figure 3B). This indicates that the presence of Nup82p is not required for the interaction between Nsp1p and Nup159p.
The Carboxyl-terminal Domain of Nup159p Is Required for the Stability of the Nup159p/Nsp1p/Nup82p Complex
To specify the domains of Nup159p involved in the interaction with Nsp1p and Nup82p, the ProtA-C-NSP1 and ProtA-NUP82 constructs were introduced into the nup159-ΔN mutant, which produces a form of Nup159p lacking its amino terminus (amino acids 1–456), and the nup159-C mutant, in which only the carboxyl-terminal third of Nup159p, containing the essential heptad repeat, remains (Del Priore et al., 1998). IgG-Sepharose chromatography was performed, and equal amounts of eluates were tested for the presence of Nup159p by immunoblotting using an antibody to the carboxyl-terminal portion of Nup159p. Both the Nup159-ΔNp and Nup159-Cp truncated proteins were detected in interactions with ProtA-C-Nsp1p (Figure 4A) and ProtA-Nup82p (Figure 4B), indicating that the heptad repeat-containing carboxyl-terminal domain of Nup159p is sufficient for its interaction with both Nsp1p and Nup82p.
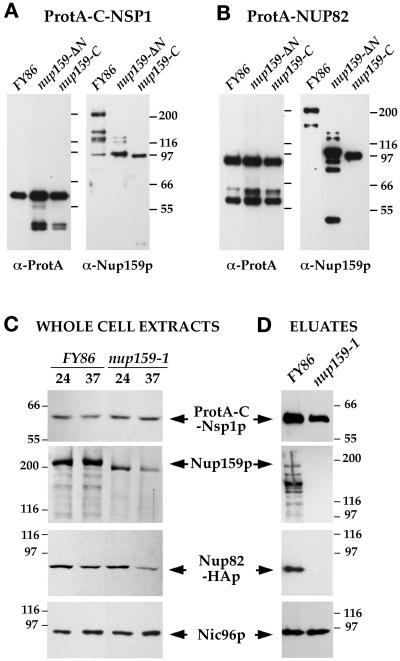
Figure 4.
A deletion within the carboxyl-terminal domain of Nup159p impairs the stability of the Nup159p/Nsp1p/Nup82p complex. (A and B) ProtA-C-Nsp1p (A) or ProtA-Nup82p (B), expressed in FY86 (NUP159-wt), a nup159ΔN, and a nup159-C strain were affinity purified using IgG-Sepharose chromatography, and the affinity-purified fractions were analyzed by Western blotting. IgG coupled to HRP was used to detect the ProtA fusion proteins, whereas the presence of Nup159p was detected using the rat7#5 antibody directed against the carboxyl-terminal domain of Nup159p. Note that full-length Nup159p and Nup159ΔNp proteins were highly sensitive to proteolysis, leading to several breakdown products recognized by the anti-Nup159p antibody, whereas no proteolysis of the Nup159-Cp protein was observed. (C) FY86 (wt) and nup159-1 strains, transformed with the pRS316-ProtA-C-NSP1 and pRS314-Nup82-HA plasmids, were grown at 24°C or shifted to 37°C for 3 h. Whole-cell extracts were prepared as described in MATERIALS AND METHODS and analyzed by Western blotting for the presence of ProtA-C-Nsp1p, Nup82-HAp, Nup159p (using the rat7#4 antibody that presents the same affinity for Nup159p and for the truncated Nup159-1p), and Nic96p. After a 3-h shift to 37°C, which induces the degradation of Nup159-1p, the apparent expression level of Nup82-HAp is also decreased. (D) Whole-cell lysates from these two strains grown at 24°C were affinity purified by IgG-Sepharose chromatography. The affinity-purified fractions (eluates) were analyzed by Western blotting and tested for the presence of ProtA-C-Nsp1p, Nup82-HAp, Nup159p, and Nic96p. Unlike wild-type Nup159p, Nup159-1p does not copurify with ProtA-C-Nsp1p. In this mutant strain, the interaction between Nup82-HAp and ProtA-C-Nsp1p was also inhibited, whereas Nic96p still copurified with ProtA-C-Nsp1p.
We subsequently analyzed the behavior of the Nup159p/Nsp1p/Nup82p subcomplex in the nup159-1 mutant strain, which produces a protein truncated for its last 96 amino acids and which is degraded after a shift to 37°C (Gorsch et al., 1995; Del Priore et al., 1998; see Figure 4C). Western blot analysis of ProtA-C-Nsp1p copurifying proteins in nup159-1 mutant cells grown at 24°C revealed that the Nup159-1p mutant protein no longer interacts with ProtA-C-Nsp1p (Figure 4D). Under those lysis conditions, the interaction between ProtA-C-Nsp1p and Nup82p was also inhibited. This suggests that the heptad repeat–containing domain of Nup159p establishes a link between Nsp1p and Nup82p, or that Nup82p can only interact with a preassembled Nsp1p/Nup159p subcomplex. In contrast, the amount of copurifying Nic96p, used as control, was not modified (Figure 4D). This result indicates that a carboxyl-terminal deletion within Nup159p impairs the stability of the Nup159p/Nsp1p/Nup82p complex but does not interfere with the assembly or stability of the other Nsp1p-containing subcomplex.
