Abstract
Two clinical strains of Escherichia coli (2138) and Enterobacter cloacae (7506) isolated from the same patient in France and showing resistance to extended-spectrum cephalosporins and low susceptibility to imipenem were investigated. Both strains harbored the plasmid-contained blaTEM-1 and blaKPC-2 genes. blaKLUC-2, encoding a mutant of the chromosomal β-lactamase of Kluyvera cryocrescens, was also identified at a plasmid location in E. cloacae 7506, suggesting the ISEcp1-assisted escape of blaKLUC from the chromosome. Determination of the KPC-2 structure at 1.6 Å revealed that the binding site was occupied by the C-terminal (C-ter) residues coming from a symmetric KPC-2 monomer, with the ultimate C-ter Glu interacting with Ser130, Lys234, Thr235, and Thr237 in the active site. This mode of binding can be paralleled to the inhibition of the TEM-1 β-lactamase by the inhibitory protein BLIP. Determination of the 1.23-Å structure of a KPC-2 mutant in which the five C-ter residues were deleted revealed that the catalytic site was filled by a citrate molecule. Structure analysis and docking simulations with cefotaxime and imipenem provided further insights into the molecular basis of the extremely broad spectrum of KPC-2, which behaves as a cefotaximase with significant activity against carbapenems. In particular, residues 104, 105, 132, and 167 draw a binding cavity capable of accommodating both the aminothiazole moiety of cefotaxime and the 6α-hydroxyethyl group of imipenem, with the binding of the former drug being also favored by a significant degree of freedom at the level of the loop at positions 96 to 105 and by an enlargement of the binding site at the end of strand β3.
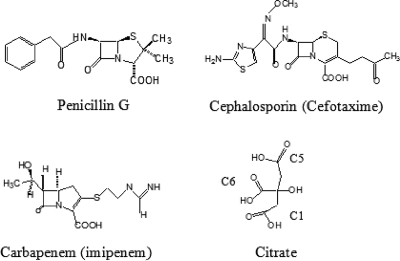
β-Lactam antibiotics include penicillins, cephalosporins, monobactams, and penems. The carbapenems, such as imipenem and meropenem, are being used with increasing frequency for the treatment of multiresistant nosocomial gram-negative pathogens (21, 30). They differ from the classical β-lactam antibiotics by the presence of a carbapenem ring fused to the four-membered β-lactam ring and by the presence of the 6α-1R-hydroxyethyl substituent instead of the acylamido group found at the 6β and 7β positions of penicillins and cephalosporins, respectively (Fig. 1). Resistance to carbapenems, while uncommon in enteric organisms, can be mediated by three unique mechanisms: first, the production of large quantities of a chromosomal AmpC cephalosporinase combined with decreased drug permeability (33, 47); second, the modification of the affinity of the target enzymes for carbapenems (the penicillin-binding proteins) (20); and third, the production of β-lactamases capable of hydrolyzing carbapenems, which can belong to Ambler molecular classes A, B, and D (12, 46). With regard to the first group, only a very limited number of class A enzymes have been found to be able to hydrolyze carbapenems efficiently (18). Most of them are chromosomally encoded (NMC-A, SME-1 to -3, IMI-1), but a new subgroup of class A carbapenemases, the KPC β-lactamases (KPC-1 to -4), correspond to plasmid-encoded enzymes that can easily disseminate. Initially identified in Klebsiella pneumoniae (2, 61), this group of potent carbapenemases is now documented for numerous pathogens, including many members of the family Enterobacteriaceae, with reports of its presence in Klebsiella oxytoca (61), Salmonella enterica (35), Escherichia coli (16, 39), Citrobacter freundii (17), Enterobacter spp. (17, 23), and Serratia marcescens (17, 63). The KPC enzymes also show an expanding geographic range (17), with recent reports of their presence in Israel (29, 39), France (37), and China (63).
FIG. 1.
Chemical structures of penicillin G, a carbapenem (imipenem), a broad-spectrum cephalosporin (cefotaxime), and a citrate molecule.
Regarding their genetic support, most of the studies published to date have described blaKPC genes on plasmids with sizes varying from 23 kb to 75 kb (37). Bratu et al. (9) reported that the blaKPC gene from a carbapenem-resistant isolate of E. coli from New York was flanked by two open reading frames (ORFs) encoding a putative transposition helper protein (GenBank accession no. AAM10642.1) and a putative transposase (GenBank accession no. AAM10644.1), as already described for S. enterica (35) and K. pneumoniae (37, 59), a result that provides further evidence that a common mobile element may be involved in the transfer of blaKPC among different species.
The substrate profile of the KPC enzymes has been investigated in previous reports (61, 62). Interestingly, they not only appear to be carbapenemases but also behave as proficient cefotaximases, since the relative catalytic efficiency for cefotaxime in the KPC family is 1.5-fold higher than that for imipenem (60). Comparatively, the other class A carbapenemases, SME-1 and NMC-A, show a detectable activity for this aminothiazoleoxime cephalosporin but demonstrate a much higher activity for imipenem (60). To date, the unique substrate specificity of KPC, which can be regarded as a cefotaximase showing a substrate range extended toward carbapenems, has yet to be understood.
At the protein level, KPC-2 displays all the conserved catalytic residues of class A β-lactamases (basically the conserved motifs Ser70-X-X-Lys and Ser130-Asp-Asn, the general base Glu166, and the β3 motif Lys234-Thr-Gly) and most of the residues that were previously proposed to be specifically involved in the catalytic activity of class A carbapenemases, including (i) Cys69 and Cys238, which establish a disulfide bridge stabilizing the active-site topology in the crystal structures of NMC-A (54) and SME-1 (50); (ii) the asparagine residue found at position 132, which occupies a peculiar position in the active site of the enzyme and provides additional space in a region of the binding site where the 6α-hydroxyethyl moiety of imipenem must be accommodated (54); and (iii) the Ser or Thr residue found at position 237, which was experimentally shown to contribute to the imipenemase activity of class A carbapenemases such as SME-1 (51). Very recently, Ke et al. determined the crystal structure of KPC-2 at a resolution of 1.85 Å (26). They highlighted more subtle structural changes likely involved in the ability of this class A carbapenemase to hydrolyze imipenem, such as a decrease in the size of the water pocket and a catalytic serine in a more shallow position in the active-site cleft, associated with a combination of active-site adjustments (Asn132 and Asn170 shifted, carbonyl group of Cys238 reoriented).
In this study, we report on an E. coli strain and an Enterobacter cloacae strain showing resistance to extended-spectrum cephalosporins and a decreased susceptibility to imipenem, which were identified from a same patient hospitalized in France. Due to the alarming worldwide spread of blaKPC genes, which have a strong dissemination potential, we investigated the genetic environments of the β-lactamase genes in the two strains, showing the presence of three plasmid-mediated bla genes coding for KPC-2, the class A β-lactamase TEM-1, and a new mutant of the chromosomally encoded class A β-lactamase KLUC from Kluyvera cryocrescens. We also characterized the KPC-2 enzyme at the biochemical and structural levels and confirmed its substrate spectrum as being that of a cefotaximase capable of hydrolyzing the carbapenem imipenem (in that sense, KPC-2 can be considered as an extremely broad-spectrum class A β-lactamase). We determined at high resolution (1.6 Å) the structure of KPC-2 and found that the C-terminal (C-ter) part of one KPC-2 monomer in the crystal was inserted in the binding site of another monomer, thus occupying the β-lactam active site. We also established the 1.23-Å structure of a KPC-2 mutant in which the five C-ter amino acids were deleted and showed the presence of a citrate molecule in the active site of the deleted enzyme. Finally, docking simulations were carried out, shedding light on the structural properties conferring to KPC-2 an extremely broad spectrum of activity, including activity against cefotaxime as well as imipenem.
MATERIALS AND METHODS
Bacterial strains.
Two strains showing decreased susceptibility to imipenem and resistance to extended-spectrum cephalosporins and aztreonam, E. coli 2138 and E. cloacae 7506, were isolated from peritoneal liquid from a 72-year-old French man hospitalized in November 2002 at the Pitié-Salpêtrière hospital (Paris, France). This patient was previously hospitalized for diffuse abdominal pains and upon suspicion of a urinary tract infection in Israel, where he received a combined ofloxacin-gentamicin regimen. One month later, he was admitted to the Pitié-Salpêtrière hospital for a psoas abscess which was the cause of a sepsis. Two strains of E. coli susceptible to β-lactams were isolated from the peritoneal fluid of the patient, who was treated with a combination of gentamicin and ceftriaxone. One week after the first sepsis, the patient developed a second sepsis and was treated with imipenem and amikacin. E. coli 2138 and E. cloacae 7506 were isolated from a peritoneal fluid culture during the second episode of sepsis.
