Abstract
The antibiotic hygromycin A (HA) binds to the 50S ribosomal subunit and inhibits protein synthesis in gram-positive and gram-negative bacteria. The HA biosynthetic gene cluster in Streptomyces hygroscopicus NRRL 2388 contains 29 open reading frames, which have been assigned putative roles in biosynthesis, pathway regulation, and self-resistance. The hyg21 gene encodes an O-phosphotransferase with a proposed role in self-resistance. We observed that insertional inactivation of hyg21 in S. hygroscopicus leads to a greater than 90% decrease in HA production. The wild type and the hyg21 mutant were comparably resistant to HA. Using Escherichia coli as a heterologous host, we expressed and purified Hyg21. Kinetic analyses revealed that the recombinant protein catalyzes phosphorylation of HA (Km = 30 ± 4 μM) at the C-2‴ position of the fucofuranose ring in the presence of ATP (Km = 200 ± 20 μM) or GTP (Km = 350 ± 60 μM) with a kcat of 2.2 ± 0.1 min−1. The phosphorylated HA is inactive against HA-sensitive ΔtolC E. coli and Streptomyces lividans. Hyg21 also phosphorylates methoxyhygromycin A and desmethylenehygromycin A with kcat and Km values similar to those observed with HA. Phosphorylation of the naturally occurring isomers of 5‴-dihydrohygromycin A and 5‴-dihydromethoxyhygromycin A was about 12 times slower than for the corresponding non-natural isomers. These studies demonstrate that Hyg21 is an O-phosphotransferase with broad substrate specificity, tolerating changes in the aminocyclitol moiety more than in the fucofuranose moiety, and that phosphorylation by Hyg21 is one of several possible mechanisms of self-resistance in S. hygroscopicus NRRL 2388.
Microorganisms that produce antibiotics are also equipped with resistance mechanisms to protect themselves from the toxic effects of their own molecules (4, 11). Three major mechanisms of resistance have been described in antibiotic-producing Streptomyces spp.: (i) antibiotic inactivation by chemical modification, (ii) target site modification, and (iii) antibiotic efflux by transporter proteins. The genes responsible for self-resistance are commonly present as components of antibiotic biosynthetic gene clusters and are often coregulated with them (14). A bacterium may make use of more than one of these intrinsic resistance strategies to prevent autotoxicity. Self-resistance by enzymatic modification of an antibiotic is primarily mediated by O phosphorylation of hydroxyl groups and N acetylation of amino groups and is quite widely seen in aminoglycoside producers (4). These bacteria often doubly modify the antibiotic by phosphorylation and acetylation for optimal resistance, as in the neomycin producer Streptomyces fradiae (20) and the paromomycin producer Streptomyces rimosus (17). Inactivation by O glycosylation has been reported in producers of macrolides such as oleandomycin (21), spiramycin (6), and pikromycin (25). Modification can occur on the final product but is also observed on pathway intermediates/precursors during antibiotic biosynthesis. For example, strA in the streptomycin biosynthesis gene cluster encodes streptomycin-6-phosphotransferase, which acts on streptidine, an intermediate in the biosynthetic pathway, and produces an inactive, phosphorylated derivative (15, 22). The remaining biosynthetic pathway utilizes this phosphorylated precursor and leads to the production of 6′-phosphorylstreptomycin. An extracellular phosphatase is responsible for the formation of the active streptomycin (13).
Antibiotic inactivation by O phosphorylation is the second most prevalent mechanism, after N acetylation, by which many pathogenic bacteria have gained resistance to a large number of drugs, especially the aminoglycosides (19). Several O-phosphotransferases from clinical isolates have been overexpressed, purified, and characterized (24), and it has been proposed that the pathogens must have acquired these antibiotic inactivating kinases, as well as the other resistance genes, from antibiotic producers themselves by horizontal gene transfer (10, 11). Consequently, an understanding of the mechanisms of self-resistance in antibiotic-producing microorganisms is an important step in gaining insights into, and thereby combating, acquired resistance in pathogenic strains.