Degradation of Nup82Δ108p at 37°C Leads to the Mislocalization of Nup159p
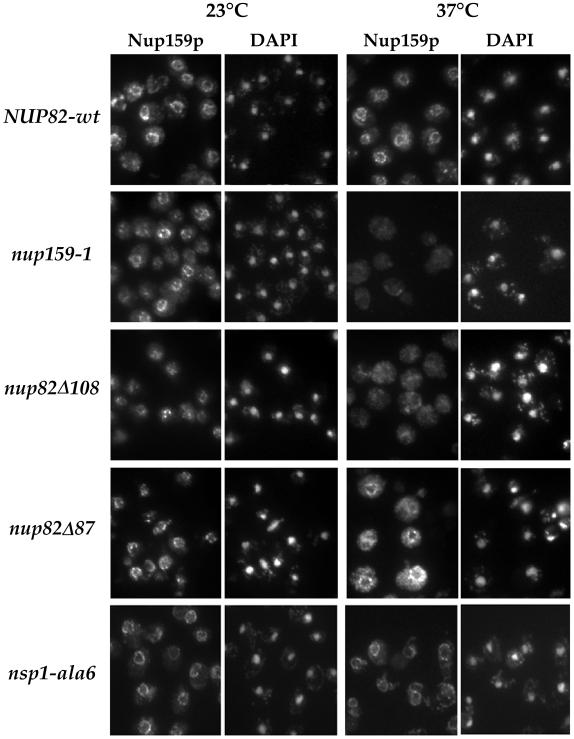
Because affinity purification studies only allow conclusions about whether protein–protein interactions are biochemically stable under specific lysis conditions, and might not accurately reflect the interactions within the living cells, we analyzed the localization of Nup159p, Nup82p, and Nsp1p in the nup159-1 and nup82Δ108 mutant strains in which the Nup159-1p and Nup82Δ108p proteins, respectively, are degraded at 37°C.
Immunofluorescence studies, using a specific anti-Nup159p antibody, revealed that in all the strains grown at 23°C, Nup159p was localized at the nuclear periphery (Figure 5). After a 3-h shift to 37°C, Nup159p was still localized at the nuclear rim in the NUP82-wt, nup82Δ87, and ala6-nsp1 strains, whereas Nup159p staining was lost from the nuclear periphery in both nup159-1 and nup82Δ108 mutant cells. Unlike in the nup159-1 strain, no specific degradation of Nup159p could be observed in the nup82Δ108 mutant strain after a 3-h shift to 37°C (see Figure 3B). Accordingly, the lack of nuclear rim staining in the nup82Δ108 cells grown at 37°C indicates that Nup159p is specifically delocalized from the nuclear pores when Nup82Δ108p is degraded. Even in the nup82Δ108 strain at 23°C, there appears to be a reduced amount of Nup159p at the nuclear rim.
Figure 5.
Nup159p is lost from the NPCs when nup159-1 or nup82Δ108 cells are shifted to 37°C. Wild-type (NUP82-wt), nup159-1, nup82Δ108, nup82Δ87, and ala6-nsp1 cells were grown at 23°C or shifted for 3 h to 37°C. The subcellular localization of Nup159p was analyzed by immunofluorescence using a polyclonal antibody directed against the repeat-containing domain of Nup159p. Cells were also stained for DNA with DAPI. At 23°C, nuclear pore labeling was observed in all five strains. After a shift to 37°C, a wild-type staining pattern was still observed in the NUP82-wt, nup82Δ87, and ala6-nsp1 strains, whereas no perinuclear staining could be detected in the nup159-1 and nup82Δ108 strains, indicating that Nup159p had been delocalized from the NPCs in these two strains.
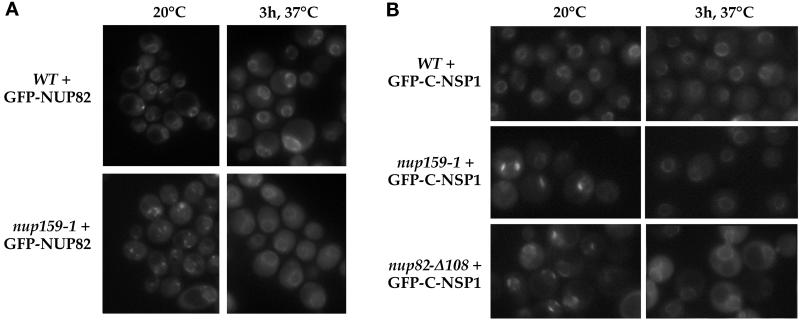
Conversely, we analyzed the localization of Nup82p in nup159-1 cells shifted to 37°C, using a GFP-Nup82p fusion protein. This protein was functional, because it could rescue the otherwise lethal phenotype of an NUP82-disrupted strain (our unpublished results). In NUP82-disrupted cells, (NUP159-wt) GFP-Nup82p gave rise to a typical punctate ring-like staining surrounding the nucleus, although some nonspecific aggregates were occasionally observed in the cytoplasm (Figure 6A). This strain was mated with the nup159-1 strain, and after sporulation, a nup159-1 strain expressing GFP-Nup82p as the only form of Nup82p was obtained (strain YV384; see Table 2). In agreement with previous immunofluorescence and electron microscopy studies demonstrating the reversible NPC clustering of the nup159-1 strain (Gorsch et al., 1995), the GFP-Nup82p chimera localized to the clustered NPCs present in nup159-1 cells grown at 23°C. After a 3-h shift at 37°C, which induces the degradation of Nup159-1p and a concomitant redistribution of the clustered NPCs (Gorsch et al., 1995), the GFP-Nup82p protein regained a ring-like pattern around the nuclear envelope, indicating that degradation of Nup159-1p at 37°C in the nup159-1 mutant strain did not lead to a complete delocalization of the GFP-Nup82p protein (Figure 6A). However, the GFP staining within the cytoplasm was somewhat enhanced at 37°C. The same result was obtained in the presence of cycloheximide, indicating that accumulation of GFP in the cytoplasm was not due to a failure to incorporate newly synthesized GFP-Nup82p into NPCs (Snay-Hodge and Cole, unpublished results). Accordingly, this cytoplasmic background could reflect either a partial delocalization of GFP-Nup82p or the degradation of a fraction of this protein. This latter hypothesis being consistent with the apparent degradation of Nup82-HAp independently observed in nup159-1 cells after a 3-h shift to 37°C (see Figure 4C).
