Abstract
During a survey conducted to evaluate the incidence of class B carbapenemase (metallo-β-lactamase [MBL])-producing Pseudomonas aeruginosa strains from hospitals in Majorca, Spain, five clinical isolates showed a positive Etest MBL screening test result. In one of them, strain PA-SL2, the presence of a new blaVIM derivative (blaVIM-13) was detected by PCR amplification with blaVIM-1-specific primers followed by sequencing. The blaVIM-13-producing isolate showed resistance to all β-lactams (except aztreonam), gentamicin, tobramycin, and ciprofloxacin. VIM-13 exhibited 93% and 88% amino acid sequence identities with VIM-1 and VIM-2, respectively. blaVIM-13 was cloned in parallel with blaVIM-1, and the resistance profile conferred was analyzed both in Escherichia coli and in P. aeruginosa backgrounds. Compared to VIM-1, VIM-13 conferred slightly higher levels of resistance to piperacillin and lower levels of resistance to ceftazidime and cefepime. VIM-13 and VIM-1 were purified in parallel as well, and their kinetic parameters were compared. The kcat/Km ratios for the antibiotics mentioned above were in good agreement with the MIC data. Furthermore, EDTA inhibited the activity of VIM-13 approximately 25 times less than it inhibited the activity of VIM-1. VIM-13 was harbored in a class 1 integron, along with a new variant (Ala108Thr) of the aminoglycoside-modifying enzyme encoding gene aacA4, which confers resistance to gentamicin and tobramycin. Finally, the VIM-13 integron was apparently located in the chromosome, since transformation and conjugation experiments consistently yielded negative results and the blaVIM-13 probe hybridized only with the genomic DNA.
Class B carbapenemases (metallo-β-lactamases [MBLs]) are the β-lactamases acquired by Pseudomonas aeruginosa that are of the most concern, since they are characterized by a very wide hydrolytic spectrum that affects all β-lactams except monobactams (17). MBLs need Zn2+ binding in their catalytic center to hydrolyze the β-lactam ring, and so they are inhibited by chelating agents like EDTA (23, 38). MBL genes are usually carried as cassettes in integrons along with other resistance determinants, such as aminoglycoside-modifying enzymes. Furthermore, the integrons involved are frequently located in plasmids or transposons, which certainly contribute to the global dissemination of these worrisome resistance mechanisms (2, 7, 11, 16, 26). Indeed, several outbreaks of MBL-producing P. aeruginosa strains have been described worldwide, and they are often amplified due to the horizontal transmission of the multiresistance determinants harbored in the cited integrons (13, 20, 21, 24, 32). There are two major groups of MBLs, IMP and VIM, which share only approximately 30% of their amino acid sequences. Among the VIM carbapenemases, there are two major phylogenetic lineages, the VIM-1 and VIM-2 clusters, whereas VIM-7 seems to be the single representative of a third phylogenetic cluster (35). The sequences of the VIM-1 and VIM-2 enzymes differ at 25 of 266 amino acids, although these differences confer remarkable changes in their functional behaviors (6, 8, 25). VIM-1 is characterized by high kcat/Km ratios (efficiency of hydrolysis [34]) for carbenicillin, cephaloridine, cephalothin, cefuroxime, cefepime, cefpirome, and imipenem and by the low level of inactivation caused by EDTA compared to those caused by other chelating agents, such as 1,10-o-phenanthroline (8). On the other hand, VIM-2 has many differences in kcat/Km ratios compared with those of VIM-1 for some antibiotics (e.g., a kcat/Km approximately 10-fold greater than that of VIM-1 for carbapenems) and seems to be much more susceptible than VIM-1 to inactivation by metal chelators, probably indicating a looser bond to the zinc ions (6). Polymorphisms in some of the active-site amino acids have been proposed as an explanation for these kinetic particularities and the reason for the differences in the substrate binding of each enzyme (6). Furthermore, the recently characterized VIM-12 enzyme, which seems to be a hybrid protein between VIM-1 and VIM-2, shows a narrow substrate specificity that is limited mainly to penicillin and imipenem, although it shows complete conservation of the active-site residues of the VIM enzymes (12, 27).
In this work, we describe a new integron-borne blaVIM gene, blaVIM-13, detected in a P. aeruginosa clinical isolate. Furthermore, VIM-13, which is found to be located closest to the VIM-1 cluster but which significantly diverges from it, was purified in parallel with VIM-1 and characterized biochemically.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
The laboratory strains and plasmids used in or constructed for this work are listed in Table 1. Five Pseudomonas aeruginosa clinical isolates resistant to imipenem and meropenem and showing a positive Etest MBL (AB Biodisk, Solna, Sweden) screening test result (imipenem and EDTA MICs at least 3 twofold dilutions lower than those of imipenem alone) were studied. These isolates were recovered during a survey conducted to evaluate the incidence of MBL-producing strains performed between August 2004 and December 2005 in two Majorcan hospitals (Hospital Son Dureta and Hospital Son Llàtzer). The MICs of piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, cefotaxime, aztreonam, imipenem, meropenem, gentamicin, tobramycin, amikacin, and ciprofloxacin were determined by the Etest method (AB Biodisk), according to the manufacturer's recommendations. Breakpoints were applied according to the recommendations if Clinical and Laboratory Standards Institute (4).
TABLE 1.