E. coli Top10 [F− mcrAΔ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG] was used for the electroporation of plasmid DNA and for cloning experiments. E. coli J53 (Rifr) was used as the recipient in conjugal mating experiments.
Antimicrobial susceptibility testing.
The following antibiotics were obtained from the indicated suppliers: ampicillin, oxacillin, and aztreonam from Bristol-Myers Squibb, France; chloramphenicol, cefuroxime, and cephalothin from Sigma Chemical Co.; cefoxitin and imipenem from Merck Sharp and Dohme-Chibret, France; cefotaxime, cefpirome, and rifampin from Hoescht-Marion-Roussel, France; ceftazidime, nitrocefin, ticarcillin, amoxicillin, and clavulanate from GlaxoSmithKline, France; piperacillin from Lederle, France; and benzylpenicillin from Sarbach, France. The MICs were determined by the Etest method (41).
Preparation of crude extracts and isoelectrofocusing.
Exponentially growing cells were harvested and resuspended in 600 μl of 50 mM phosphate sodium buffer (pH 7.0). The suspensions were disrupted by sonication and the crude extracts were used in β-lactamase detection. Isoelectric focusing was performed with an LKB Multiphor apparatus using pH 3.5-to-9.5 polyacrylamide gel plates (Pharmacia Biotech, Saint Quentin en Yvelines, France). Gels were focused at 30 W for 90 min at 10°C. β-Lactamase activity was revealed by staining the gel with the chromogenic β-lactam nitrocefin (42).
Mating-out assays and plasmid content analysis.
Transfer of β-lactam resistance to E. coli J53 was attempted by liquid and solid mating-out assays. The recipient and donor cells were mixed at 1:1 and 4:1 ratios and incubated in brain heart infusion with moderate shaking at 37°C for 3 h. After incubation, 200 μl of each mixture was plated out on a Millipore filter disc onto brain heart infusion plates and incubated for 18 h at 37°C. Transconjugants were selected on LB agar containing ampicillin (100 μg/ml), ticarcillin (50 μg/ml), and rifampin (1000 μg/ml). The plasmid content of each transconjugant strain was examined by the procedures of Birnboim and Doly (4) and Takahashi et al. (55). Plasmid DNAs prepared from E. coli 2138 and E. cloacae 7506 were electroporated into E. coli Top10. Transformants were selected on LB agar containing ampicillin at 100 μg/ml.
DNA amplification.
DNA amplification of β-lactamase genes was carried out with the various specific primers (Eurogentec, Belgium) listed in Table 1. The DNA amplifications were performed on 100-μl samples containing DNA (5 μl), deoxynucleoside triphosphate (250 μM), primers (0.4 μM each), Taq DNA polymerase (1 U), and its buffer. The following cycles were used: 10 min of denaturation at 94°C (1 cycle) and 1 min of denaturation at 94°C, 1 min of annealing (see temperatures in Table 1), and 1 min of polymerization at 72°C (35 cycles) followed by a 10-min extension at 72°C.
TABLE 1.
Nucleotide sequences of primers
| Primer | Sequencea | Gene | Annealing temp (°C) | Reference or source |
|---|---|---|---|---|
| SHV rev | 5′-GCGTTGCCAGTGCTCGATCAG-3′ | blaSHV | 62 | 3 |
| SHV bis | 5′-ATGCGTTATATTCGCCTGTGTATT-3′ | |||
| TEM H | 5′-TGAGATCGAAGGGCCGTT-3′ | blaTEM | 52 | 53 |
| TEM rev | 5′-GGTCTGACAGTTACCAAT-3′ | |||
| SmeA | 5′-CGGTCCTGA GGGGATGAC-3′ | blaSme-1 | 53 | 38 |
| SmeB | 5′-CGTGATGCTTCCGCAATA-3′ | |||
| KPC 1S | 5′-ATGTCACTGTATCGCCGT-3′ | blaKPC-1 | 48 | 61 |
| KPC 1AS | 5′-CCTTACTGCCCGTTGACG-3′ | |||
| KPCINT S | 5′-ATGACTATCCCGTCGGC-3′ | blaKPC-1 | 52 | 61 |
| KPCINT AS | 5′-CATCGCGTACACACCGATGGA-3′ | |||
| KLUC-1 S | 5′-ATGGTTAAAAAATCATTA-3′ | blaKLUC-1 | 47 | 15 |
| KLUC-1 AS | 5′-CTATAATCCCTCAGTGAC-3′ | |||
| CTX-M1A | 5′-CTTCCAGAATAAGGAATC-3′ | blaCTX-M | 48 | 6 |
| CTX-M1B | 5′-CCGTTTCCGCTATTACAA-3′ | |||
| CTX-MA | 5′-CGCTTTGCGATGTGCAG-3′ | |||
| ISEcpI 1A | 5′-AATCTAACATCAAATGCA-3′ | 50 | 45 | |
| ISEcpI 1B | 5′-TTTTGCTGCAAGAAATACATA-3′ | |||
| IS1 S | 5′-ATCGCCTTGGGTTACAAG-3′ | 49 | 62 | |
| IS1 AS | 5′-CTTGTCCTTCAAGCGGTA-3′ | |||
| KPC-EcoRI | 5′-CCGGAATTCTTACTGCCCGTTGACGCCCAA-3′ | blaKPC-1 | 60 | Present study |
| KPC-NdeI | 5′-GAATTCCATATGTCACTGTATCGCCGTCTAGTT-3′ | |||
| Tase 4 | 5′-ATGGCTGCCAGTTACCTT-3′ | 54 | 59 | |
| Tase 1 | 5′-CCTTGGCATCTTCACGTCCCT-3′ | |||
| KPdelS | 5′-CGAGGGATTGGGCTAAGAATTCGAGC-3′ | 54 | Present study | |
| KPdelAS | 5′-GCTCGAATTCTTAGCCCAATCCCTCG-3′ |
Where present, underlining indicates the restriction site for the endonuclease included in the primer designation.
Plasmid profile analysis and probing.
Plasmid DNA from E. coli 2138, E. cloacae 7506, E. coli Top10 transformants, and E. coli J53 transconjugants was extracted using a Qiagen plasmid midi-prep kit. The DNA preparations were electrophoresed on 0.8% agarose gels in the presence of 0.5× TBE buffer (45 mM Tris-HCl, 45 mM boric acid, and 1.25 mM EDTA, pH 8.3) at a constant voltage for 16 h at 4°C. After Southern transfer onto a nylon membrane (Hybond-N+; Amersham), the DNA was cross-linked on the membrane by 5 min of UV exposure. Probe labeling and hybridization experiments were performed according to the manufacturer's instructions (ECL kit; Amersham) by use of internal fragments obtained by PCR from blaKPC or blaKLUC as probes.
Nucleic acid techniques and sequence analysis.
Genomic DNA from E. coli 2138 and E. cloacae 7506 was extracted as described previously (49). For cloning experiments, the extracted DNA was partially digested with Sau3AI. The fragments were ligated into the dephosphorylated vector pBKCMV previously digested with BamHI. The ligations were done at 4°C for 16 h with 100 ng of chromosomal DNA, 200 ng of digested plasmid vector pBKCMV, and 1 U of T4 DNA ligase (Amersham). After purification and concentration with the High Pure PCR product purification kit (Boehringer Mannheim), the ligation mixture was transformed by electroporation into E. coli Top10. Transformants resistant to β-lactam antibiotics were selected on LB agar plates supplemented with 50 μg/ml of ampicillin.
Recombinant plasmid DNA was extracted using the rapid procedure of Birnboim and Doly (4). The inserted DNA fragments were sequenced on both strands by primer walking using the Big Dye version 3.0 sequencing kit (Applied Biosystems) and an Applied Biosystems ABI 3100 sequencer. Sequence analysis was performed with the software available at the National Center of Biotechnology Information website. The program ORF finder was used to determine all the putative ORFs, which were analyzed using the BLASTP program (1).
β-Lactamase purification.