Figure 6.
In vivo localization of GFP-Nup82p and GFP-Nsp1p in the nup159-1 or nup82Δ108 mutant strains. (A) The GFP-NUP82 fusion gene was expressed in a NUP82-disrupted strain and in a nup82::HIS3/nup159-1 strain (strain YV384). The GFP-Nup82p staining was observed by fluorescence microscopy in living cells grown at 20°C or shifted for 3 h to 37°C as described in MATERIALS AND METHODS. In wild-type NUP159 cells, the GFP-Nup82p fusion protein was localized at the NPCs and gave a ring-like staining at both 20 and 37°C. In nup159-1 mutant cells grown at 20°C, the GFP-Nup82p was less homogeneously distributed around the nuclear envelope because of the NPC clustering phenotype of the nup159-1 strain. After a 3-h shift to 37°C that leads to a redistribution of the NPCs, the GFP-Nup82p was still localized at the NPC in the nup159-1 strain, although enhanced cytoplasmic staining is also evident. (B) The GFP-C-NSP1 construct was introduced in a wild-type (WT) strain and the nup159-1 and nup82Δ108 strains. Cells were grown at 20°C or shifted for 3 h at 37°C. In wild-type cells, GFP-Nsp1p gave a ring-like staining at both 20 and 37°C. In nup159-1 and nup82Δ108 strains, the GFP-Nsp1p labeling was restricted to a few clusters at 20°C and gave rise to a homogeneous ring staining after a 3-h shift to 37°C.
The loss of Nup159p from the NPCs in nup82Δ108 cells grown at 37°C, together with the persistence of a perinuclear GFP-Nup82p staining in nup159-1 cells shifted to 37°C, therefore indicates that Nup82p most likely anchors Nup159p at the NPC even though the stabilization of this complex may require the presence of the three proteins. Because our affinity purification studies revealed that in the nup82Δ108 strain, ProtA-C-Nsp1p stably interacts with Nup159p even at 37°C (Figure 3B), we determined whether a fraction of Nsp1p might be delocalized from the NPCs in nup82Δ108 and nup159-1 cells after a 3-h shift to 37°C. Therefore, the in vivo localization of GFP-tagged Nsp1p was analyzed in wild-type, nup159-1, and nup82Δ108 cells (Figure 6B). At 20°C, GFP-C-Nsp1p was localized at NPCs that were homogeneously distributed in the wild-type strain and clustered in the nup159-1 strain and unexpectedly also in the nup82Δ108 strain. After a 3-h shift to 37°C, the nuclear envelope was uniformly stained, reflecting redistribution of clustered NPCs. Most of the GFP-C-Nsp1p protein remained at the nuclear envelope in both the wild-type and the two mutant strains, with no significant increase of the cytoplasmic GFP labeling (Figure 6B). Accordingly, if as anticipated, a fraction of GFP-Nsp1p involved in the Nsp1p/Nup82p/Nup159p complex is delocalized from the NPCs, this result indicates that only a minor fraction of Nsp1p is found in this complex compared with the fraction of Nsp1p in the Nsp1p/Nup49p/Nup57p/Nic96p complex (also see DISCUSSION).
Carboxyl-terminal Truncations of Nup159p and Nup82p Lead to Similar Defects in NPC Distribution and Nucleocytoplasmic Transport
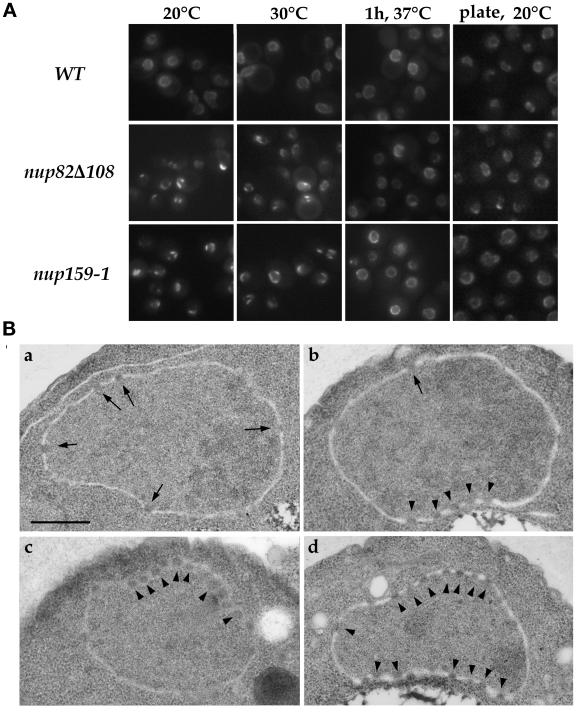
To assess the specificity of the clustering phenotype observed with the GFP-C-NSP1 construct in nup82Δ108 cells grown at 20°C, we introduced the GFP-NUP49 construct (Belgareh and Doye, 1997) into the nup159-1 and nup82Δ108 strains and examined the in vivo localization of the GFP-Nup49p chimera at various temperatures. In nup82Δ108 cells grown to early log phase at temperatures between 16 and 20°C, a mild NPC clustering phenotype, similar to the one observed in nup159-1 cells, was observed (Figure 7A). However, this clustering phenotype was not as striking as the one observed in other clustering mutants, such as an nup133− mutant strain (Belgareh and Doye, 1997), because a faint fluorescent signal was still detected around the nuclear envelope. Thin section electron microscopy confirmed this nonhomogeneous NPC distribution and further revealed that this mild clustering phenotype was not associated with major alterations in the structure of the nuclear envelope (Figure 7B). This clustering phenotype was less pronounced when nup82Δ108 cells were grown at 30°C, and a wild-type distribution of the nuclear pores was restored when cells grown at 20°C were shifted for 1 h to 37°C (Figure 7A). This result thus demonstrates that nup82Δ108 cells, like nup159-1 cells, display a reversible clustering of nuclear pores. Interestingly, the ability to analyze NPC distribution in living yeast cells further revealed that nup159-1 and nup82Δ108 cells grown on plates, or reaching late log phase in liquid culture, did not display any major defect in nuclear pore distribution (Figure 7A).