Laboratory strains and plasmids used or constructed in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| P. aeruginosa PAO1 | Completely sequenced reference strain | Laboratory collection |
| E. coli | ||
| XL1-Blue | F′::Tn10 proA+B+lacIq Δ(lacZ)M15 recA1 endA1 gyrA96 (Nalr) thi hsdR17(rK− mK−) mcrB1 | Laboratory collection |
| BL21 | F−ompT hsdSB (rB− mB−) gal dcm | Laboratory collection |
| Plasmids | ||
| pGEM-T | PCR cloning vector | Promega |
| pUCP-24 | Gmr; pUC18-based Escherichia-Pseudomonas shuttle vector | 39 |
| pGEX-6P-1 | Apr; GST-fusion purification vector | Amersham |
| pGTVIM-1 | pGEM-T containing blaVIM-1 gene from PA-SD2 strain | This work |
| pGTVIM-13 | pGEM-T containing blaVIM-13 gene from strain PA-SL2 | This work |
| pUCPVIM-1 | Gmr; pUCP-24 containing blaVIM-1 gene | This work |
| pUCPVIM-13 | Gmr; pUCP-24 containing blaVIM-13 gene | This work |
| pGEXVIM-1 | pGEX-6P-1 containing blaVIM-1 gene without signal peptide; used for purification | This work |
| pGEXVIM-13 | pGEX-6P-1 containing blaVIM-13 gene without signal peptide; used for purification | This work |
| pGTA4 | pGEM-T containing aacA4 gene from strain PA-SD2 | This work |
| pGTA4A108T | pGEM-T containing aacA4 gene from strain PA-SL2 (Ala108Thr variant) | This work |
Characterization of MBL-encoding genes.
Following total DNA extraction with a DNeasy tissue kit (Qiagen, Hilden, Germany), the strains were evaluated for the presence of MBL-encoding genes by PCR amplification with primers (Table 2) specific for blaIMP-1, blaIMP-2, blaVIM-1, and blaVIM-2 or closely related genes, followed by DNA sequencing (9). Sequencing reactions were performed with a BigDye Terminator kit (PE Applied Biosystems, Foster City, CA), and the sequences were analyzed on an ABI Prism 3100 DNA sequencer (PE Applied Biosystems). The resulting sequences were then compared with those available in the GenBank database (www.ncbi.nih.gov/BLAST). Multiple-sequence alignments were performed and MBL phylograms were prepared with the ClustalW program (version 1.83).
TABLE 2.
Primers used in this work
| Primer | Sequence (5′-3′)a | PCR product size (bp) | Use |
|---|---|---|---|
| VIM1-F | ATGTTAAAAGTTATTAGTAGTTTATTG | 801 | Amplification and sequencing of blaVIM-1 and related genes |
| VIM1-R | CTACTCGGCGACTGAGC | ||
| VIM2-F | ATGTTCAAACTTTTGAGTAAG | 801 | Amplification and sequencing of blaVIM-2 and related genes |
| VIM2-R | CTACTCAACGACTGAGCG | ||
| IMP1-F | ATGAGCAAGTTATCTGTATTC | 741 | Amplification and sequencing of blaIMP-1 and related genes |
| IMP1-R | TTAGTTGCTTGGTTTTGATGG | ||
| IMP2-F | ATGAAGAAATTATTTGTTTTATG | 741 | Amplification and sequencing of blaIMP-2 and related genes |
| IMP2-R | TTAGTTACTTGGCTGTGATG | ||
| VIM-F | AGTGGTGAGTATCCGACAG | Sequencing of blaVIM | |
| VIM-R | ATGAAAGTGCGTGGAGAC | ||
| INT-F | CTCTCACTAGTGAGGGGC | 1,010 | Amplification and sequencing of intI |
| INT-R | ATGAAAACCGCCACTGCG | ||
| INT-R-I | CGCAGTGGCGGTTTTCAT | 210 | Amplification and sequencing of gene(s) between intI and blaVIM |
| VIM1-F-I | CAATAAACTACTAATAACTTTTAAC | ||
| qacE-F | GAAAGGCTGGCTTTTTCTTG | 210 | Amplification of qacEΔ1 |
| qacE-R | ATTATGACGACGCCGAGTC | ||
| qacE-F-I | CAAGAAAAAGCCAGCCTTTC | Variable | Amplification and sequencing of gene(s) between blaVIM and qacEΔ1 |
| VIM1-R-I | GCTCAGTCGCCGAGTAG | ||
| aacA4-R | TTAGGCATCACTGCGTGTTC | 697 | Cloning of aacA4 gene |
| V1-F-BHI | TCGGATCCGGGGAGCCGAGTGGTGAG | 723 | Cloning of blaVIM-1 and blaVIM-13 (without signal peptide) for purification |
| V13-F-BHI | TCGGATCCGGGGAGTCGAGAGGTGAG | ||
| V1/13-R-ERI | TCGAATTCCTACTCGGCGACTGAGCG |
The underlined sequences represent sites for restriction enzymes.
Characterization of genetic elements harboring class B carbapenemases.
The possible locations of MBL-encoding genes in self-transferable plasmids were evaluated in conjugation and transformation experiments performed by previously described procedures (9). To ascertain the location of blaVIM-13, Southern blotting and hybridization experiments with a blaVIM-13-specific PCR probe were performed with the chromosomal and plasmid DNA of strain PA-SL2 by using an enhanced chemiluminescence kit (GE Healthcare, Little Chalfont, United Kingdom), according to the manufacturer's instructions. The integrons harboring the MBL-encoding genes were characterized by PCR, followed by DNA sequencing with specific primers (Table 2), to amplify intI1, qacEΔ1, and the DNA regions located between intI1 or qacEΔ1 and the corresponding MBL-encoding gene (9).
