Abstract
The ISCR elements ISCR3, ISCR4, ISCR5, ISCR14, and ISCR16 all share a percent G+C of 68 to 69%. They also share between 75% and 97% identity in their transposase open reading frames. Furthermore, with the exception of ISCR5, they are all found adjacent to sections of groEL that display the highest identity to the same gene from Xanthomonas spp. The combined information is consistent with the descent from an ancestral ISCR element in a Xanthomonas-like organism.
ISCR elements are an unusual group of insertion sequences that have similarities to the IS91 family in both structure and function (7, 8). At present there are 16 members of the ISCR family (http://www.cardiff.ac.uk/medic/aboutus/departments/medicalmicrobiology/genetics/iscr/iscr_elements.html), and all are found adjacent to genes that are not the normal complement of the host cell, the vast majority being antibiotic resistance genes (8). They are thus implicated in the acquisition of these genes by the host bacterium via plasmids. Previously we noted that ISCR elements vary in percent G+C from 54 to 69%, indicating different origins (8). We also noted that both ISCR4 and ISCR3 are found adjacent to partial groEL genes (8). Here we extend this analysis to new members of the ISCR family, i.e., ISCR14 and ISCR16, and provide an explanation of the evolution of the ISCR3 group of ISCR elements.
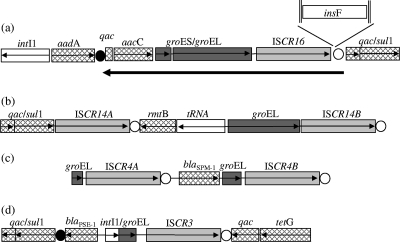
Searches of the EMBL databases at EMBL-EBI (using the FASTA protein similarity search at http://www.ebi.ac.uk/fasta33) with ISCR elements ISCR5 and ISCR3 revealed high identities with several recent additions to the database. These included identical sequences (Fig. 1a) found in the following two separate plasmids: pSN254 from Salmonella enterica serovar Newport (GenBank accession number CP000604) and pAPEC-01-R from avian pathogenic Escherichia coli (GenBank accession number DQ517526). This new ISCR element has been named ISCR16. A further sequence, that of ISCR14 (Fig. 1b), has been found both in a panresistant Pseudomonas aeruginosa isolate from Brazil (GenBank accession number DQ914960) (2) and in a Klebsiella pneumoniae isolate (GenBank accession number EU269034). The genetic loci of these new ISCR elements together with the genetic loci of the closely related ISCR elements ISCR3 and ISCR4 are drawn for comparison, shown in Fig. 1.
FIG. 1.
Schematic of the genetic loci of ISCR elements found adjacent to groEL sequences: (a) ISCR16; (b) ISCR14; (c) ISCR4; (d) ISCR3. Open reading frames are depicted as open boxes with arrows indicating the direction of transcription. Open reading frames encoding resistance are hatched. groEL genes are dark gray and ISCR elements are light gray. Filled circles depict the 59 base elements of the various gene cassettes. The origin of replication of the various ISCR elements are shown as open circles and the inverted repeats found at the ends of the insertion sequence insF are shown as vertical parallel lines. The large arrow underneath panel a shows the amount of DNA accumulated upstream of ISCR16 due to subsequent replicative transposition events. The sequences of ISCR14A and ISCR14B are identical, as are the sequences of ISCR4A and ISCR4B.