Figure 7.
The nup82Δ108 mutant exhibits a mild and reversible NPC clustering phenotype. (A) To analyze NPC distribution in vivo, wild-type (WT), nup82Δ108, and nup159-1 strains were transformed with the pFL38-GFP-NUP49 plasmid. Yeast cells were grown in liquid SD medium lacking uracil to early log phase at 20 or 30°C, grown at 20°C, and shifted for 1 h to 37°C or grown on plates at 20°C for 2 d. These cells were examined for GFP fluorescence as described in MATERIALS AND METHODS. Wild-type cells displayed a homogeneous labeling pattern around the nuclear envelope at all temperatures. In contrast, a mild clustering phenotype was observed in nup82Δ108 cells and nup159-1 cells at 20°C and to a lesser extent at 30°C. This clustering phenotype is reversed after a 1-hour shift to 37°C. Strikingly, nup82Δ108 and nup159-1 cells grown on plates at 20°C exhibited a homogeneous staining of the nuclear envelope. (B) Electron micrographs of nuclei from wild-type (a) and nup82Δ108 (b–d) strains grown at 16°C. Wild-type cells have nuclei with evenly distributed NPCs, whereas nup82Δ108 cells exhibited grouped or clustered NPCs. Bar, 0.5 μm. Arrows point to single NPCs, and arrowheads point to clustered nuclear pores.
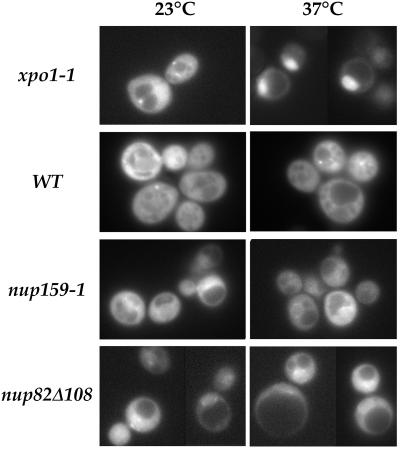
Besides this highly reversible alteration in NPC distribution, both nup159-1 and nup82Δ108 mutant strains display major defects in poly(A)+ RNA export (Gorsch et al., 1995; Hurwitz and Blobel, 1995). Because a mutation in Crm1p/Xpo1p, a shuttling protein that shows homology to importin β–like transport factors, leads to defects in export of both NES-bearing proteins and poly(A)+ RNA, it was suggested that these pathways might be tightly coupled in Saccharomyces cerevisiae (Stade et al., 1997). This result therefore prompted us to investigate nuclear protein export in nup82Δ108 and nup159-1 mutant cells expressing the NLS-GFP2-NES fusion protein. Strikingly, although xpo1-1 mutant cells rapidly accumulated the NLS-GFP2-NES fusion protein inside the nucleus after a shift to 37°C, no major alteration in the localization of NLS-GFP2-NES was detected in the nup82Δ108 and nup159-1 cells, even after a shift to 37°C for 3 h. In both cases, only a few cells displayed a modest accumulation of the NLS-GFP2-NES reporter at the nuclear periphery (Figure 8). This stands in contrast to the rapid inhibition of poly(A)+ RNA export that was previously reported in these two mutant strains (Gorsch et al., 1995; Hurwitz and Blobel, 1995) and thus indicates that export of poly(A)+ RNA and NES-bearing proteins can be uncoupled.
Figure 8.
nup159-1 and nup82Δ108 cells do not display a major defect in NES-mediated protein export at 37°C. xpo1-1, wild-type (FY86), nup159-1, and nup82Δ108 cells were transformed with the NES-GFP2-NLS plasmid. Cells were maintained at 23°C or shifted to 37°C for 3 h. In xpo1-1 cells shifted to 37°C, an intranuclear accumulation of the NES-GFP2-NLS reporter and a concomitant decrease of the cytoplasmic staining was observed. In contrast, only a slight perinuclear accumulation of the reporter fusion protein could be observed in nup159-1 and nup82Δ108 cells shifted to 37°C.