Cloning and characterization of the new VIM-13.
The blaVIM-1 and blaVIM-13 genes were amplified in parallel by PCR with previously described primers VIM1-F and VIM1-R (Table 2). The purified PCR products were then ligated to plasmid pGEM-T to obtain plasmids pGTVIM-1 and pGTVIM-13, respectively, which were transformed into strain Escherichia coli XL1-Blue made competent with CaCl2 (31). Transformants were selected in 50 μg/ml ampicillin MacConkey agar plates, and their sequences were checked by PCR amplification. The cloned blaVIM-1 and blaVIM-13 genes were sequenced to confirm that no mutations were produced during PCR amplification. The cloned blaVIM-1 and blaVIM-13 genes were then liberated from the corresponding pGEM-T plasmids by restriction with EcoRI and were further ligated to plasmid pUCP24 (digested with the same enzyme) to obtain pUCPVIM-1 and pUCPVIM-13, respectively. E. coli XL1-Blue transformants were then selected on 10 μg/ml gentamicin MacConkey agar plates. In both cases, recombinant plasmids with DNA inserts with the same orientation as the lacZ promoter (checked by PCR and sequencing) were selected for further study. Additionally, plasmids pUCPVIM-1 and pUCPVIM-13 were also electroporated as described previously (33) into strain P. aeruginosa PAO1, and transformants were selected in 50 μg/ml gentamicin Luria-Bertani agar plates. The MICs of piperacillin, piperacillin-tazobactam, ceftazidime, cefepime, cefotaxime, aztreonam, imipenem, and meropenem were determined by Etest for the E. coli XL1-Blue and P. aeruginosa PAO1 transformants harboring pUCPVIM-1 or pUCPV-13. The Compute pI/Mw tool from www.expasy.org was used to predict the molecular mass of the new enzyme, VIM-13. To determine the pI of VIM-13 in parallel with that of VIM-1, isoelectric focusing of crude sonic extracts of strains PA-SL2 and PA-SD2 with Phast gels (pH gradient, 3 to 9) was performed in a Phast system apparatus (Pharmacia AB, Uppsala, Sweden).
Antibiotics and other chemicals.
Ampicillin, benzylpenicillin, piperacillin, cephalothin, cefotaxime, ceftazidime, cefuroxime, and the chelating agents (EDTA, dipicolinic acid, and 1,10-o-phenanthroline) were purchased from Sigma-Aldrich Chemie GmbH (Steinheim, Germany). Cefepime (Maxipime) was purchased from Bristol-Myers Squibb, imipenem was a gift from Merck (Whitehouse Station, NJ), and meropenem was a gift from AstraZeneca (London, United Kingdom).
Purification of VIM-1 and VIM-13 enzymes.
To purify the VIM-1 and VIM-13 enzymes, the blaVIM-1 and blaVIM-13 genes were cloned into the pGEX-6P-1 vector (BamHI and EcoRI restriction sites) with the primers V1-F-BHI or V13-F-BHI (depending on the amplified blaVIM gene) and V1/13-R-ERI (Table 2). Cloning of the blaVIM genes allowed the creation of a fusion protein between glutathione S-transferase (GST) and the VIM-type enzymes lacking the signal peptide. The GST tag was then cleaved off and the β-lactamases were purified to homogeneity with the GST gene fusion system (Amersham Pharmacia Biotech, Europe GmbH), in accordance with the manufacturer's instructions. E. coli BL21 was used for the expression and the purification of the VIM-1 and VIM-13 proteins (from plasmids pGEXVIM-1 and pGEXVIM-13, respectively). The purified proteins were electrophoresed in sodium dodecyl sulfate-polyacrylamide gels and appeared as a band of 26 kDa (purity, >99%). The concentrations of the purified proteins were determined by a protein assay (Bio-Rad, Richmond, CA). Finally, enzyme samples were equilibrated by dialyzing them overnight at 4°C in 50 mM HEPES (pH 7.5)-50 μM ZnSO4. Purified samples of VIM-1 and VIM-13 supplemented with 20 μg/ml of bovine serum albumin (BSA) were stored at −80°C.
Kinetic parameters.
The VIM-1 and VIM-13 MBLs were further used in biochemical studies with the β-lactam antibiotics mentioned above. The experiments were carried out at 25°C in a Nicolet Evolution 300 spectrophotometer (Thermo Electron Corporation, Waltham, MA), and the data obtained were analyzed with Vision Pro software (Thermo Electron Corporation). The wavelengths and molar extinction coefficients used were described previously (15). The tests were repeated three times in 50 mM HEPES (pH 7.5) with 50 μM ZnSO4 and 20 μg/ml BSA. The representation of the substrate concentration ([S]) against the hydrolysis velocity (V) showed a Michaelis-Menten curve, and its Lineweaver-Burk conversion obtained by using 1/V and 1/[S] provided a straight line with an intersection at the abscissa of −1/Km. The Km and kcat parameters were calculated from at least three independent experiments with 6 to 12 substrate concentrations, depending on the antibiotic (3, 29, 34).
Inactivation of the VIM-13 and VIM-1 enzymes by chelating agents.
VIM-1 and VIM-13 inactivation by Zn2+ removal was studied at 20°C in 50 mM HEPES (pH 7.5) buffer supplemented with 20 μg/ml of BSA in the presence of different concentrations of EDTA, dipicolinic acid, and 1,10-o-phenanthroline. The reporter substrate was 100 μM imipenem, and the measurements were obtained after 10 min of incubation of the buffer mentioned above containing the enzyme and the corresponding chelating agent. The inhibitor concentration that gave a 50% reduction of the hydrolytic activity of each enzyme (IC50) was determined for the chelating agents mentioned above (25).