In order to purify KPC-2 on a large scale for kinetic and crystallographic experiments, we cloned blaKPC-2 in the expression vector pET29a. The bla gene encoding KPC-2 was obtained by DNA amplification from the clinical E. cloacae strain 7506. The PCR was performed using a reverse primer, KPC-EcoRI, containing the restriction site EcoRI (Table 1) and a forward primer, KPC-NdeI, designed to anneal at the beginning of the blaKPC-2 gene and containing the restriction site NdeI (Table 1). The PCR product obtained using these two primers was purified from agarose gel with the Prepagen kit (Bio-Rad) and was first cloned in a pCRScript vector. The insert containing the blaKPC-2 gene was then recovered by digestion with EcoRI and NdeI and ligated by T4 DNA ligase into the EcoRI- and NdeI-restricted sites of the pET29a plasmid. To create the KPC-2 mutant with a C-ter deletion (C-ter-deleted KPC-2 mutant), site-directed mutagenesis was made on the pET29-blaKPC-2 plasmid by use of the QuikChange kit (Stratagene) and the primers KPdelS and KPdelAS (Table 1). The last residue of the C-ter-deleted KPC-2 protein was Gly291. The recombinant plasmids were introduced by transformation into E. coli BL21(DE3), which was used as the host cell type for protein expression.
The KPC-2 enzyme was purified from a culture of 6 × 500 ml of LB supplemented with 50 μg/ml of kanamycin. Protein expression was induced by adding 0.4 mM of IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture. Bacterial cells were pelleted, resuspended in 60 ml of Bis-Tris at 40 mM and a pH of 5.9, and lysed by ultrasonic treatment. After nucleic acid precipitation by spermine (0.2 M) at 4°C, the supernatant was dialyzed against 3 liters of 40 mM Bis-Tris, pH 5.9, and applied onto a 2.5- by 10-cm Q Sepharose fast flow column (Pharmacia Co. Ltd., Sweden) previously equilibrated with the dialysis buffer. The unadsorbed active fractions, detected with the chromogenic cephalosporin nitrocefin (42), were pooled and loaded onto a Bioscale S cation-exchange column (Bio-Rad) previously equilibrated in Bis-Tris at 40 mM and a pH of 5.9. The active fractions, eluted by a linear gradient of 0 to 1 M NaCl in 40 mM Bis-Tris, pH 5.9, were loaded onto a gel filtration Superdex 75 (Pharmacia Co. Ltd., Sweden) previously equilibrated with Bis-Tris at 40 mM and a pH of 5.9. Finally, the enzyme was concentrated onto Microcon 3 (Millipore) to a final concentration of 30 mg/ml. The same purification protocol was used to purify the C-ter-deleted KPC-2 protein to a final concentration of 20 mg/ml.
Kinetic studies.
The kinetic parameters Km and kcat were determined spectrophotometrically at 37°C in 50 mM phosphate buffer (pH 7.0) by use of an Uvikon 940 spectrophotometer. The absorption coefficients used were those previously described (7). Kinetic parameters were determined by fitting the Henri-Michaelis-Menten equation to the experimental data by use of the regression analysis program LEONORA, written by Cornish-Bowden (14). The values of kcat and Km were estimated using a nonlinear least-squares regression method with dynamic weights (14).
Crystallization and structure determination.
KPC-2 crystals were grown in sitting drops against a well solution containing 20% of polyethylene glycol 6000, 0.1 M of KSCN, and 0.1 M of sodium acetate (pH 5.5). Crystals reached a size of 0.2 mm by 0.3 mm by 0.05 mm within 72 h. The crystals belong to space group P21 and had the following unit-cell parameters: a was 56.3 Å, b was 91.4 Å, c was 73.1 Å, α was 90°, β was 112.63°, and γ was 90°. Crystals of the C-ter-deleted KPC-2 protein were obtained in 20% of polyethylene glycol 4000, 0.1 M of KSCN, and 0.1 M of citrate (pH 4.0). The crystals belong to the space group P212121 and had the following unit-cell parameters: a was 47.8 Å, b was 66 Å, c was 72.1 Å, and α equaled β equaled γ equaled 90°.
Data were collected at the Institut Biologie Physique et Chimie (IBPC, Paris, France) by use of the generator RIGAKU model Micro7 (to 2.1 Å) and on the beamline FIP-BM30A (to 1.6 Å for KPC-2 and 1.23 Å for the C-ter-deleted mutant) at the ESRF (European Synchrotron Radiation Facility, Grenoble, France) (48). These data were processed with XDS (25). With the data collected at the IBPC, an initial structure was obtained by molecular replacement with the program PHASER (34) using the SME-1 structure (PDB accession number 1DY6) as a search model. Refinement was performed using CNS (11) and Refmac (57) using anisotropic refinements for the structure at 1.23 Å.
The refinement data for the two structures are summarized in Table 2. For KPC-2, the asymmetric unit contained two independent molecules. In the final model, the electron density in the N-ter part was not sufficiently defined to place the first residues Leu25 to Val29 and Leu25 to Ala30 of molecules A and B, respectively. After 14 cycles of refinement with CNS and two cycles using Refmac, the model showed an Rcrystal factor of 18.8% and an Rfree value of 21%, calculated for 5% of randomly selected data. For the C-ter-deleted KPC-2 mutant, a single monomer was found in the asymmetric unit. In the final model obtained with an Rcrystal factor of 16.5% and an Rfree value of 18.3%, only the first residue, Leu25, was lacking.
TABLE 2.
Crystallographic analysis
| Statistic | Value fora:
|
|
|---|---|---|
| Native KPC-2 | C-ter-deleted KPC-2 | |
| Data collection statistics | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a (Å) | 56.3 | 47.8 |
| b (Å) | 91.4 | 66 Å |
| c (Å) | 73.1 | 72.1 |
| β (°) | 112.63 | 90 (= α = γ) |
| Wavelength (Å) | 0.9791 | 0.9791 |
| Resolution (Å) | 19.76-1.6 | 26.4-1.23 |
| Highest-resolution shell (Å) | 1.79-1.6 | 1.31-1.23 |
| No. of unique reflections | 86,682 | 65,188 |
| Redundancy | 8.9 (6.8) | 13.6 (11.6) |
| Rsym (%) | 8.4 (41.3) | 4.8 (19.7) |
| I/σ (I) | 22.5 (3.7) | 23.7 (6.6) |
| Completeness (%) | 96.5 (99.6) | 98 (92) |
| Refinement statistics | ||
| Rcrystal (%) | 18.8 | 16.5 |
| Rfree (%) | 21 | 18.3 |
| No. of protein atoms | 3,953 | 1,985 |
| No. of solvent atoms | 306 | 184 |
| RMSD bond distance (Å) | 0.005 | 0.006 |
| RMSD angle (°) | 1.2 | 1.1 |
| Average B value | 10.9 | 11.8 |
Values in parentheses refer to statistics in the highest-resolution shell (1.79-1.6 Å) for native KPC-2 and (1.31-1.23 Å) for C-ter-deleted KPC-2.
Docking.
The β-lactam structures were constructed with a closed β-lactam ring using the module Modeler in InsightII (Accelrys). Hydrogen atoms were added and each molecule was energy minimized using the CHARMM program (10) and the force field PARAM22 (31) by using the steepest-descent algorithms. Thereafter, the ligands were docked into KPC-2 and TEM-1 structures employing either the Gold program (58) using the Goldscore scoring function (40) or the Autodock program (36). For Gold, the ligands were docked within a sphere of 12 Å around the catalytic proton of Ser70. For Autodock (version 4), the grid of the docking simulation was defined by a 60-Å by 60-Å by 60-Å cube centered on the Ser70 within the active site. The docking simulation was performed using a Lamarckian genetic algorithm search routine. The 10 best solutions for each docked ligand were conserved for analysis. The figures were prepared using Pymol (Delano Scientific LLC), available at http://pymol.sourceforge.net/.
Nucleotide sequence accession numbers and data deposition.
The GenBank accession numbers are EF057432 for the blaKLUC-2 gene from E. cloacae 7506 and DQ989639 and DQ989640 for the blaKPC-2 gene from E. coli 2138 and E. cloacae 7506, respectively. The KPC-2 and the C-ter-deleted KPC-2 mutant coordinates and structure factors have been deposited in the Protein Data Bank; the access codes are 3DWO and 3C5A, respectively.
RESULTS AND DISCUSSION
Antimicrobial susceptibility patterns and β-lactamase production in E. coli 2138 and E. cloacae 7506.