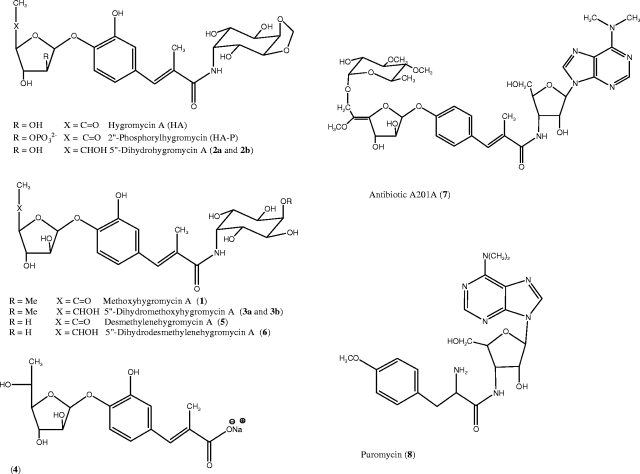
Streptomyces hygroscopicus NRRL 2388 produces the antibiotics hygromycin A (HA) and methoxyhygromycin A (compound 1) (Fig. 1). These antibiotics are structurally unrelated to the more widely known hygromycin B (9). The antimicrobial activity of HA arises from binding to the 50S ribosomal subunit and inhibition of the peptidyl transferase reaction of protein synthesis (7, 9). The structure of HA is composed of three distinct moieties, a 5-dehydro-α-l-fucofuranose (A subunit) attached by a glycosidic bond to (E)-3-(3,4-dihydroxyphenyl)-2-methylacrylic acid (B subunit), which in turn is attached by a peptide bond to 2L-2-amino-2-deoxy-4,5-O-methylene-neo-inositol (C subunit). Feeding experiments with precursor compounds revealed that the biosynthetic route to HA has three convergent branches made of a combination of pentose sugar (A subunit), polyketide (B subunit), and aminocyclitol (C subunit) syntheses (9). The biosynthetic gene cluster of HA, which has recently been reported (9, 16), is 31.5 kb long and comprises 29 open reading frames (ORFs). Putative functions have been assigned to 26 of the hyg genes by sequence analysis. Hyg26 has been shown to be responsible for generation of the 5-dehydro-α-l-fucofuranose moiety by oxidation of α-l-fucofuranose. A Δhyg26 mutant has been shown to produce 5‴-dihydrohygromycin A (compound 2a), 5‴-dihydromethoxyhygromycin A (compound 3a), and (E)-3-(3-hydroxy-4-O-α-fucofuranosylphenyl)-2-methylacrylic acid (compound 4, 5‴-dihydrohygromycin A lacking the aminocyclitol) (see Fig. 1 for structures) (16).
FIG. 1.
Structures of HA and related compounds.
With regard to self-resistance, analysis of the hyg gene cluster suggests that all three mechanisms outlined above may operate in S. hygroscopicus NRRL 2388. The hyg21 gene encodes a putative phosphotransferase that may inactivate HA by O phosphorylation. The hyg6 and hyg29 are methyltransferase homologs, and one of these may confer resistance by methylation of rRNA, the target for HA, while the other is presumed to be responsible for introduction of the methylene group in the aminocyclitol moiety. The hyg19 and hyg28 genes putatively encode a transmembrane protein and an ABC transporter, respectively, which may play a role in antibiotic efflux. There has been no experimental verification of these proposed roles for Hyg19, Hyg21, and Hyg28.
In the present study, we report a genetic and biochemical investigation of hyg21 and its gene product. We have shown that hyg21 is not an absolute requirement for hygromycin biosynthesis and that Hyg21 is a phosphotransferase that catalyzes the transfer of the γ-phosphoryl group of ATP to the 2‴-hydroxyl group of HA, abolishing its antibacterial property. We have also determined the steady-state kinetic parameters for turnover of HA and its analogs and have shown that Hyg21 can phosphorylate a range of substrates (Fig. 1) bearing a fucofuranose moiety, including compounds 1, 2, 3, 4, desmethylenehygromycin A (compound 5), 5‴-dihydrodesmethylenehygromycin A (compound 6), and antibiotic A201A (compound 7).
MATERIALS AND METHODS
Antibiotics and chemicals.