Cloning and characterization of a new AAC(6′)-Ib variant (Ala108Thr).
Primers VIM1-R-I and aacA4-R (Table 2) were used to amplify in parallel aacA4 from the total DNA of strain PA-SD2 (which harbors the aacA4 gene described by Lambert et al. [14] downstream of blaVIM-1) and strain PA-SL2 (which harbors the new Ala108Thr variant). The PCR products were ligated to plasmid pGEM-T to obtain plasmids pGTA4 and pGTA4A108T, respectively, which were transformed into E. coli XL1-Blue made competent with CaCl2 (31). Transformants were selected in 50 μg/ml ampicillin MacConkey agar plates, and their sequences were checked by PCR amplification. The cloned aacA4 genes were sequenced to confirm that no mutations were produced during PCR amplification. In both cases, recombinant plasmids with DNA inserts with the same orientation as that of the lacZ promoter (checked by PCR and sequencing) were selected for further study. The spectra of aminoglycosides affected by both AAC(6′)-Ib enzymes were assessed by determination of the MICs (Etest) of gentamicin, tobramycin, and amikacin for the selected transformants.
Nucleotide sequence accession numbers.
The nucleotide sequences described in this work have been deposited in the GenBank database under accession numbers DQ365886 (blaVIM-13), EF577407 (the VIM-13 integron from strain PA-SL2), and EF577408 (the VIM-1 integron from clone PA-SD2).
RESULTS
MBL-producing P. aeruginosa isolates from Majorcan hospitals: detection of the new blaVIM-13.
Five P. aeruginosa clinical isolates resistant to imipenem and meropenem and showing a positive Etest MBL screening test result were studied. These isolates were recovered during a survey to evaluate the incidence of MBL-producing strains performed between August 2004 and December 2005 in two Majorcan hospitals (Hospital Son Dureta and Hospital Son Llàtzer). The strains were evaluated for the presence of MBL-encoding genes by PCR amplification, followed by DNA sequencing. Three of the isolates showed positive PCR amplification with blaVIM-2-specific primers, and sequencing of the PCR products confirmed the presence of blaVIM-2. On the other hand, isolates PA-SD2 and PA-SL2 showed positive PCR amplification with blaVIM-1-specific primers. Sequencing of the PCR products confirmed the presence of blaVIM-1 in PA-SD2 but, interestingly, revealed the presence of a new blaVIM derivative, designated blaVIM-13, in PA-SL2. blaVIM-13-producing isolate PA-SL2 was further characterized in this work, along with blaVIM-1-producing isolate PA-SD2 for comparative purposes. Isolate PA-SL2 was recovered on 15 October 2005 from a skin ulcer of a patient admitted to the Nephrology Ward of Hospital Son Llàtzer. In addition to its high level of resistance to carbapenems, this isolate showed resistance to the other β-lactams tested (except aztreonam), gentamicin, tobramycin, and ciprofloxacin (Table 3). Isolate PA-SD2 was recovered from the peritoneal fluid of a patient admitted to the ICU of Hospital Son Dureta and showed a resistance phenotype similar to that of isolate PA-SL2, although it was additionally resistant to aztreonam (Table 3).
TABLE 3.
MICs of E. coli and P. aeruginosa strains with cloned genes and MICs of clinical strains used in this work
| Strain | Plasmid | Cloned gene | MIC (μg/ml)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | PIP | PIP-TZ | AZM | CAZ | FEP | CTX | IMP | MER | GEN | TOB | AMK | |||
| PA-SD2 (harboring blaVIM-1) | >32 | >256 | >256 | 24 | >256 | >256 | >256 | >32 | >32 | 48 | 16 | 3 | ||
| PA-SL2 (harboring blaVIM-13) | >32 | >256 | >256 | 4 | 32 | 32 | >256 | 24 | 12 | >256 | 48 | 16 | ||
| P. aeruginosa PAO1 | pUCPVIM-1 | blaVIM-1 | 64 | 48 | 1.5 | >256 | >256 | >256 | >32 | >32 | ||||
| P. aeruginosa PAO1 | pUCPVIM-13 | blaVIM-13 | 192 | 192 | 1.5 | 128 | 64 | >256 | >32 | >32 | ||||
| P. aeruginosa PAO1 | 0.125 | 3 | 3 | 1 | 1 | 1 | 16 | 1.5 | 0.38 | |||||
| E. coli XL1-Blue | pUCPVIM-1 | blaVIM-1 | 6 | 6 | 0.125 | 32 | 12 | >256 | 1 | 0.25 | ||||
| E. coli XL1-Blue | pUCPVIM-13 | blaVIM-13 | 8 | 8 | 0.125 | 6 | 1 | >256 | 1 | 0.19 | ||||
| E. coli XL1 Blue | pGTA4 | aacA4 | 0.064 | 1 | 2 | 0.75 | ||||||||
| E. coli XL1-Blue | pGTA4A108T | aacA4 variant (Ala108Thr) | 0.064 | 1 | 2 | 0.75 | ||||||||
| E. coli XL1-Blue | 0.064 | 1.5 | 1 | 0.19 | 0.38 | 0.19 | 0.19 | 0.25 | 0.064 | 0.19 | 0.25 | 0.75 | ||
CIP, ciprofloxacin; PIP, piperacillin; PIP-TZ, piperacillin-tazobactam; AZM, aztreonam; CAZ, ceftazidime; FEP, cefepime; CTX, cefotaxime; IMP, imipenem; MER, meropenem; GEN, gentamicin; TOB, tobramycin; AMK, amikacin.