In some mutant strains such as mtr10 or kap104 knockout strains, inhibition of mRNA export appeared as an indirect consequence of a defect in the import of specific heterogeneous nuclear ribonucleoproteins (hnRNPs) (Aitchison et al., 1996; Senger et al., 1998). We therefore analyzed Npl3p and Nab2p localization in nup159-1 and nup82Δ108 cells shifted for 1 h to 37°C (a time point at which, as previously published, poly(A)+ RNA export is greatly inhibited in these strains). Immunofluorescence studies revealed that these two hnRNPs remained entirely nuclear in the nup159-1 and nup82Δ108 mutant strains, whereas under the same conditions, Npl3p and Nab2p were virtually entirely cytoplasmic in mtr10 and kap104 knockout strains, respectively (Snay-Hodge and Cole, unpublished results). This demonstrates that unlike in these two karyopherin mutant strains, the defect in poly(A)+ RNA export observed in nup159-1 and nup82Δ108 cells is not an indirect consequence of a defect in Npl3p or Nab2p import, even though one cannot exclude that transport of another hnRNP or a specific mRNA transport factor might be inhibited in these strains.
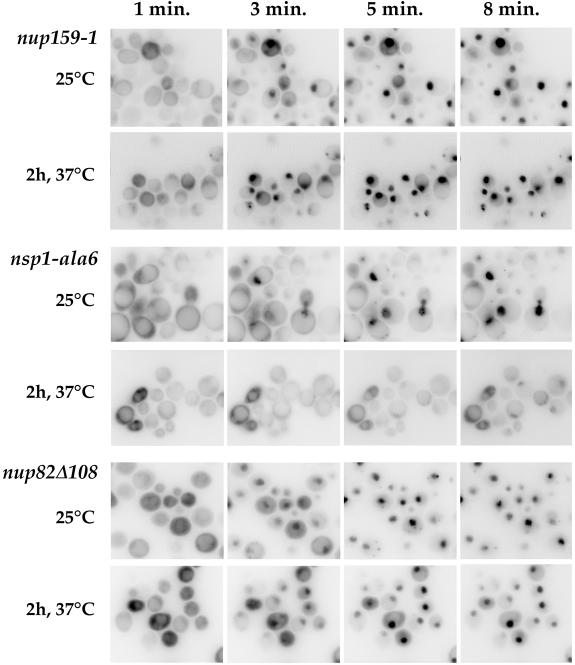
Although nup159 and nup82 mutant strains display major defects in poly(A)+ RNA export (Gorsch et al., 1995; Grandi et al., 1995a; Hurwitz and Blobel, 1995; Del Priore et al., 1998), all of the nsp1 mutant strains so far described were defective mainly for nuclear protein import (Mutvei et al., 1992; Nehrbass et al., 1993). Previous studies using the NLS-β-galactosidase reporter did not allow conclusions as to whether nup82Δ108 cells were competent for nuclear protein import at the restrictive temperature because the level of expression of the reporter was extremely low, most likely as a consequence of the mRNA export defect (Hurwitz and Blobel, 1995). We therefore reinvestigated nuclear protein import in the nup82Δ108 mutant strain, using the recently characterized Mig1p-GFP-β-galactosidase reporter (De Vit et al., 1997). The nuclear import of this reporter can be induced by addition of glucose, which enables one to follow the in vivo import of a nondiffusible protein without any additional cell treatment. As controls, we used the nup159-1 mutant strain that is not defective for nuclear protein import (Gorsch et al., 1995; Del Priore et al., 1998) and the ts nsp1-ala6 mutant strain (Wimmer et al., 1992b). In nup159-1 mutant cells, full nuclear import of the reporter was achieved within 5–8 min after addition of glucose, even when cells had already been shifted to 37°C for 2 h (Figure 9). In contrast, the nuclear import of Mig1p-GFP-β-galactosidase was somewhat defective in nsp1-ala6 mutant cells grown at permissive temperature and clearly inhibited when these cells were shifted to 37°C for 2 h (Figure 9). At this temperature, only a fraction (∼30%) of nsp1-ala6 cells displayed a faint accumulation of the reporter 10 min after addition of glucose. As shown in Figure 9, nup82Δ108 mutant cells did not display any major alteration in the rate of import of this reporter, even after a 2-h shift to 37°C, a time at which nuclear export of poly(A)+ RNA is drastically inhibited in this strain (Hurwitz and Blobel, 1995). We conclude that unlike the ts nsp1 mutant strains characterized to date, the nup82Δ108 and nup159-1 strains have little if any defect in nuclear protein import.
Figure 9.
In vivo analysis of the nuclear import of Mig1-GFP-lacZ in ts nup159, nsp1, and nup82 mutant strains. Temperature-sensitive nup159, nsp1, and nup82 mutant cells expressing the Mig1-GFP-LacZ reporter protein were grown in glycerol-containing medium at the indicated temperatures. At this stage, glucose was added, and living cells were examined and recorded at 1, 3, 5, and 8 min after glucose addition. A rapid nuclear import of the reporter was observed in the nup159-1 and nup82Δ108 mutant strains at both 23 and 37°C. In contrast, a modest defect in import was seen in nsp1-ala6 mutant cells grown at 25°C, and only a few nsp1-ala6 mutant cells shifted for 2 h to 37°C displayed any import of reporter protein, even 8 min after glucose addition. Note that to improve the detection of the faint nuclear signal, the images have been converted to negative images using the Adobe Photoshop program.