All antibiotics and chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise indicated. HA was kindly supplied by Pfizer, Inc. Analogs of HA and compounds 1, 2a, 3a, and 4 (Fig. 1) were isolated from the wild type and a Δhyg26 blocked mutant strain of S. hygroscopicus (16). Compounds 5 and 6 were isolated from a hyg6 deletion mutant (unpublished data). A diastereomeric mixture of 5‴-dihydrohygromycin A (compounds 2a and 2b) was prepared by reduction of HA as described previously (16). A diastereomeric mixture of 5‴-dihydromethoxyhygromycin A (compounds 3a and 3b) was prepared from methoxyhygromycin A (compound 1) in the same way. Antibiotic A201A was a generous gift from A. Jimenez, Centro de Biologia Molecular Severo Ochoa, Madrid, Spain. [2-3H]myo-inositol (20.0 Ci/mmol) was procured from Moravek Biochemicals (Brea, CA). Primers for PCR were obtained from Integrated DNA Technologies (Coralville, IA). Enzymes for DNA manipulations were purchased from New England Biolabs (Beverly, MA). The prestained protein molecular weight markers in the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis were obtained from Bio-Rad (Hercules, CA).
Bacterial strains, plasmids, and culture conditions.
Cosmid 17E3, based upon the SuperCos1 vector, was the source for hyg21 gene and has been described previously (16). The pCR4-TOPO vector and One-Shot Mach1-T1 chemically competent Escherichia coli cells from Invitrogen (Carlsbad, CA) were used for cloning the hyg21 PCR product. The pET-15b vector from EMD Biosciences (San Diego, CA) was used for the expression of N-terminal His-tagged Hyg21 protein. The pET-15b vector with 0.5-kb hyg21 ORF was designated pET15b-hyg21. E. coli BL21-CodonPlus (DE3), obtained from Stratagene (La Jolla, CA), was the expression host for His-tagged Hyg21 protein. E. coli BW25113/pIJ790 (a strain with the λ-RED recombination plasmid), E. coli DH5α/pIJ773 [a strain with the aac(3)IV apramycin resistance cassette plasmid], and E. coli ET12567/pUZ8002 (a nonmethylating plasmid donor strain for intergeneric conjugation) have been described previously (8) and were provided by the John Innes Institute (United Kingdom). S. hygroscopicus NRRL 2388 is the wild-type HA producer strain used to generate the Δhyg21 mutant in which hyg21 was replaced by the apramycin resistance cassette. The E. coli ΔtolC strain was procured from the E. coli Genetic Stock Center at Yale University. E. coli strains were normally grown at 37°C in Luria-Bertani (LB) medium supplemented with the required antibiotics. S. hygroscopicus wild-type and deletion strains were propagated on ISP2 medium (0.4% yeast extract, 1.0% malt extract, 0.4% dextrose, 2.0% agar [pH 7.2]) at 30°C. Ampicillin (100 μg/ml, final concentration), apramycin (50 μg/ml), carbenicillin (100 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (50 μg/ml) were used as required.
DNA manipulations and analyses.
Plasmid DNA was isolated by using a QIAprep Spin Miniprep kit from Qiagen (Valencia, CA). DNA fragments from agarose gels were isolated by using Qiagen's QIAquick gel extraction kit. The QIAquick PCR purification kit was used for cleaning up the PCR product. Genomic DNA from Streptomyces strains was obtained by using the Wizard Genomic DNA purification kit from Promega (Madison, WI). Automated DNA sequencing was performed at the DNA core facilities at Virginia Commonwealth University and Oregon Health Sciences University. The DNA sequences were analyzed with Accelrys DS Gene software. The Hyg21 amino acid sequence was compared to sequences in the public domain by using the BLASTP program at the NCBI server (1). A motif search was carried out against the PROSITE database at the Expasy proteomics server (12).
Cloning of hyg21 and construction of the expression plasmid.
The 552-bp hyg21 gene was amplified from cosmid 17E3 by using the GC-Rich PCR system from Roche Diagnostics according to the manufacturer's instructions. Primers (5′-CATATGCCGGAAACTTCGTTCCAGGGC-3′ and 5′-GGATCCTCAGCCGTTCGCCAGGAT-3′) were designed to create an NdeI restriction site (boldface) at the initiation codon and a BamHI restriction site (in boldface) 3′ to the termination codon. Amplification was performed with a GeneAmp PCR System 2700 (Applied Biosystems) using the following conditions: initial denaturation at 95°C for 5 min, 30 cycles of amplification (45-s denaturation at 95°C, 45-s annealing at 58°C, and 45-s extension at 72°C), and a final 7-min extension at 72°C. The PCR product was cloned into pCR4-TOPO vector and was sequenced to ensure that no mutations occurred during amplification. The hyg21 from the recombinant TOPO vector was then subcloned into the NdeI and BamHI restriction sites of pET15b. The resulting pET15b-hyg21 plasmid was introduced into the E. coli BL21-CodonPlus (DE3) expression host by transformation.