Characterization of the new VIM-13.
The degrees of identity of blaVIM-13 with blaVIM-1, blaVIM-2, and blaVIM-7 were 93%, 88%, and 78%, respectively. The amino acid sequence of the enzyme (VIM-13) exhibited identities of 93%, 88%, and 76% with the amino acid sequences of VIM-1, VIM-2 and VIM-7, respectively. Figure 1 shows the multiple-sequence alignment of VIM-type MBLs, and Fig. 2 shows their phylograms (obtained with the ClustalW program, version 1.83). VIM-13 was found to be located the closest to the VIM-1 cluster, but it significantly diverged from it. In order to characterize this new VIM enzyme, blaVIM-13 and blaVIM-1 were cloned in parallel into plasmid pUCP24. The MICs conferred by the resulting plasmids in both E. coli XL1-Blue and P. aeruginosa PAO1 backgrounds are shown in Table 3. Compared to the levels of resistance conferred by VIM-1, VIM-13 conferred lower levels of resistance to ceftazidime and cefepime but higher levels of resistance to piperacillin. Additionally, VIM-13 showed a molecular mass of 28,220 Da. The results of isoelectric focusing showed a pI of approximately 5.1 for VIM-13, which is almost identical to that previously described for VIM-1 (8).
FIG. 1.
Multiple-sequence alignment (performed with the ClustalW program, version 1.83) of the VIM-1, VIM-2, VIM-7, VIM-12, and VIM-13 carbapenemases. The signal peptide is shown in underlined boldface, and loop L3 (the active site of the enzyme [6]) is shown with a black background. Dashes indicate gaps. Asterisks, colons, and periods indicate identical, conserved, and semiconserved residues, respectively.
FIG. 2.
Phylogram (performed with the ClustalW program, version 1.83) of VIM-type MBLs.
Characterization of the genetic element harboring blaVIM-13.
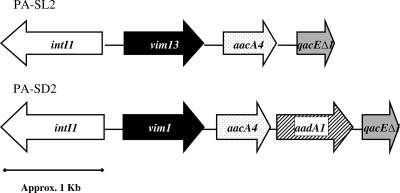
Several attempts to transfer the blaVIM-13 determinant by conjugation to P. aeruginosa PAO1 or E. coli HB101 and by electroporation to PAO1 consistently failed. Furthermore, the results of Southern blotting showed the hybridization of the blaVIM-13-specific PCR probe with strain PA-SL2 genomic DNA but not with plasmid DNA (data not shown). Therefore, blaVIM-13 is apparently located in the chromosome of PA-SL2. The integron harboring blaVIM-13 in isolate PA-SL2 was characterized by a set of five PCR amplifications, followed by DNA sequencing, with specific primers for the amplification of intI1, qacEΔ1, and the DNA regions located between intI1 or qacEΔ1 and the VIM-13-encoding gene. Figure 3 shows the resulting integron structure. According to the intI1 sequence, it is a class 1 integron and contains the 3-bp insertion (GGG) between the −10 and −35 boxes known to activate the P2 promoter, which was otherwise conserved in the integron (5). The sequence of the downstream region of blaVIM-13, including the 59-bp element, showed only weak similarity to the sequences previously reported for blaVIM-1 and blaVIM-2 (data not shown) (16). The integron from VIM-13-producing strain PA-SL2 contained an aacA4 gene cassette right after the VIM-13-encoding gene. Another remarkable finding was that the deduced amino acid sequence for the aacA4 gene from PA-SL2 showed a polymorphism (Ala108Thr) not previously described for this aminoglycoside-modifying enzyme. With the exception of this change, this aacA4 showed an amino acid sequence identical to that previously described by Lambert et al. (14). The integron harboring blaVIM-1 of isolate PA-SD2 was characterized in parallel for comparative purposes. As shown in Fig. 3, the structure was similar, but in this case, and additional gene (aadA1) was detected after aacA4. Furthermore, the aacA4 gene did not contain the polymorphism described above.
FIG. 3.
Structure of the blaVIM-1 (strain PA-SD2) and blaVIM-13 (strain PA-SL2) carrying the integrons described in this work.
To explore whether the unique polymorphism detected in the VIM-13 integron has an effect on the spectra of aminoglycosides affected, aacA4 and its Ala108Thr variant were cloned from PA-SD2 and PA-SL2, respectively. As shown in Table 3, the enzymes from both strains yielded the same MICs and conferred resistance to gentamicin and tobramycin but did not affect resistance amikacin, as described by Lambert et al. (14).
Kinetic parameters of VIM-13.