The MICs of a variety of antimicrobial agents tested against E. coli 2138 and E. cloacae 7506 are shown in Table 3. The isolates showed reduced susceptibility to imipenem (MICs = 3 μg/ml). They were also resistant to extended-spectrum cephalosporins and aztreonam, with E. cloacae 7506 displaying the highest level of resistance (Table 3). When tested in the presence of clavulanic acid, the MICs of amoxicillin decreased from >256 μg/ml to 96 and 24 μg/ml for E. cloacae 7506 and E. coli 2138, respectively. In contrast, tazobactam addition did not modify the MICs of piperacillin (>256 μg/ml).
TABLE 3.
MICs of β-lactam antibiotics for E. coli 2138, E. cloacae 7506, E. coli Top10 transformant from E. coli 2138, E. coli J53 transconjuguant from E. cloacae 7506, and E. coli Top10 and J53 reference strains
| β-Lactam(s)a | MIC (μg/ml) determined by Etests for:
|
|||||
|---|---|---|---|---|---|---|
| E. coli 2138 (parent strain producing blaKPC-2 and blaTEM-1) | E. coli Top10 transformant (containing blaKPC-2 and blaTEM-1) | E. coli Top10 | E. cloacae 7506 (parent strain producing blaKPC-2, blaKLUC-2, and blaTEM-1) | E. coli J53 transconjugant (containing blaKPC-2 and blaTEM-1) | E. coli J53 | |
| Amoxicillin | >256 | >256 | 3 | >256 | >256 | 3 |
| Amoxicillin + Cla | 24 | 48 | 2 | 96 | 16 | 2 |
| Ticarcillin | >256 | >256 | 1.5 | >256 | >256 | 1.5 |
| Piperacillin | >256 | >256 | 0.75 | >256 | >256 | 0.38 |
| Piperacillin + Taz | >256 | >256 | 0.75 | >256 | >256 | 0.38 |
| Cefotaxime | 16 | 12 | 0.047 | >32 | 4 | 0.032 |
| Ceftazidime | 8 | 6 | 0.19 | 32 | 2 | 0.064 |
| Aztreonam | 64 | 24 | 0.047 | >256 | 8 | <0.016 |
| Imipenem | 3 | 1.5 | 0.19 | 3 | 0.5 | 0.094 |
Cla, clavulanic acid; Taz, tazobactam.
Isoelectric focusing analysis of a crude extract of E. coli 2138 revealed two bands with pI values of 5.4 and 6.7 (data not shown), suggesting the presence of two β-lactamases in the clinical isolate. For this strain, no DNA amplification was obtained with blaSHV-, blaSME-, and blaCTX-M-specific primers, whereas positive results were obtained for blaTEM- and blaKPC-specific primers. DNA sequence analysis identified the genes as blaTEM-1 and blaKPC-2, with the amino acid sequence of KPC-2 showing a single amino acid difference, Ser175→Gly, compared with KPC-1, the first carbapenem-hydrolyzing β-lactamase of class A identified from K. pneumoniae 1534 (61). Regarding E. cloacae 7506, the isoelectric focusing results suggested the production of at least three β-lactamases, with pIs of 5.4, 6.7, and 7.4. The corresponding amplified bla genes were identified by DNA sequence analysis as being blaTEM-1, blaKPC-2, and a variant of blaKLUC-1, the chromosomal β-lactamase of K. cryocrescens (15). This new variant, termed KLUC-2, showed 86% amino acid identity with a subgroup of plasmid-mediated CTX-M-type extended-spectrum β-lactamases (CTX-M-1, -3, -10, -11, and -12) and was characterized by a single amino acid difference, Gly115→Arg, compared with KLUC-1. The Gly115→Arg amino acid modification is located outside of the β-lactam active site, so it is likely that it does not alter the substrate profile of the enzyme. The identity of this new point mutant β-lactamase was confirmed by cloning and sequencing from the recombinant plasmid pBK-KLUC2, a BamHI genomic DNA fragment harboring blaKLUC-2 from E. cloacae 7506.
Horizontal transfer of a plasmid carrying blaKPC-2 and blaTEM-1.
The transfer of β-lactam resistance by conjugation to E. coli J53 could be obtained for E. cloacae 7506 but not for E. coli 2138. The transconjugants obtained from E. cloacae 7506 exhibited a phenotype of resistance to β-lactams very similar to that of the parent strain, except in the cases of the extended-spectrum β-lactams cefotaxime, ceftazidime, and aztreonam, which exhibited values that were 8- to at least 32-fold lower (Table 3). Regarding E. coli 2138, transformants could be obtained by the electroporation of extracted plasmid DNA into E. coli Top10. They exhibited a phenotype of resistance to β-lactam antibiotics very similar to that of the parent strain E. coli 2138, as well as to the E. coli J53 transconjugant from E. cloacae 7506 (Table 3). By using PCR amplification, we identified the presence of blaKPC-2 and blaTEM-1 in both the E. coli 2138 transformant and the E. cloacae 7506 transconjugant, whereas no amplification of blaKLUC-2 was obtained for either. It is worth noting here that KLUC-2 production undoubtedly contributes to a significant increase in the level of resistance to aminothiazoleoxime cephalosporins, since the MICs for extended-spectrum β-lactams measured for the strains producing only KPC-2 and TEM-1 (E. coli 2138 and its transformant, the E. coli J53 transconjugant of E. cloacae 7506) were found to be 4- to 32-fold lower than the values found for E. cloacae 7506, producing KPC-2, TEM-1, and KLUC-2 (Table 3).
Analysis of the plasmid profile of E. cloacae 7506 revealed the presence of at least three plasmids: HMWP-1, showing a very high molecular weight; HMWP-2, with an estimated size of 40 kb; and LMWPs, corresponding to low-molecular-weight plasmid forms (data not shown). For the transconjugants obtained from E. cloacae 7506, as well as the E. coli 2138 clinical isolate and the corresponding E. coli Top10 transformants, plasmid profile analysis revealed the presence of only one plasmid, showing an electrophoretic mobility identical to that of HMWP-2 as observed for E. cloacae 7506. Southern analysis of the extracted plasmids indicated that the blaKPC probe hybridized to a single band corresponding to HMWP-2 shared by the four strains, namely, E. coli 2138, E. cloacae 7506, and their respective transformants/transconjugants, while the blaKLUC probe hybridized to HMWP-1, present only in the clinical strain E. cloacae 7506 (data not shown).
Genetic environment of the blaTEM-1, blaKLUC-2, and blaKPC-2 genes from the E. coli 2138 and E. cloacae 7506 strains.
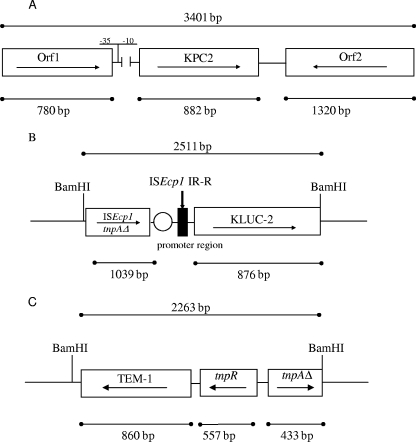
Since the upstream and downstream regions of several blaKPC-2 genes from various clinical isolates were previously reported to be identical (35, 61, 62), we designed two pairs of primers, KPC 1AS and KPCINT S on one hand and Tase 1 and Tase 4 on the other (Table 1), to amplify the upstream and downstream regions of blaKPC-2, respectively. Analysis of the PCR products by DNA sequencing revealed that the nucleotide sequences flanking blaKPC-2 in E. coli 2138 and E. cloacae 7506 are identical and closely resemble those previously reported for K. pneumoniae 1534 (61), Salmonella strain 4707 (35), and K. oxytoca 3127 (62), comprising two ORFs found upstream (Orf1; putative transposition helper protein; GenBank accession no. AAM10642.1) and downstream (Orf2; putative transposase; GenBank accession no. AAM10644.1) of blaKPC-2 (Fig. 2A). The only difference that could be noticed in the genetic environment of blaKPC-2 in our two strains from what was reported for previously studied sequences was the absence of a region 216 bp upstream from blaKPC-2 (Fig. 2A) containing the −10 and −35 promoter sequences previously determined for blaKPC-1 by mRNA primer extension (61). Nevertheless, we performed a computational search for transcription sites upstream from blaKPC-2 that indicated the presence of a highly conserved putative promoter 41 bp upstream from the deletion, with a perfectly conserved −35 consensus sequence (TTGACA) and a well-conserved −10 box (TATCTT) located 18 bp from the −35 site (Fig. 2A).