Expression and purification of His-tagged Hyg21 from E. coli.
E. coli BL21 cells harboring the pET15b-hyg21 plasmid were grown overnight in LB medium supplemented with 100 μg of ampicillin/ml and 50 μg of chloramphenicol/ml. A 1,000-ml portion of LB broth plus ampicillin and chloramphenicol, divided into two 4-liter fermentation flasks, was inoculated to 5% (vol/vol) with an overnight BL21 seed culture. The cells were allowed to grow at 37°C for ∼2 h until an A600 of 0.6 was reached. Expression of His-tagged protein was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), followed by incubation at 30°C for 5 h. The cells were harvested by centrifugation and resuspended in 25 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). His-tagged protein was purified under native conditions following the procedure outlined in Qiagen's Ni-NTA spin protocol. All of the protein purification steps were carried out at 4°C. Cells were disrupted by sonication, and the soluble fraction was applied to 1 ml of Ni-NTA agarose preequilibrated with lysis buffer. The column was washed with 50 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]), and the protein was eluted with elution buffer containing 250 mM imidazole in fractions of 500 μl. Each fraction was analyzed by SDS-PAGE using a 13% acrylamide gel for the presence of Hyg21. The eluted protein was dialyzed for 14 h against a buffer containing 50 mM Tris-HCl (pH 7.5), 20% glycerol and 5 mM 2-mercaptoethanol, with a replacement with fresh buffer after 4 h. The protein concentration was determined by Bio-Rad protein assay using bovine serum albumin as standard, and 50-μl aliquots of the purified protein were stored at −80°C until further use.
Phosphorylation assay conditions.
Initial enzyme assays were performed with ∼10 μg of purified enzyme in a 100-μl reaction mixture of 50 mM Tris-HCl (pH 7.5), 5 mM 2-mercaptoethanol, 10 mM MgCl2, 200 μM substrate, and 1 mM ATP. The reaction mixture was incubated at 30°C for 2 h and frozen at −20°C. Hyg21 activity was studied by reverse-phase high-pressure liquid chromatography (HPLC) analysis by monitoring the appearance of phosphorylated product, which had a shorter retention time than the substrate. For kinetics studies, the reaction volume was scaled up to 600 μl with ∼11 μg of enzyme. The assays were carried out for 15 min, during which phosphorylation activity was linear. The ATP concentration was fixed at 400 μM, and the antibiotic concentration was varied from 5 to 100 μM. To determine kinetic parameters for ATP and GTP, the concentrations of these were varied, and the concentration of HA was fixed at 90 μM. In all cases, the samples were analyzed by HPLC, and the amount of product formed was determined from its peak area using an HA standard curve as reference. The rate of phosphorylation was reported as the nanomoles of product formed per minute. The data were plotted in GraFit 4 to determine the kinetic parameters. The pH profile was obtained using 100 μM HA and 1,000 μM ATP at 30°C for 15 min in the following buffers: 50 mM potassium acetate, pH 5; 50 mM Tris-maleate, pH 6 or 6.5; and 50 mM Tris-HCl, pH 7, 7.5, 8, or 9. The effect of temperature on Hyg21 activity was checked using 100 μM HA and 1,000 μM ATP in 50 mM Tris-HCl (pH 7.5) at room temperature (23°C) and at 30, 37, and 50°C in a 15-min assay.
Purification and characterization of HA-P.