Table 4 shows the kinetic parameters of VIM-13 with various β-lactams in comparison with those of VIM-1, which were obtained in parallel experiments. VIM-13 showed higher kcat/Km ratios (and, therefore, a greater efficiency of hydrolysis) than VIM-1 for all antibiotics tested, with the exceptions of ceftazidime and cefepime. It is remarkable that VIM-13 showed a poorer efficiency of hydrolysis (kcat/Km) for ceftazidime and cefepime than VIM-1: approximately 2- and 10-fold lower, respectively. Interestingly, there are two reasons for the lower kcat/Kms of VIM-13 for ceftazidime and cefepime (in fact, the two lowest values among the antibiotics studied). While VIM-13 showed a low kcat for ceftazidime in comparison to that of VIM-1 (turnover rates, 10 and 42 s−1, respectively), it showed an extremely high Km (and, therefore, a lower affinity) for cefepime (1,870 and 337 μM, respectively). This lower efficiency of hydrolysis of VIM-13 for ceftazidime and cefepime was also in good agreement with the MIC data (Table 3), which show that either the XL1-Blue or the PAO1 pUCPVIM-13 transformants have lower levels of resistance than the pUCPVIM-1 transformants, particularly to cefepime. For the rest of the β-lactams tested, although VIM-13 tended to show a better efficiency of hydrolysis than VIM-1 (including the doubling of its efficiency of hydrolysis for imipenem and meropenem), no significant differences in the MICs were observed in either the E. coli or the P. aeruginosa background. Moreover, as has been observed for VIM-2 and the very recently characterized VIM-11, VIM-13 showed a higher catalytic efficiency for imipenem than for meropenem (6, 18).
TABLE 4.
Kinetic parameters for VIM-13 enzyme with various β-lactams in comparison with those for VIM-1a
| β-Lactam |
kcat (s−1)
|
Km (μM)
|
kcat /Km (M−1 s−1)
|
|||
|---|---|---|---|---|---|---|
| VIM-1 | VIM-13 | VIM-1 | VIM-13 | VIM-1 | VIM-13 | |
| Penicillins | ||||||
| Penicillin G | 418 ± 116 | 757 ± 146 | 1,303 ± 393 | 1,127 ± 172 | 3.2E5 | 6.7E5 |
| Ampicillin | 307 ± 67 | 67 ± 9.5 | 1,737 ± 376 | 197 ± 48 | 1.8E5 | 3.4 E5 |
| Piperacillin | 374 ± 107 | 362 ± 15 | 1,377 ± 340 | 729 ± 96 | 2.7E5 | 4.9E5 |
| Cephalosporins | ||||||
| Cephalothin | 449 ± 133 | 656 ± 143 | 135 ± 29 | 76 ± 16 | 3.3E6 | 8.6E6 |
| Cefuroxime | 239 ± 22 | 283 ± 99 | 148 ± 36 | 56 ± 5 | 1.6E6 | 5E6 |
| Cefotaxime | 204 ± 26 | 612 ± 120 | 285 ± 25 | 233 ± 46 | 7.18E5 | 2.6E6 |
| Ceftazidime | 42 ± 9.6 | 10 ± 1 | 1,132 ± 320 | 509 ± 120 | 3.7E4 | 1.9E4 |
| Cefepime | 119 ± 17 | 61 ± 21 | 337 ± 110 | 1,870 ± 569 | 3.5E5 | 3.3E4 |
| Carbapenems | ||||||
| Imipenem | 3.4 ± 1.2 | 54 ± 17 | 4 ± 1.9 | 18.5 ± 2.2 | 8.38E5 | 2.92E6 |
| Meropenem | 20 ± 0.2 | 9 ± 1.5 | 72 ± 7.7 | 15.5 ± 2.3 | 2.8E5 | 5.92E5 |
The kcat and Km values represent the means of three measurements ± standard deviations.
Interaction of VIM-13 with metal-chelating agents.
Data on the inhibition of hydrolysis activity of VIM-13 (compared to that of VIM-1) after incubation with EDTA, dipicolinic acid, or 1,10-o-phenanthroline are shown in Table 5. No significant differences between VIM-1 and VIM-13 were observed when they were incubated with dipicolinic acid or 1,10-o-phenanthroline, although dipicolinic acid showed a slightly greater ability to inactivate both enzymes. Interestingly, the data for EDTA revealed that this chelator inhibited the activity of VIM-13 approximately 25 times less than it inhibited the activity of VIM-1, which indicates a stronger binding of Zn ions in VIM-13 than in VIM-1.
TABLE 5.
IC50s of various chelating agents for VIM-1 and VIM-13 enzymes
| Chelating agent | IC50 (μM)a
|
|
|---|---|---|
| VIM-1 | VIM-13 | |
| Dipicolinic acid | 41.6 ± 2.9 | 48.0 ± 8.5 |
| 1,10-ο-Phenanthroline | 73.0 ± 7.2 | 69.3 ± 4.0 |
| EDTA | 9.3 ± 2.1 | 252.6 ± 42.4 |
The IC50s represent the means of three measurements ± standard deviations.
DISCUSSION
The isolation of MBL-producing gram-negative bacilli, particularly P. aeruginosa, is a problem of growing concern in the nosocomial environment. Indeed, integrons encoding carbapenemases plus aminoglycoside-modifying enzymes are increasingly being reported (1, 13, 21, 22, 30, 37). Moreover, we are facing a marked increase in the diversity of MBLs: up to 25 IMP-type MBLs and 14 VIM-type MBLs have been reported so far (10, 23, 27, 38). In this work, we describe a new VIM-type MBL, designated VIM-13, that exhibits 93% and 88% amino acid sequence identities with VIM-1 and VIM-2, respectively. Additionally, we also report here for the first time the detection of a VIM-1-producing P. aeruginosa strain in Spain (VIM-2 had been found in P. aeruginosa and VIM-1 has been found only in members of the family Enterobacteriaceae [28, 36]).