FIG. 2.
(A) Schematic representation of the blaKPC-2-containing fragment (GenBank accession numbers DQ989639 and DQ989640) obtained by several amplifications with the primers IS1 S, IS1 AS, Tase 4, and Tase 1 (Table 1). The transcriptional orientations of the two flanking ORFs, Orf1 and Orf2, are indicated by arrows. The broken line upstream from blaKPC-2 indicates a lacking stretch of nucleotides compared with previously reported sequences surrounding blaKPC-2. The putative promoter found 41 bp upstream from the deletion is indicated (−35 and −10 sites). (B) Schematic representation of the BamHI segment containing the blaKLUC-2 gene (GenBank accession number EF057432). The truncated transposase gene tnpA of the ISEcp1 element (tnpAΔ) is found upstream from blaKLUC-2. The blaKLUC-2 promoter region is indicated by an open circle. The black box represents the right inverted repeat of ISEcp1 (IR-R). (C) Schematic representation of the BamHI fragment containing the blaTEM-1 gene and the genes coding for the resolvase (tnpR) and a part of the transposase (tnpAΔ), respectively (GenBank accession number AB187515).
The sequencing of the pBK-KLUC2 BamHI fragment encoding KLUC-2 (2,511 bp) showed the presence of an ISEcpI insertion sequence upstream from blaKLUC-2 comprising a truncated tnpA gene. A perfect 24-bp inverted repeat characteristic of the ISEcpI element (accession no. EF057432) (32) was found 48 bp upstream of blaKLUC-2, and the promoter sequence driving blaKLUC-2 transcription from the 3′ noncoding sequence of ISEcpI (44) was found to be present (Fig. 2B). In contrast, the search for the presence of ISEcp1 in the total DNA of the E. coli 2138 strain by specific amplification with the primers ISEcpI-1A and -1B, described in Table 1, revealed the absence of this element from E. coli 2138 (data not shown). Interestingly, our sequence analysis also showed that the 48-bp sequence upstream from blaKLUC-2 has 87% nucleotide identity with the nucleotide sequences located upstream of the blaCTX-M-3, blaCTX-M-10, and blaCTX-M-15 genes. Lartigue et al. previously reported that (i) the genetic environment of blaKLUC-1 in K. cryocrescens was similar to that observed for the blaCTX-M-3, blaCTX-M-15, and blaCTX-M-10 genes in enterobacterial strains from various geographical origins (28) and (ii) that the mobilization of the Kluyvera ascorbata blaKLUA gene on plasmids is mediated by an ISEcp1 genetic mobile element (27). Hence, it is very likely that the mobilization of blaKLUC-2 on the E. cloacae 7506 plasmid is attributable to the presence of the ISEcp1 element found upstream from the β-lactamase gene.
Finally, the sequencing of the fragments containing TEM-1 from E. coli 2138 and E. cloacae 7506 revealed the presence of two genes coding for a resolvase, TnpA, and its regulator, TnpR, present upstream from blaTEM-1 in both strains (Fig. 2C), as previously described for other blaTEM-1-containing plasmids of clinical origin (19).
Kinetic parameters.
The kinetic parameters for the KPC-2 β-lactamase from E. coli 2138 and the derived mutant in which the C-ter part of the protein was deleted were determined. The catalytic constants obtained from the two proteins were nearly identical and are summarized in Table 4. The enzyme displayed a broad substrate spectrum including the β-lactam antibiotics from the penicillin, cephalosporin, carbapenem, and monobactam groups. The highest catalytic activity was measured with cephalothin, which demonstrated a kcat value approximately 35 times higher than that for aztreonam, which was the antibiotic showing the lowest rate of hydrolysis (Table 4). The kcat values for penicillin G, piperacillin, cefuroxime, and cefotaxime were similar and approximately 1.5 to 2 times lower than that found for cephalothin. As expected, KPC-2 showed a significant hydrolytic activity against the carbapenem imipenem (kcat = 20 s−1), which was, however, turned over at a rate approximately three times lower than that for cefotaxime (kcat = 66 s−1). Strikingly, no significant hydrolytic activity could be measured for ticarcillin and ceftazidime, as previously reported by Yigit et al. (62).
TABLE 4.
Kinetic parameters of various β-lactam antibiotics for the KPC-2 β-lactamase
| Substrate | kcat (s−1)a | Km (μM)a | kcat/Km (mM−1·s−1) |
|---|---|---|---|
| Benzylpenicillin | 45 ± 2 | 44 ± 3 | 1,000 |
| Ampicillin | 20 ± 1 | 29 ± 2 | 700 |
| Ticarcillin | NDb | ND | ND |
| Piperacillin | 40 ± 3 | 51 ± 1 | 800 |
| Cephalothin | 106 ± 5 | 141 ± 3 | 750 |
| Cefuroxime | 66 ± 6 | 160 ± 2 | 410 |
| Cefotaxime | 66 ± 3 | 174 ± 1 | 380 |
| Ceftazidime | ND | ND | ND |
| Aztreonam | 3 ± 0.1 | 35 ± 2 | 900 |
| Cefpirome | 12 ± 0.1 | 251 ± 20 | 500 |
| Imipenem | 20 ± 1 | 88 ± 8 | 230 |
Standard deviation values are indicated after the kinetic parameter values.
ND, not determinable.
KPC-2 had the lowest Km values for ampicillin and aztreonam (29 μM and 35 μM, respectively) and the highest for cefpirome (251 μM). Other substrates, such as imipenem and cefotaxime, had intermediate Km values (from 88 to 174 μM). Thus, the kcat/Km ratios were very similar for all of the β-lactam antibiotics tested in the present study, ranging from 230 to 1,000 mM−1·s−1. The hydrolytic efficiency of KPC-2 for imipenem (230 mM−1·s−1) was lower than that measured for cefotaxime (380 mM−1·s−1) and other β-lactam antibiotics (410 to 1,000 mM−1·s−1). These data confirm that KPC-2 has a significant activity against the carbapenem imipenem, but the turnover rate for this antibiotic is approximately 1.5 to 3 times lower than that measured for cefotaxime, as previously reported by Yigit et al. (62). Thus, KPC-2 can be regarded as better as a cefotaximase than as a carbapenemase. Accordingly, the production of KPC-2 in E. coli 2138 and its E. coli Top10 transformant accounts for a low level of resistance to imipenem (technically, the strains remain susceptible), contrasting with the MICs for cefotaxime, which were found to be approximately 10-fold higher than those for imipenem (Table 3). It is also worth mentioning here that, in contrast to the fact that KPC-2 lacks any significant catalytic activity for ceftazidime, the MICs of this drug measured for the two clinical isolates and the corresponding transformant/transconjugant strains producing KPC-2 were roughly comparable to those found for cefotaxime (Table 3). Yigit et al. reported similar conclusions for an E. coli DH5α clone producing only KPC-2, which displayed a MIC for ceftazidime of 32 μg/ml, while the catalytic activity measured for this antibiotic was very weak (<0.12 s−1) and the Km value not determinable (62). Taken altogether, these data might indicate that the resistance to ceftazidime observed for KPC-2 producers could be associated with other mechanisms that have yet to be elucidated.
Determination of the crystal structure of KPC-2 and its C-ter-deleted mutant: peculiar crystallographic packing.