Interestingly, groEL gene sections of various lengths have now been found upstream of four different ISCR elements (Fig. 1). While ISCR16 has complete copies of groEL and groES immediately upstream, ISCR14, ISCR4, and ISCR3 have 5′ truncated versions of the groEL gene which are missing 189 bp, 1,266 bp, and 1,287 bp, respectively. Furthermore, all groEL genes have the highest identity with the following various Xanthomonas groEL genes: 86.6% and 85.9% identity to groEL and groES genes over 1,700 bp from Xanthomonas axonopodis pv. citri and Xanthomonas campestris pv. citri for ISCR16, respectively; 86.9% and 86.7% identity over 1,450 bp to groEL genes from Xanthomonas axonopodis pv. citri and Xanthomonas campestris pv. vesicatoria for ISCR14, respectively; 92.6% and 92% identity over 160 bp to groEL from Xanthomonas axonopodis pv. citri and Xanthomonas campestris pv. citri for ISCR4, respectively; and 83.1% and 79.3% identity over 640 bp to groEL genes from Xanthomonas campestris pv. vesicatoria and Xanthomonas campestris pv. citri for ISCR3, respectively. Importantly, groEL genes are found only upstream of the ISCR transposase gene, adjacent to the terminus of each ISCR element. The other end of the ISCR element includes the oriIS sequence, and therefore, replicative transposition of each ISCR element, as shown in Fig. 1, proceeds from the right-hand (oriIS) end to the left-hand (terIS) end of each element. ISCR elements function by initially transposing next to a target gene or section of DNA. In a second or subsequent transposition event(s), the adjacent gene or genes are cotransposed. For the related element IS1294, this cotransposition of adjacent DNA happens at approximately 10% of each transposition event (6) and the sequence cotransposed is always adjacent to the terIS of the element. Where subsequent replicative transposition events mobilize larger sections at each movement event, they have the effect of accumulating a sequence of a different origin at the terIS end of the element. Therefore, analysis of this sequence can provide a history of the movement of the ISCR element. For example, immediately upstream of ISCR16 are complete groES and groEL genes that share most identity with the same genes from various Xanthomonas species. Further upstream is an aacC gene that is not in the form of a gene cassette, i.e., it is not of integron origin, as well as a small section of qacEΔ1 (110 bp) and then an aadA1 gene cassette, followed by an integrase gene (Fig. 1a). Therefore, the likely history of this element is that once it was in a position adjacent to groEL and groES genes in a Xanthomonas-like organism, a second transposition event moved the groEL and groES genes next to an aacC gene, a qacEΔ1 gene, and subsequently into another integron adjacent to an aadA1 gene, with each transposition event having the effect of accumulating additional DNA sequences. The final insertion adjacent to the aadA1 gene is consistent both with replicative transposition events as described above or homologous recombination as suggested previously (4). However, these elements can also transpose just their own DNA, as seen in Fig. 1b for ISCR14A, or a smaller section of DNA found adjacent to them, as can be seen in Fig. 1b, c, and d where truncated sections of groEL have been mobilized.
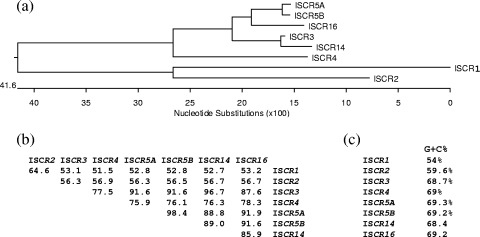
A phylogenetic tree based on an alignment of these ISCR transposase sequences with other known ISCR transposases using DNAStar software reveals some more-interesting observations regarding the possible evolution of these ISCR elements. The alignment shows that ISCR3, ISCR4, ISCR5A&B, ISCR14, and ISCR16 are closely related, with identities ranging from 76% to 97%, while ISCR1 and ISCR2 are significantly different (Fig. 2a and b). Furthermore, while ISCR3-5, ISCR14, and ISCR16 all share a percent G+C of 68% to 69%, ISCR1 and ISCR2 have a percent G+C of 54% and 59%, respectively (Fig. 2).
FIG. 2.
Comparison of ISCR elements. (a) Phylogenetic tree of ISCR3 group elements with ISCR1 and ISCR2. (b) Sequence identity comparisons, based on a Clustal alignment with the PAM250 matrix prepared using Lasergene DNAStar software. Alignments were undertaken on the available complete ISCR transposase sequences. (c) Percent G+C comparisons of the DNA sequences of various complete ISCR elements.