Several genes encoding aminoglycoside-modifying enzymes have been detected in integrons harboring blaVIM (11, 19, 26, 30, 37). Strain PA-SL2, which is resistant to gentamicin and tobramycin, harbored an Ala108Thr variant of the previously described acetyltransferase encoded by aacA4 (14). Its Leu83Ser change modifies its spectrum, affecting tobramycin, netilmicin, and gentamicin but not amikacin (14). As shown in this work, the Ala108Thr polymorphism that was detected seems not to affect further the spectrum of the enzyme.
The locations of several VIM-producing integrons have been traced to plasmids, although a chromosomal location has been suggested in other cases (38). The VIM-13 integron was apparently located in the chromosome of strain PA-SL2. Nevertheless, the potential location of this integron on a mobilizable transposon, which would therefore facilitate its dissemination, still needs to be explored.
Regarding the biochemical properties of VIM-13, the results of MIC and kinetics experiments clearly indicated that VIM-13 hydrolyzed piperacillin better than VIM-1 and hydrolyzed ceftazidime and cefepime worse than VIM-1. Furthermore, VIM-13 showed higher kcat/Km ratios than VIM-1 for the rest of the β-lactams, although no differences in MICs were documented, since pUCPVIM-1 already conferred very high levels of resistance (MICs > 256 μg/ml) to cefuroxime, cephalothin, cefotaxime, and ampicillin even in the E. coli background. Interesting particular properties were also observed when the individual kinetic parameters (kcat or Km) of both enzymes were compared. For instance, the high Km of cefepime for VIM-13 appeared to be responsible for its low efficiency of hydrolysis. On the other hand, with other substrates, such as ampicillin or meropenem, notable differences in individual kinetic parameters between VIM-1 and VIM-13 balanced out, finally yielding similar efficiencies of hydrolysis, as observed by Docquier et al. when they compared VIM-1 and VIM-2 (6). The 19-amino-acid difference between the two enzymes should explain the kinetic data for VIM-1 and VIM-13. As indicated by Docquier et al. (6), amino acid changes in loop L3 (the active enzyme site) could explain the distinct biochemical behaviors of VIM-1 and VIM-2 (6) because these changes affect substrate binding. Interestingly, VIM-13 showed two amino acid replacements, His201Leu and Ser205Arg, in the loop cited by Docquier et al. (6). Furthermore, the Ser205Arg polymorphism is also observed in VIM-2, whereas the histidine residue at position 201 is also modified in this enzyme, although in this case it is replaced by a tyrosine (Fig. 1). At position 205, VIM-13, like VIM-2, has an arginine instead of a serine. It has previously been speculated that this replacement may favor the interaction of the charged guanidinium group directly with the benzylpenicillin carboxylate (O-NH2), as the distance of Ser is too short to create any interaction with the substrate (6). In agreement with that hypothesis, VIM-13 showed higher kcat/Km values than VIM-1 for penicillin G. At position 201, VIM-2 and VIM-13 showed the replacements His to Tyr and His to Leu, respectively (Fig. 1). The fact that these enzymes showed lower catalytic efficiencies against cefepime is interesting; therefore, it can be hypothesized that the His-imidazole group is somehow important in the direct interaction of this antibiotic with bulky substituents, although this hypothesis needs further confirmation. Hence, further site-directed mutagenesis studies are needed in order to demonstrate the involvement of the specific residues in the efficiencies of hydrolysis. Moreover, the recently characterized VIM-12 enzyme shows no amino acid changes in this loop in comparison with the amino acid sequence of VIM-1, although clear kinetic differences have been found for VIM-12 (12). This fact clearly indicates that the changes in the active center are not the only changes responsible for the different biochemical behaviors (18). Moreover, amino acid differences have also been shown to be responsible for the worse inhibitory power shown by EDTA against VIM-13 in comparison to that against VIM-1. Nevertheless, no amino acid changes were observed in VIM-13 zinc-ligand residues in comparison with the amino acid sequence of VIM-1 or VIM-2 (6). Interestingly, the IC50 of EDTA for VIM-2 from the work of Poirel et al. (25) was 50 μM, a value intermediate between our IC50 for VIM-1 (9.3 μM) and our IC50 for VIM-13 (252.6 μM). This fact seems to indicate that differences in the amino acid compositions and not only the differences related to the zinc-ligand residues are also responsible for the different levels of inhibition caused by the chelating agents against MBLs.
In summary, we describe a new integron-borne blaVIM gene, blaVIM-13, detected in a P. aeruginosa clinical isolate in Majorca, Spain. Although the encoded VIM-13 enzyme is found to be located the closest to the VIM-1 cluster, it shows remarkable differences from VIM-1 in its biochemical and microbiological behaviors.
Acknowledgments
This work was supported by the Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III, through the Spanish Network for the Research in Infectious Diseases (grants REIPI C03/14 and RD06/0008) and grant PI061368 and by the Govern de les Illes Balears (grant PROGECIC-4C).
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Adrian, P. V., C. J. Thomson, K. P. Klugman, and S. G. Amyes. 2000. New gene cassettes for trimethoprim resistance, dfr13, and streptomycin-spectinomycin resistance, aadA4, inserted on a class 1 integron. Antimicrob. Agents Chemother. 44:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, Y., M. Murakami, K. Suzuki, H. Ito, R. Wacharotayankun, S. Ohsuka, N. Kato, and M. Ohta. 1995. A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39:1612-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beceiro, A., L. Domínguez, A. Ribera, J. Vila, F. Molina, R. Villanueva, J. M. Eiros, and G. Bou. 2004. Molecular characterization of the gene encoding a new AmpC β-lactamase in a clinical strain of Acinetobacter genomic species 3. Antimicrob. Agents Chemother. 48:1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, vol. 26, no. 3, 16th informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Docquier, J. D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J. M. Frère, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Fluit, A. C., and F. J. Schmitz. 1999. Class 1 integrons, gene cassettes, mobility, and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 18:761-770. [DOI] [PubMed] [Google Scholar]
- 8.Franceschini, N., B. Caravelli, J. D. Doquier, M. Galleni, J. M. Frère, G. Amicosante, and G. M. Rossolini. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutiérrez, O., C. Juan, E. Cercenado, F. Navarro, E. Bouza, P. Coll, J. L. Pérez, and A. Oliver. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 51:4329-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson, N. D., A. Hossain, L. L. Buck, E. S. Moland, and K. S. Thomson. 2005. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-291.