DISCUSSION
In this study the use of protease-deficient strains expressing various protein A-tagged nucleoporins enabled us to demonstrate that Nup159p can be isolated as part of a subcomplex that also contains both Nsp1p and Nup82p. Our data, together with previous studies demonstrating the physical association of a fraction of Nsp1p with Nup82p (Grandi et al., 1995a), indicate that Nsp1p, Nup82p, and Nup159p constitute a nuclear pore subcomplex. The fact that Nup159p had not been detected previously in physical association with Nsp1p or Nup82p (Grandi et al., 1993, 1995a) is most likely due to its sensitivity to proteolysis (Gorsch et al., 1995; Kraemer et al., 1995) and its poor labeling upon silver staining (our unpublished results). Coomassie blue staining of ProtA-Nup82p affinity-purified fractions further indicates that Nup82p belongs to a unique NPC subcomplex. This stands in contrast to Nsp1p, which appears to be part of two NPC subcomplexes (Grandi et al., 1995a). Because immunogold electron microscopy previously located Nup159p at the cytoplasmic side of the nuclear pore complex (Kraemer et al., 1995), the Nup82p/Nsp1p/Nup159p subcomplex itself most likely has the same location. In agreement with this hypothesis, Nup82p has been localized recently by immunogold electron microscopy to the cytoplasmic side of the NPCs (Fahrenkrog et al., 1998; Hurwitz et al., 1998).
Analysis of the interactions among these three nucleoporins in various mutant strains demonstrated that the Nup159p essential carboxyl-terminal domain, previously shown to contain sufficient information for the targeting to NPCs (Del Priore et al., 1998) is essential for its interactions with Nsp1p and Nup82p. In addition, our studies indicate that the carboxyl-terminal domains of Nsp1p and Nup82p, containing heptad repeats, are sufficient for their interaction with Nup159p. Together with previous studies characterizing the interaction between Nsp1p and Nup82p (Grandi et al., 1995a), these results therefore demonstrate that within this NPC subcomplex, interactions are mediated through heptad repeat regions, most likely forming intermolecular coiled coils. Further analysis of the Nsp1p/Nup82p/Nup159p subcomplex using the nup159-1 mutant strain that produces a protein truncated within its carboxyl-terminal domain revealed that under conditions in which the Nup159-1p mutant protein no longer efficiently interacts with ProtA-C-Nsp1p, the interaction between ProtA-C-Nsp1p and Nup82p was also inhibited. In contrast, a 108-amino acid deletion at the carboxyl terminus of Nup82p prevented its interaction with Nsp1p but did not affect the interaction between Nup159p and Nsp1p. Similarly, it was reported previously that deletions within the heptad repeat domain of Nic96p impair the interaction of Nic96p with the Nsp1p/Nup57p/Nup49p subcomplex (Grandi et al., 1995b). These results therefore suggest a common organization within the Nup82/Nsp1p/Nup159p and Nic96p/Nsp1p/Nup49p/Nup57p subcomplexes, with a heptad repeat containing nucleoporin (respectively Nup82p or Nic96p) acting as a docking site for an NPC subcomplex composed of nucleoporins containing both coiled-coil domains and nucleoporin repeat motifs.
In agreement with this hypothesis, immunofluorescence analysis effectively demonstrated that Nup159p was delocalized from the NPC in nup82Δ108 cells shifted to 37°C, a temperature at which the Nup82Δ108p mutant protein becomes degraded. In addition, affinity purification studies demonstrating that Nsp1p and Nup159p are stably associated in the nup82Δ108 strain even at 37°C suggest that Nup82p might anchor both Nup159p and Nsp1p at the NPC. However, in vivo analysis of GFP-C-Nsp1p localization in the nup82Δ108 mutant cells did not allow the identification of a major cytosolic fraction of Nsp1p delocalized at 37°C. Because Nsp1p is part of two distinct subcomplexes, the most likely explanation is that only a minor, not clearly detectable fraction of Nsp1p is associated with Nup82p and Nup159p. Even though this study does not allow precise quantitation of this fraction, the fact that a minor fraction of Nsp1p is engaged in the Nsp1p/Nup159p/Nup82p complex is in agreement with the reproducible lower fraction of Nsp1p that could be copurified with ProtA-Nup82p compared with the fraction that copurified with ProtA-Nup49p (Figure 1B, compare the amount of Nsp1p detected by Coomassie blue staining and by Western blotting using an anti-Nsp1p antibody).
All the ts mutants of NSP1 analyzed so far (including the nsp1-ala6 allele) map within the first heptad repeat region of its carboxyl-terminal domain, a region required for complex formation with Nup49p and Nup57p (Wimmer et al., 1992a; Nehrbass et al., 1993; Schlaich et al., 1997). Our biochemical and fluorescence data, demonstrating that the interaction between Nsp1-ala6p and both Nic96p and Nup82p is similarly compromised in the presence of competing wild-type Nsp1p, suggest that identical or overlapping domains within the heptad-repeat sequence of Nsp1p might be involved in interactions with both the Nsp1p/Nup159p/Nup82p and the Nsp1p/Nup49p/Nup57p/Nic96p NPC subcomplexes. Because all of the characterized mutants of NSP1 display major defects in nuclear protein import (Mutvei et al., 1992; Nehrbass et al., 1993), we therefore examined whether the nup82Δ108 mutant, which may alter the localization of a fraction of Nsp1p, would also be defective for nuclear protein import. However, in vivo analyses of nuclear protein import, using Mig1-GFP-β-galactosidase as a reporter, although confirming and extending to this reporter the role of Nsp1p in nuclear protein import (Mutvei et al., 1992; Nehrbass et al., 1993), did not reveal any nuclear protein import defect in the nup82Δ108 mutant strain at times at which nuclear export of poly(A)+ RNA is drastically inhibited. This stands in contrast to the defect in the rate of nuclear import of an SV40-NLS-GFP fusion protein that has been previously reported for the ts nup82-3 mutant allele (Shulga et al., 1996). Although one cannot exclude that nup82 mutant cells might have an import rate defect for the SV40-NLS substrate but not for Mig1p, a pleiotropic defect of the nup82-3 mutant allele, which has so far not been further characterized, cannot be excluded. One can thus speculate that the fraction of Nsp1p that associates with Nup82p and Nup159p on the cytosolic face of the NPC is not required for proper nuclear protein import. This fraction of Nsp1p could thus either perform a nonessential function or be involved in other transport processes.