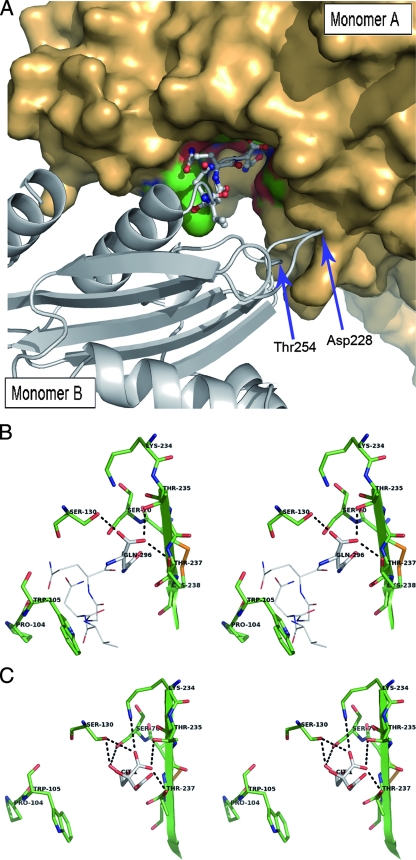
Compared with what is seen for other common class A β-lactamases, including the TEM and CTX-M groups and the two carbapenemases SME-1 and NMC-A, the C-ter end of the KPC-2 protein is characterized by five additional amino acid residues (VNGQQ). Interestingly, these additional residues are involved in contacts between neighboring monomers in the crystal. As shown in Fig. 3, the C-ter residues of one KPC-2 monomer (molecule B) in a given asymmetric unit clearly establish direct contacts with the active-site residues of its nearest neighbor (molecule A) in the adjacent asymmetric unit (Fig. 3A), such that the last C-ter amino acid (Gln296) of molecule B is deeply anchored in the active site of the neighboring molecule A, thereby blocking the accessibility of the KPC-2 active site to any other antibiotic molecule (Fig. 3B). Several hydrogen-bonding interactions link the carboxylate moiety and the side chain of Gln296 (molecule B) to Ser70, Ser130, Thr235, Thr237, and Asn170 in molecule A (Fig. 3B). In addition, Gln295 in the C-ter part of the symmetric B molecule also establishes two hydrogen bonds with the Tyr129 and Trp105 main-chain atoms from the neighboring A molecule (data not shown). Finally, three H-bonding interactions contribute to stabilize the A-B interface at the level of Asp228 and Thr254 in molecule B on one hand (shown in Fig. 3A) and Lys273 and His274 in molecule A on the other (not visible in Fig. 3A). This mode of binding can be paralleled to the inhibition of the TEM-1 β-lactamase by the β-lactamase inhibitory protein BLIP, which is a 17-kDa protein produced by Streptomyces clavuligerus (52). Indeed, BLIP inhibits TEM-1 by inserting a β-hairpin turn into the active site of the β-lactamase, the BLIP residue Asp49, forming strong hydrogen bonds to the four conserved residues Ser130, Lys234, Ser235, and Arg244, found in the catalytic cavity of TEM-1 (52).
FIG. 3.
(A) View of the interactions between the crystallographically observable C-terminally extended peptide of molecule B in one asymmetric unit and the active-site cleft of molecule A from an adjacent asymmetric unit. The monomer A surface is represented in bronze, with the active-site residues colored in green and red. Monomer B is shown in a cartoon representation colored in gray, with the C-ter amino acids anchored in the active site of molecule A being represented in a ball-and-stick configuration. Asp228 and Thr254, two other residues also interacting by H-bonds with the monomer A residues Lys273 and His274, are indicated by arrows. (B) Stereo view depicting the active site of KPC-2 (monomer A, shown as green sticks) containing the last C-ter residues from a symmetric molecule (monomer B, shown with Corey-Pauling-Koltun [CPK] lines). The last residue in monomer B (Gln296) is indicated with CPK sticks. Hydrogen bonds are shown as dotted lines. (C) Stereo view showing the active site of the C-ter-deleted mutant of KPC-2 (green sticks) with a bound citrate molecule (CPK sticks).
As expected, the active site in the structure of the KPC-2 C-ter-deleted protein did not contain any of the amino acids coming from of a neighbor β-lactamase molecule in the crystal. Nevertheless, a well-defined electron density was still present in the catalytic cleft. This additional density could clearly be attributed to a citrate molecule very likely provided by the crystallization buffer. The citrate anion, which is characterized by three carboxylate groups (Fig. 1), was found to interact at the level of the C-1, C-5, and C-6 carboxylate moieties with the two hydroxyl oxygen atoms of Ser70 and Ser130, the side chains of residues Ser130, Thr235, Thr237, and Lys234 (Fig. 3C), and the main-chain atoms of Thr216 (not visible in Fig. 3C). Despite these multiple interactions, citrate was found to have no significant inhibitory activity against KPC-2 (data not shown), likely because of the high desolvation energy of the molecule. It has to be highlighted here that Ke et al. also observed in their KPC-2 structure that the catalytic pocket is filled by a bicine molecule interacting with conserved active-site residues through a set of hydrogen-bonding and electrostatic bonding interactions resembling those reported here for the C-6 carboxylate moiety of the citrate molecule (26). Even if the protein-protein or protein-citrate/bicine interactions observed for KPC-2 are not necessarily relevant from a biological point of view, the existence of such interactions may indicate that the occupancy of the KPC-2 active site by a carboxylate group is somewhat important, possibly to stabilize the enzyme in an active conformation. Similar interactions are also observed for β-lactamase binding sites, generally involving a sulfate anion (22, 24, 56).
Analysis of the KPC-2 structures and comparison of KPC-2 with other class A β-lactamases.
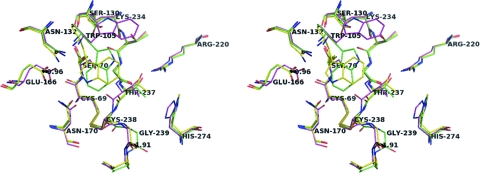
The structures of KPC-2 (1.6 Å) and its C-ter-deleted mutant (1.23 Å) determined in the present study and the 1.85-Å KPC-2 structure recently reported by Ke et al. (26) superimposed well with each other (overall root mean square deviation [RMSD] of 0.42 Å for the α carbons). However, significant displacements mostly affecting elements of the active site could be observed in the superimposition. One of the most marked structural changes was observed at the end of strand β3 at the level of residues Thr237-Gly239, which were found to be shifted outward from the active-site center in our structures, particularly in the deleted form (up to 1.91 Å) (Fig. 4). The very pronounced displacement of the 237-to-239 region in the C-ter-deleted KPC-2 mutant, which reflects the flexibility of the polypeptide chain in this part of the protein, seems to stem from a direct crystallographic contact between Gly239 in a given KPC-2 polypeptide chain and Gly235 in a neighbor chain in the crystal (data not shown). This steric contact is stabilized by a water molecule establishing an H-bond bridge between the main-chain carbonyl oxygen atoms of Cys238 in one polypeptide chain and Thr254 in the adjacent chain (data not shown). Concomitantly to the shift observed at the level of residues 237 to 239, a marked shift in the positioning of the catalytic residue Glu166 was observed in our two structures, by up to 0.96 Å outward from the catalytic center (Fig. 4). Overall, these adjustments corresponded to a significant enlargement of the active site in our structures, with the distance between the catalytic Glu166 residue on one hand and the β3 residue Thr237 on the other being significantly increased in our structures (by up to 1.36 Å) relative to the one reported by Ke et al.
FIG. 4.
Superimposition at the level of the binding cavity of the three KPC-2 structures determined in the present study (KPC-2 in yellow and the C-ter-deleted mutant in green) and by Ke et al. (26) (in magenta). Oxygen and nitrogen atoms are in red and blue, respectively. The distances are indicated by dashed lines and are given in Å.
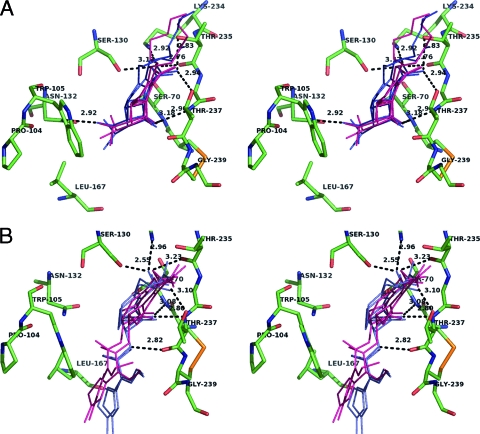
The two KPC-2 structures reported in the present study also readily superimposed on those of the other class A carbapenemases, SME-1 (RMSD = 0.69 Å) and NMC-A (RMSD = 0.97 Å), and showed at the level of the active site the same conformational adjustments that have previously been proposed to be important for carbapenemase activity (26, 50, 54), such as the more shallow conformation of the catalytic active-site Ser70 and the disulfide bond formed between Cys69 and Cys238, as shown in Fig. 4; Cys69 and Cys238 are two residues strictly conserved in class A carbapenemases. Moreover, the positioning of the side chains of the essential amino acid residues in the active site of our KPC-2 structures (Lys73, Ser130, Asn132, Lys234, Thr237) appeared to be similar to that found for the other two carbapenemases (50, 54). Accordingly, docking results obtained with imipenem (Fig. 5A) suggested that this drug fits very well in the KPC-2 binding site. On one side, the 6α-hydroxyethyl group is anchored in the cavity drawn by residues 105, 167, and Asn132, with the latter residue making an H bond with the hydroxyethyl moiety of imipenem. On the other side, the carboxylate moiety of the drug strongly interacts with the conserved active-site residues Ser130, Thr235, Thr237 (H-bonding interactions), and Lys234 (salt bridge interaction) (Fig. 5A), such that the β-lactam carbonyl oxygen is ensconced in the oxyanion hole (from 2.87 to 3.25 Å and from 2.89 to 3.14 Å from the backbone amides of Ser70 and Thr237, respectively, in the examples of docking solution shown in Fig. 5A).