- 11.Jones, M. E., E. Peters, A. M. Weersink, A. Fluit, and J. Verhoef. 1997. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet 349:1742-1743. [DOI] [PubMed] [Google Scholar]
- 12.Kontou, M., S. Pournaras, I. Kristo, A. Ikonomidis, A. N. Maniatis, and C. Stathopoulos. 2007. Molecular cloning and biochemical characterization of VIM-12, a novel hybrid VIM-1/VIM-2 metallo-beta-lactamase from a Klebsiella pneumoniae clinical isolate, reveal atypical substrate specificity. Biochemistry 46:13170-13178. [DOI] [PubMed] [Google Scholar]
- 13.Lagatolla, C., E. Edalucci, L. Dolzani, M. L. Riccio, F. De Luca, E. Medessi, G. M. Rossolini, and E. A. Tonin. 2006. Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeruginosa in a nosocomial setting of high-level endemicity. J. Clin. Microbiol. 44:2348-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, T., M. C. Ploy, and P. Courvalin. 1994. A spontaneous point mutation in the aac(6′)-Ib′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 115:297-304. [DOI] [PubMed] [Google Scholar]
- 15.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frère, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livermore, D. M. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 34:634-640. [DOI] [PubMed] [Google Scholar]
- 18.Marchiaro, P., P. E. Tomatis, M. A. Mussi, F. Pasteran, A. M. Viale, A. S. Limansky, and A. J. Vila. 2008. Biochemical characterization of metallo-β-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob. Agents Chemother. 52:2250-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Freijo, P., A. C. Fluit, F. J. Schmitz, J. Verhoef, and M. E. Jones. 1999. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob. Agents Chemother. 43:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes, R. E., M. A. Toleman, J. Ribeiro, H. S. Sader, R. N. Jones, and T. R. Walsh. 2004. Integron carrying a novel metallo-beta-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistant gene aac(6′)-30/aac(6′)-Ib′: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 48:4693-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendes, R. E., M. Castanheira, M. A. Toleman, H. S. Sader, R. N. Jones, and T. R. Walsh. 2007. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene (aac(6′)-31), and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob. Agents Chemother. 51:2611-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nemec, A., L. Dolzani, S. Brisse, P. van den Broek, and L. Dijkshoorn. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233-1240. [DOI] [PubMed] [Google Scholar]
- 23.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 24.Peña, C., C. Suárez, F. Tubau, O. Gutiérrez, A. Domínguez, A. Oliver, M. Pujol, F. Gudiol, and J. Ariza. 2007. Nosocomial spread of Pseudomonas aeruginosa producing the metallo-β-lactamase VIM-2 in Spanish hospital: clinical and epidemiological implications. Clin. Microbiol. Infect. 13:1026-1029. [DOI] [PubMed] [Google Scholar]
- 25.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing beta-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pournaras, S., A. Ikonomidis, L. S. Tzouvelekis, D. Tokatlidou, N. Spanakis, A. N. Maniatis, N. J. Legakis, and A. Tsakris. 2005. VIM-12, a novel plasmid-mediated metallo-beta-lactamase from Klebsiella pneumoniae that resembles a VIM-1/VIM-2 hybrid. Antimicrob. Agents Chemother. 49:5153-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prats, G., E. Miró, B. Mirelis, L. Poirel, S. Bellais, and P. Nordmann. 2002. First isolation of a carbapenem-hydrolyzing beta-lactamase in Pseudomonas aeruginosa in Spain. Antimicrob. Agents Chemother. 46:932-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn, J. P., D. Miyashiro, D. Sahm, R. Flamm, and K. Bush. 1989. Novel plasmid-mediated beta-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 33:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Preparation and transformation of competent E. coli using calcium chloride, p. 1.116-1.118. In Molecular cloning. a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Senda, K., Y. Arakawa, K. Nakashima, H. Ito, S. Ichiyama, K. Shimokata, N. Kato, and M. Ohta. 1996. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 40:349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sykes, R. B., D. P. Bonner, K. Bush, and N. H. Georgopapadakou. 1982. Aztreonam (SQ 26,776) a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob. Agents Chemother. 21:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-beta-lactamase gene in Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tórtola, M. T., S. Lavilla, E. Miró, J. J. González, N. Larrosa, M. Sabaté, F. Navarro, and G. Prats. 2005. First detection of a carbapenem-hydrolyzing metalloenzyme in two Enterobacteriaceae isolates in Spain. Antimicrob. Agents Chemother. 49:3492-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsakris, A., A. Ikonomidis, N. Spanakis, A. Poulou, and S. Pournaras. 2007. Characterization of In3Mor, a new integron carrying VIM-1 metallo-beta-lactamase and sat1 gene, from Morganella morganii. J. Antimicrob. Chemother. 59:739-741. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]