Although the function of Nsp1p within the Nsp1p/Nup159p/Nup82p NPC subcomplex remains to be clarified, the nup159-1 and nup82Δ108 mutant strains display very similar phenotypes. In both cases, a deletion within the carboxyl-terminal domain of these proteins leads to a major defect in poly(A)+ RNA export at 37°C, suggesting that the defect in poly(A)+ RNA export observed in the nup82Δ108 strain could be due to the loss from NPCs of the repeat-containing nucleoporin Nup159p. However, as discussed by Ohno et al. (1998), this apparent specific defect in poly(A)+ RNA export may reflect a structural defect, serious enough to prevent export of large hnRNP particles but not import or export of smaller transport substrates. In addition to this transport defect that occurs at restrictive temperature, the nup159-1 and nup82Δ108 mutant strains both display a mild and highly reversible clustering of the NPCs. In particular, this clustering phenotype appeared to depend on the growth conditions, because nup159-1 or nup82Δ108 cells from dense liquid culture or grown on plates showed a normal NPC distribution. Whether this redistribution could be somehow linked to the induction of specific stress response genes that can be turned on under such growth conditions remains to be determined. Moreover, this particular phenotype gives a specificity to the essential Nup159p and Nup82p nucleoporins, which seem to have a different role in NPC distribution than the nonessential components of the Nup84p/Nup85p/Nup120p/C-Nup145p subcomplex, whose deletion individually led to a constitutive clustering of the NPCs associated with structural alterations of the nuclear envelope (Wente and Blobel, 1994; Aitchison et al., 1995a; Heath et al., 1995; Goldstein et al., 1996; Siniossoglou et al., 1996; Teixeira et al., 1997).
Although the overall structure of the nuclear pore and its functions have been conserved from yeast to vertebrates, few yeast nucleoporins show a very high degree of homology to any metazoan nucleoporins over their entire length. In addition, in only one case has it been possible to substitute a metazoan nucleoporin for a yeast nucleoporin (Aitchison et al., 1995b). Therefore, it is not yet possible to determine directly whether yeast and metazoan nucleoporins that share sequence homology perform homologous roles. However, evidence is beginning to accumulate that suggests that the organization of nucleoporins into functional subcomplexes and, at a higher level, the organization of subcomplexes within NPCs may well be conserved. For instance, p62, which is considered the vertebrate homologue of yeast Nsp1p, has been shown to be associated with p58 and p54 (Finlay et al., 1991; Guan et al., 1995; Hu et al., 1996), which are related to (and may also be homologues of) Nup57p and Nup49p, respectively (Hu et al., 1996; Doye and Hurt, 1997; Schlaich et al., 1997). In addition, a fraction of Nup93, a probable vertebrate homologue of yeast Nic96p, physically interacts with p62, just as yeast Nic96p interacts with Nsp1p (Grandi et al., 1997). Interestingly, yeast Nup159p and vertebrate Nup214/CAN share a similar location on the cytoplasmic side of the NPCs and closely related degenerate FG repeat motifs (Kraemer et al., 1994, 1995). In addition, both Nup159p and Nup214/CAN contain a coiled-coil domain (located respectively within the carboxyl-terminal part of Nup159p and within the central region of Nup214/CAN) that has been shown to be necessary and sufficient for their nuclear pore association (Fornerod et al., 1995; Del Priore et al., 1998). Recently, Nup214/CAN has been shown to associate through its coil-coiled domain with a novel nuclear pore protein referred to as Nup88 or Nup84 (Bastos et al., 1997; Fornerod et al., 1997). Although the sequence homology between Nup88/Nup84 and Nup82p is marginal (Fornerod et al., 1996), Nup88/Nup84, like yeast Nup82p, contains a carboxyl-terminal coiled-coil domain (Bastos et al., 1997; Fornerod et al., 1997). These data suggest that the yeast Nup82p/Nup159p and mammalian Nup88/Nup214 associations could be functionally homologous.
In yeast, a fraction of Nsp1p coprecipitates with Nup159p and Nup82p. Similarly, a minor fraction of Xenopus p62 was found in association with a p200 N-acetylglucosaminylated protein, the putative Xenopus homologue of mammalian Nup214/CAN (Macaulay et al., 1995). Furthermore, a protein of 66 kDa (referred to as CC66) coprecipitates, although less consistently than Nup88, with the coiled domain of Nup214/CAN in mammalian cells (Fornerod et al., 1996). Although the difference in molecular masses is significant, the fact that CC66 could be identical to p62 has not been clearly excluded, leaving open the possibility that a fraction of p62 could interact with the Nup214/Nup88 subcomplex in mammalian cells. If this is the case, the fact that p62 was found to be located on both sides of the NPC at or near the ends of the central plug (Guan et al., 1995) might reflect the low abundance of the fraction of p62 associated with Nup214 in Xenopus (Macaulay et al., 1995). Finally, hCRM1 was found to be associated with CAN/Nup214 (Fornerod et al., 1997). Our preliminary data from two-hybrid analyses suggest that Crm1p/Xpo1p associates with Nup159p (Stafford and Cole, personal communication), extending the similarity between yeast and metazoan cells. However, Crm1p/Xpo1p must associate with other nucleoporins as well, because protein export is maintained in cells in which Nup159p has been lost from nuclear pore complexes.