FIG. 5.
(A) Binding cavity of KPC-2 (in green) with imipenem docked into the active site (showed as thin lines). Four docking solutions obtained by using either Gold (in pink) or Autodock (in blue) are represented. H-bonding interactions and the corresponding distances (in Å) are indicated by dotted lines for one of the four docking solutions. (B) Docking of cefotaxime into the active sites of KPC-2 (in green). Four docking solutions for cefotaxime are shown by using either pink lines (Gold solutions) or blue lines (Autodock solutions).
Another striking structural specificity was observed for KPC-2 at the level of the loops at positions 96 to 105 (96-105 loops) which superimposed with a RMSD value of 1.34 Å in our two models, compared to 0.42 Å for the rest of the protein (data not shown), indicating that this region is characterized by a significant flexibility, as confirmed by a local rise of the B factor values for the corresponding atoms (the average B factors were 25 Å2 compared to 11 Å2 for the rest of the polypeptide chain). Interestingly, a proline and a tryptophan specifically occupy positions 104 and 105 of the loop, respectively. These two residues modulate the shape of the entrance of the active site, with the space close to Pro104 being occupied by the side chain of the omega loop residue Leu167, which thus participates directly in the formation of the edge of the binding cavity (Fig. 5). Taken altogether, these data indicate that two structural elements important for the shape of the binding cavity in KPC-2, i.e., the 237-to-239 region at the end of strand β3 and the 96-105 loop facing β3, display a significant degree of freedom and probably represent important determinants of the substrate specificity of KPC-2 for C3G. To investigate this hypothesis further, we used the minimized structure of cefotaxime to carry out docking simulations with the C-ter-deleted KPC-2 structure which corresponds to the most open conformation of the enzyme. The docking simulations made with cefotaxime strongly suggested that the bulky side chain of the aminothiazole moiety binds in the region of Leu167, while the oxyimine moiety of the antibiotic lies on the other side, at the level of the pocket formed by the shifted 237-to-239 region in the C-ter part of strand β3 (Fig. 5B). The adjustment of the positioning of the CTX aminothiazoleoxyme group in this configuration seems to favor tight contacts between the β-lactam carbonyl oxygen of the antibiotic and the backbone amides of Thr237 and Ser70 in the KPC-2 oxyanion hole (from 2.68 to 2.80 Å and from 3.08 to 3.41 Å, respectively, in the docking solutions shown in Fig. 5B). Previous structural studies of the CTX-M-type cefotaximases have suggested that the extended hydrolytic spectra of these enzymes could rely at least in part on (i) specific amino acid complementarities involving most particularly amino acids such as residue 104 and (ii) an increased flexibility in the C-ter part of strand β3 (5, 13). In KPC-2, Pro104 seems to be important in providing additional room in the vicinity of the side chain of the Ω-loop residue Leu167, which thus can adopt a position is favorable to the adjustment of the aminothiazol moiety of CTX in the binding site (Fig. 5B). On the other hand, the fact that the 237-to-239 region in the β3 strand was experimentally shown in the present study to exhibit a high degree of freedom strengthens the idea that an increased mobility at the level of this important secondary structure element also could contribute to the accommodation of C3G in KPC-2.
Conclusions.
In the present report, we describe for the first time the direct transfer of KPC-2 in two strains, E. coli 2138 and E. cloacae 7506, which were recovered from the same patient. The blaKPC-2 gene, which is carried by a large plasmid in a genetic environment nearly identical to that previously reported for blaKPC-2 in other epidemic strains (61, 62), encodes a class A carbapenemase which displays an extremely broad substrate range, including the carbapenems and the broad-spectrum cephalosporins such as cefotaxime, and which is characterized by a combination of remarkable structural features, such as the Pro104-Trp105-Asn132-Leu167 motif and a significant flexibility at the level of the 96-105 loop and the 237-to-239 region in β3. In France, KPC-2 was first described for a K. pneumoniae isolate in 2005, and it had a U.S. origin (37). In the present study, the patient was first hospitalized in Israel before being admitted in our hospital in France, suggesting the transfer of the KPC producer between the two countries. These worrying observations confirm that KPC-2 is a class A carbapenemase having a high interspecies and intercontinental dissemination potential, a specificity that accounts for the fact that this enzyme has spread in the United States (8), Israel, France, China, and Colombia (29, 37, 39, 63) in a relatively short time.
The potential of diffusion of KPC-2 is undoubtedly reinforced by its location on transferable plasmids that can cohabit with other resistance determinants, such as plasmid-encoded extended-spectrum β-lactamase KLUC-2 described for the first time in the present study. This is a very worrisome evolution, since it could lead to bacterial infections that are very difficult to treat because of the high level of resistance to broad-spectrum cephalosporins due to the associated extended-spectrum β-lactamase activities plus the decreased susceptibility to imipenem caused by the carbapenemase activity of KPC-2. Finally, our investigations of the blaKLUC-2 gene, reported here for the first time as being at a plasmid location in an Enterobacteriaceae clinical isolate, clearly confirm that the blaKLU genes from K. ascorbata, K. georgiana, and K. cryocrescens can escape their initial chromosomal location following mobilization events involving ISEcp1 insertion sequence elements (43, 45).
Acknowledgments
We thank Ines Gallay for assistance with the generator at the IBPC. We thank Michel Pirocchi at FIP beamline (ESRF, Grenoble, France) for help with use of the facilities. We thank Xavier Henry, Roland Bismuth, and Liliane Bodin, who contributed to the collection and the identification of the clinical strains. We thank Jeremie Piton for help with Autodock.
This work was supported by the European Community (COBRA, contract LSHM-CT-2003-503335, 6th PCRD).
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. F., D. R. Lonsway, J. K. Rasheed, J. Biddle, B. Jensen, L. K. McDougal, R. B. Carey, A. Thompson, S. Stocker, B. Limbago, and J. B. Patel. 2007. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J. Clin. Microbiol. 45:2723-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthelemy, M., J. Peduzzi, and R. Labia. 1988. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1). Biochem. J. 251:73-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., C. Recule, R. Baraduc, C. Chanal, D. Sirot, C. De Champs, and J. Sirot. 2003. Effect of D240G substitution in a novel ESBL CTX-M-27. J. Antimicrob. Chemother. 52:29-35. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet, R., J. L. Sampaio, R. Labia, C. De Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouthors, A. T., J. Delettre, P. Mugnier, V. Jarlier, and W. Sougakoff. 1999. Site-directed mutagenesis of residues 164, 170, 171, 179, 220, 237 and 242 in PER-1 β-lactamase hydrolysing expanded-spectrum cephalosporins. Protein Eng. 12:313-318. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 9.Bratu, S., D. Landman, M. Alam, E. Tolentino, and J. Quale. 2005. Detection of KPC carbapenem-hydrolyzing enzymes in Enterobacter spp. from Brooklyn. New York. Antimicrob. Agents Chemother. 49:776-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187-217. [Google Scholar]
- 11.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 12.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Y., J. Delmas, J. Sirot, B. Shoichet, and R. Bonnet. 2005. Atomic resolution structures of CTX-M β-lactamases: extended spectrum activities from increased mobility and decreased stability. J. Mol. Biol. 348:349-362. [DOI] [PubMed] [Google Scholar]
- 14.Cornish-Bowden, A. 1995. Analysis of enzyme kinetic data. Oxford University Press, New York, NY.