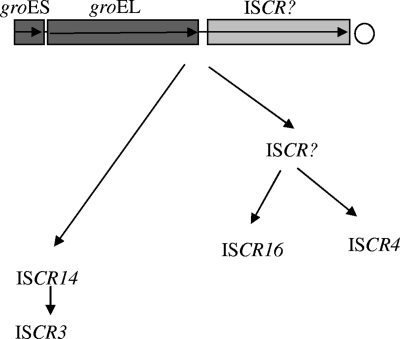
A further alignment of the sequence found between the stop codon of groEL and the start codon of the various ISCR transposases reveals that the ISCR3- and ISCR14-associated sequences are identical. Their transposases also share 96.7% identity, which suggests that ISCR3 is a direct descendant of ISCR14 or that they share a recent common ancestor. The sequences found between groEL and ISCR4 and between groEL and ISCR16 are both approximately 35 bp shorter than those of the respective sequences from ISCR3 and ISCR14 and are only 75% identical (Fig. 3). This suggests that ISCR16 and ISCR4 also have a common ancestor but that it is not as recent as ISCR3 and ISCR14. The data therefore indicate that the ISCR3 group of ISCR elements has originated from an ancestral ISCR element that was at one time found adjacent to a groES-groEL operon in a Xanthomonas-like organism. A hypothetical model of the evolution of the ISCR3 group of ISCR elements is shown in Fig. 3.
FIG. 3.
Schematic of the hypothetical evolution of ISCR3 group elements.
The sequences of the ISCR3 group of ISCR elements have diverged nearly 25% since their original ancestor adjacent to groEL. This is also mirrored by a similar divergence in sequence of the associated groEL genes of between 74 and 98% (data not shown). The fact that ISCR14 and ISCR4 were discovered in the same Pseudomonas isolate (3, 9) suggests that homologous recombination plays a role in the divergence of ISCR and groEL sequences; this is especially due to the fact that groEL sequences are well conserved.
Finally, it is interesting to note that groEL sequences show the highest identity to Xanthomonas spp. often of the pathovar citri, a fruit pathogen. ISCR16 was found on plasmids in an avian pathogenic E. coli isolate from a turkey in Iowa (5) as well as a similar plasmid in Salmonella enterica serovar Newport (10). ISCR3 is found in Salmonella enterica phage type DT104 and several other Salmonella enterica pathovars. ISCR14 and ISCR4 have been found in a panresistant strain of P. aeruginosa causing serious infection control problems in Brazil (1, 3). Thus, it appears that the ISCR3 group of ISCR elements is mobilizing genes from environmental organisms to clinically relevant pathogens. One possible route could be small birds feeding both on fruit and in turkey sheds and then from Salmonella strains of avian origin into prevalent human-pathogenic Salmonella species such as Salmonella enterica phage type DT104.
Acknowledgments
This work was funded by the Wellcome Trust project grant no. [084627/Wellcome Trust].
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Castanheira, M., T. R. Fritsche, H. S. Sader, R. N. Jones, Y. Doi, D. de Oliveira Garcia, and D. L. Paterson. 2008. RmtD 16S RNA methylase in epidemiologically unrelated SPM-1-producing Pseudomonas aeruginosa isolates from Brazil. Antimicrob. Agents Chemother. 52:1587-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doi, Y., D. de Oliveira Garcia, J. Adams, and D. L. Paterson. 2007. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob. Agents Chemother. 51:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doi, Y., A. C. Ghilardi, J. Adams, D. de Oliveira Garcia, and D. L. Paterson. 2007. High prevalence of metallo-β-lactamase and 16S rRNA methylase coproduction among imipenem-resistant Pseudomonas aeruginosa isolates in Brazil. Antimicrob. Agents Chemother. 51:3388-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall, R. M. 2007. Antibiotic resistance gene cluster of pAPEC-O1-R. Antimicrob. Agents Chemother. 51:3461-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson, T. J., Y. M. Wannemeuhler, J. A. Scaccianoce, S. J. Johnson, and L. K. Nolan. 2006. Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3929-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavakoli, N., A. Comanducci, H. M. Dodd, M. C. Lett, B. Albiger, and P. Bennett. 2000. IS1294, a DNA element that transposes by RC transposition. Plasmid 44:66-84. [DOI] [PubMed] [Google Scholar]
- 7.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 58:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toleman, M. A., D. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Global perspective of CR elements associated with metallo-β-lactamase producing isolates: report from the SENTRY surveillance program, p. 110. Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 10.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. Leclerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS ONE 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]