ACKNOWLEDGMENTS
We thank Michael Hurwitz (Rockefeller University, New York, NY), Ed Hurt (University of Heidelberg, Heidelberg, Germany), Michael De Vit (Washington University School of Medicine, St. Louis, MO), and Karsten Weis (University of California, San Francisco, CA) for plasmids, yeast strains, and antibodies. We thank Michael Hurwitz, Günter Blobel and Nelly Panté for communicating results on the localization of Nup82p before publication. We also thank Michel Bornens (Institut Curie, Paris, France) for constant support, Paola Grandi (Institut Curie), Emmanuelle Fabre, and Ulf Nehrbass (Institut Pasteur, Paris, France) for critical reading of the manuscript and the members of our laboratories for helpful discussions and advice. This work was supported by a Centre National de la Recherche Scientifique grant (UMR144), the Institut Curie, and a grant from the Association pour la Recherche contre le Cancer (all to V.D.) and by grant GM33998 from the National Institute of General Medical Sciences, National Institutes of Health (to C.N.C.). N.B. received a fellowship from the Ministère de l’Enseignement et de la Recherche Supérieure. S.D. was supported by National Research Service Award fellowship AR07576 from the National Institute of Arthirtis and Musculoskeletal Diseases, National Institutes of Health.
Abbreviations used:
- CCD
charge-coupled device
- GFP
green fluorescent protein
- HA
hemagglutinin
- hnRNP
heterogeneous nuclear ribonucleoprotein
- HRP
horseradish peroxidase
- NES
nuclear export signal
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- NUP
nucleoporin
- ProtA
protein A
- ts
temperature-sensitive
- wt
wild-type
REFERENCES
- Aitchison JD, Blobel G, Rout MP. Nup120: a yeast nucleoporin required for NPC distribution and mRNA export. J Cell Biol. 1995a;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995b;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Doye V. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J Cell Biol. 1997;136:747–759. doi: 10.1083/jcb.136.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges T, Petfalski E, Tollervey D, Hurt EC. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behaviour of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VV, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V, Heath CV, Snay CA, MacMillan A, Gorsch LC, Dagher S, Cole CN. A structure/function analysis of the Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae. J Cell Sci. 1998;110:2987–2999. doi: 10.1242/jcs.110.23.2987. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., Hurt, E.C., Aebi, U., and Panté, N. (1998). Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Boer J, Vanbaal S, Jaegle M, Vonlindern M, Murti KG, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxyterminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Interaction of cellular proteins with the leukemia specific fusion proteins DEK-CAN and SET-CAN and their normal counterpart, the nucleoporin CAN. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Gopal Murti K, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with “GLFG” nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Emig S, Weise C, Hucho F, Pohl T, Hurt EC. A novel nuclear pore protein Nup82p which specifically binds to a fraction of Nsp1p. J Cell Biol. 1995a;130:1263–1273. doi: 10.1083/jcb.130.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995b;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Muller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Strambio-de-Castillia C, Blobel G. Two yeast nuclear pore complex proteins involved in mRNA export form a cytoplasmically oriented subcomplex. Proc Natl Acad Sci USA. 1998;95:11241–11245. doi: 10.1073/pnas.95.19.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Silver P. Use of A. victoria green fluorescent protein to study protein dynamics in vivo. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. 34, unit 9.7. Brooklyn, NY: Greene Publishing Associates/Wiley-Interscience; 1996. pp. 9.7.22–9.7.28. [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch ET, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Matunis MJ, Blobel G. Biochemical and molecular analysis of the mammalian nuclear pore complex and the regulation of nuclear transport. Mol Biol Cell. 1996;7:95a. (abstract). [Google Scholar]
- Mutvei A, Dihlmann S, Herth W, Hurt EC. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation. Eur J Cell Biol, 1992;59:280–295. [PubMed] [Google Scholar]
- Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt EC. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of the nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol. 1996;133:1153–1162. doi: 10.1083/jcb.133.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Toward the molecular dissection of protein import into the nucleus. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- Pemberton L, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton L, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich NL, Häner M, Lustig A, Aebi U, Hurt EC. In vitro reconstitution of a heterotrimeric nucleoporin complex consisting of recombinant Nsp1p, Nup49p and Nup57p. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA- binding protein Np13p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter R. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Siniossoglou S, Podtelejnikov S, Benichou JC, Mann M, Dujon B, Hurt E, Fabre E. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 1997;16:5086–5097. doi: 10.1093/emboj/16.16.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Gasser SM, Caplan AJ. The nucleus and nucleocytoplasmic transport in Saccharomyces cerevisiae. In: Broach JR, Jones E, Pringle J, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 471–542. [Google Scholar]
- Wimmer C, Doye V, Grandi P, Nehrbass U, Hurt E. A new subclass of nucleoporins that functionally interacts with nuclear pore protein NSP1. EMBO J. 1992a;11:5051–5061. doi: 10.1002/j.1460-2075.1992.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer C, Doye V, Nehrbass U, Schlaich N, Hurt EC. Approaches towards a genetic analysis of the nuclear pore complex in yeast. In: Brown AJP, Tuite MF, McCarthy JEG, editors. Protein Synthesis and Targeting in Yeast, NATO ASI series H. Vol. 71. Berlin: Springer-Verlag; 1992b. pp. 269–281. [Google Scholar]
- Wright R, Rine J. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G. Cloning and characterization of human karyopherin beta 3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]