- 15.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshpande, L. M., R. N. Jones, T. R. Fritsche, and H. S. Sader. 2006. Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program (2000-2004). Microb. Drug Resist. 12:223-230. [DOI] [PubMed] [Google Scholar]
- 17.Deshpande, L. M., P. R. Rhomberg, H. S. Sader, and R. N. Jones. 2006. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999-2005). Diagn. Microbiol. Infect. Dis. 56:367-372. [DOI] [PubMed] [Google Scholar]
- 18.Drusano, G. L., H. Lode, and J. R. Edwards. 2000. Meropenem: clinical response in relation to in vitro susceptibility. Clin. Microbiol. Infect. 6:185-194. [DOI] [PubMed] [Google Scholar]
- 19.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 20.Edwards, R., and D. Greenwood. 1996. Mechanisms responsible for reduced susceptibility to imipenem in Bacteroides fragilis. J. Antimicrob. Chemother. 38:941-951. [DOI] [PubMed] [Google Scholar]
- 21.Hawkey, P. M. 1997. Resistance to carbapenems. J. Med. Microbiol. 46:451-454. [PubMed] [Google Scholar]
- 22.Herzberg, O. 1991. Refined crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2.0 A resolution. J. Mol. Biol. 217:701-719. [DOI] [PubMed] [Google Scholar]
- 23.Hossain, A., M. J. Ferraro, R. M. Pino, R. B. Dew III, E. S. Moland, T. J. Lockhart, K. S. Thomson, R. V. Goering, and N. D. Hanson. 2004. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents Chemother. 48:4438-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelsch, C., L. Mourey, J. M. Masson, and J. P. Samama. 1993. Crystal structure of Escherichia coli TEM1 β-lactamase at 1.8 Å resolution. Proteins 16:364-383. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch, W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 26.Ke, W., C. R. Bethel, J. M. Thomson, R. A. Bonomo, and F. van den Akker. 2007. Crystal structure of KPC-2: insights into carbapenemase activity in class A β-lactamases. Biochemistry 46:5732-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lartigue, M. F., L. Poirel, D. Aubert, and P. Nordmann. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50:1282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of bla(CTX-M) genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 29.Leavitt, A., S. Navon-Venezia, I. Chmelnitsky, M. J. Schwaber, and Y. Carmeli. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livermore, D. M. 1995. Bacterial resistance to carbapenems. Adv. Exp. Med. Biol. 390:25-47. [DOI] [PubMed] [Google Scholar]
- 31.MacKerell, A. D., D. Bashford, M. Bellot, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiorkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586-3616. [DOI] [PubMed] [Google Scholar]
- 32.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mainardi, J. L., P. Mugnier, A. Coutrot, A. Buu-Hoi, E. Collatz, and L. Gutmann. 1997. Carbapenem resistance in a clinical isolate of Citrobacter freundii. Antimicrob. Agents Chemother. 41:2352-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miriagou, V., L. S. Tzouvelekis, S. Rossiter, E. Tzelepi, F. J. Angulo, and J. M. Whichard. 2003. Imipenem resistance in a Salmonella clinical strain due to plasmid-mediated class A carbapenemase KPC-2. Antimicrob. Agents Chemother. 47:1297-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 37.Naas, T., P. Nordmann, G. Vedel, and C. Poyart. 2005. Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob. Agents Chemother. 49:4423-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naas, T., L. Vandel, W. Sougakoff, D. M. Livermore, and P. Nordmann. 1994. Cloning and sequence analysis of the gene for a carbapenem-hydrolyzing class A β-lactamase, Sme-1, from Serratia marcescens S6. Antimicrob. Agents Chemother. 38:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Navon-Venezia, S., I. Chmelnitsky, A. Leavitt, M. J. Schwaber, D. Schwartz, and Y. Carmeli. 2006. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob. Agents Chemother. 50:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nissink, J. W., C. Murray, M. Hartshorn, M. L. Verdonk, J. C. Cole, and R. Taylor. 2002. A new test set for validating predictions of protein-ligand interaction. Proteins 49:457-471. [DOI] [PubMed] [Google Scholar]
- 41.Novak, S., and S. Munro. Routine tests in many clinical laboratories, p. 5.5.8. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, 2nd ed. ASM Press, Washington, DC.
- 42.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson, A. B., M. Silverman, D. A. Boyd, A. McGeer, B. M. Willey, V. Pong-Porter, N. Daneman, and M. R. Mulvey. 2005. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob. Agents Chemother. 49:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podglajen, I., J. Breuil, and E. Collatz. 1994. Insertion of a novel DNA sequence, 1S1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105-114. [DOI] [PubMed] [Google Scholar]
- 45.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raimondi, A., A. Traverso, and H. Nikaido. 1991. Imipenem- and meropenem-resistant mutants of Enterobacter cloacae and Proteus rettgeri lack porins. Antimicrob. Agents Chemother. 35:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth, M., P. Carpentier, O. Kaikati, J. Joly, P. Charrault, M. Pirocchi, R. Kahn, E. Fanchon, L. Jacquamet, F. Borel, A. Bertoni, P. Israel-Gouy, and J. L. Ferrer. 2002. FIP: a highly automated beamline for multiwavelength anomalous diffraction experiments. Acta Crystallogr. D 58:805-814. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Sougakoff, W., G. L'Hermite, L. Pernot, T. Naas, V. Guillet, P. Nordmann, V. Jarlier, and J. Delettre. 2002. Structure of the imipenem-hydrolyzing class A β-lactamase SME-1 from Serratia marcescens. Acta Crystallogr. D 58:267-274. [DOI] [PubMed] [Google Scholar]
- 51.Sougakoff, W., T. Naas, P. Nordmann, E. Collatz, and V. Jarlier. 1999. Role of ser-237 in the substrate specificity of the carbapenem-hydrolyzing class A β-lactamase Sme-1. Biochim. Biophys. Acta 1433:153-158. [DOI] [PubMed] [Google Scholar]
- 52.Strynadka, N. C., S. E. Jensen, P. M. Alzari, and M. N. James. 1996. A potent new mode of β-lactamase inhibition revealed by the 1.7 Å X-ray crystallographic structure of the TEM-1-BLIP complex. Nat. Struct. Biol. 3:290-297. [DOI] [PubMed] [Google Scholar]
- 53.Sutcliffe, J. G. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. USA 75:3737-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaren, P., L. Maveyraud, X. Raquet, S. Cabantous, C. Duez, J. D. Pedelacq, S. Mariotte-Boyer, L. Mourey, R. Labia, M. H. Nicolas-Chanoine, P. Nordmann, J. M. Frere, and J. P. Samama. 1998. X-ray analysis of the NMC-A β-lactamase at 1.64-Å resolution, a class A carbapenemase with broad substrate specificity. J. Biol. Chem. 273:26714-26721. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi, S., and Y. Nagano. 1984. Rapid procedure for isolation of plasmid DNA and application to epidemiological analysis. J. Clin. Microbiol. 20:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tranier, S., A. T. Bouthors, L. Maveyraud, V. Guillet, W. Sougakoff, and J. P. Samama. 2000. The high resolution crystal structure for class A β-lactamase PER-1 reveals the bases for its increase in breadth of activity. J. Biol. Chem. 275:28075-28082. [DOI] [PubMed] [Google Scholar]
- 57.Vagin, A. A., R. A. Steiner, A. A. Lebedev, L. Potterton, S. McNicholas, F. Long, and G. N. Murshudov. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60:2184-2195. [DOI] [PubMed] [Google Scholar]
- 58.Verdonk, M. L., J. C. Cole, M. J. Hartshorn, C. W. Murray, and R. D. Taylor. 2003. Improved protein-ligand docking using GOLD. Proteins 52:609-623. [DOI] [PubMed] [Google Scholar]
- 59.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, and J. P. Quinn. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walther-Rasmussen, J., and N. Hoiby. 2007. Class A carbapenemases. J. Antimicrob. Chemother. 60:470-482. [DOI] [PubMed] [Google Scholar]
- 61.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yigit, H., A. M. Queenan, J. K. Rasheed, J. W. Biddle, A. Domenech-Sanchez, S. Alberti, K. Bush, and F. C. Tenover. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob. Agents Chemother. 47:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, R., H. W. Zhou, J. C. Cai, and G. X. Chen. 2007. Plasmid-mediated carbapenem-hydrolysing β-lactamase KPC-2 in carbapenem-resistant Serratia marcescens isolates from Hangzhou, China. J. Antimicrob. Chemother. 59:574-576. [DOI] [PubMed] [Google Scholar]