Ten reaction mixtures (5 ml each) with a final composition of 200 μM HA, 1,000 μM ATP, 10 mM MgCl2, 5 mM 2-mercaptoethanol, and 0.5 mg of enzyme in 50 mM Tris-HCl (pH 7.5) were incubated overnight at 30°C. The assay mixtures were pooled and filtered by using Millex-LCR 0.45-μm-pore-size filter unit from Millipore, and the phosphorylated HA (HA-P) was purified by semipreparative reversed-phase HPLC. Fractions containing the purified product were pooled and dried by rotary evaporation. The dried sample was dissolved in deuterium oxide for structural elucidation by nuclear magnetic resonance (NMR) analysis. 1H NMR spectra were recorded on Nicolet NM-500 MHz (modified with a Tecmag Libra interface) instruments calibrated using residual undeuterated solvent as an internal reference. Two-dimensional 1H correlation spectra were recorded on a Bruker AMX-400 NMR spectrometer. A standard HA sample was run under identical conditions for comparison. Coupling constants (J) were expressed in hertz. Abbreviations for multiplicities are as follows: s, singlet; d, doublet; t, triplet; q, quartet; and m, multiplet.
HA-P in D2O.
Values for HA-P in D2O were as follows: 1H NMR (500 MHz, CDCl3) δ 7.34 (d, J = 8.5 Hz, 1H), 7.16 (s, 1H), 7.08 (s, 1H), 7.06 (s, 1H), 5.95 (d, J = 2.5 Hz, 1H), 5.31 (s, 1H), 4.97 (s, 1H), 4.69 (m, 1H), 4.63 (t, J = 4.5 Hz, 1H), 4.58 (d, J = 6.5 Hz, 1H), 4.36-4.31 (m, 2H), 4.20 (dd, J = 3.5, 9.5 Hz, 1H), 4.08 (dd, J = 5.0, 9.5 Hz, 1H), 3.95 (t, J = 4.5 Hz, 1H), 2.25 (s, 3H), and 2.12 (s, 3H).
HA in D2O.
Values for HA in D2O were as follows: 1H NMR (500 MHz, CDCl3) δ 7.23 (d, J = 8.5 Hz, 1H), 7.10 (s, 1H), 7.03 (s, 1H), 7.01 (s, 1H), 5.82 (d, J = 4.5 Hz, 1H), 5.25 (s, 1H), 4.92 (s, 1H), 4.73 (m, 1H), 4.58 (t, J = 4.5 Hz, 1H), 4.50-4.46 (m, 1H), 4.37-4.35 (m, 1H), 4.31-4.25 (m, 2H), 4.14 (dd, J = 3.5, 9.5 Hz, 1H), 4.02 (dd, J = 5.5, 9.5 Hz, 1H), 3.90 (dd, J = 3.5, 6.0 Hz, 1H), 2.15 (s, 3H), and 2.07 (d, J = 1.5 Hz, 3H).
HPLC and LC-mass spectrometry (MS) conditions.
Reversed-phase HPLC analyses of the phosphorylation assays and the fermentation broths were carried out by using an Agilent 1100 Series HPLC system with either an analytical 5-μm Eclipse XDB-C18 column (4.6 by 150 mm) or a semipreparative 5-μm Phenomenex C18 column (250 by 10 mm). The solvents used were A (10% methanol, 90% water, 0.05% formic acid) and B (90% methanol, 10% water, 0.05% formic acid). Gradient conditions for the analytical column were 100% A for 2 min, 100 to 36% A for 16 min, 36 to 0% A for 2 min, 0% A for 8 min, 0 to 100% A for 2 min, and 100% A for 10 min at a flow rate of 1 ml/min. Gradient conditions for the semipreparative column were 100% A for 2 min, 100 to 44% A for 16 min, 44 to 0% A for 2 min, 0% A for 6 min, 0 to 100% A for 1 min, and 100% A for 5 min at a flow rate of 3 ml/min. HA and related compounds were detected at 272 nm. Analysis of radiolabeled HA production by feeding [2-3H]myo-inositol was carried out on a Beckman System Gold instrument equipped with a radiodetector. MS characterization was performed by using a Perkin-Elmer SCIEX API 2000 mass spectrometer.
Insertional inactivation of hyg21.
The hyg21 gene of S. hygroscopicus NRRL 2388 was replaced by the apramycin resistance cassette using the PCR-targeted Streptomyces gene replacement method (8). The Expand High Fidelity kit from Roche was used for PCR amplification of apramycin resistance cassette from pIJ773 with the primers hyg21_KO_Forw (5′-ACCGTTGACCGAGAATGGGTAAAGGAGCAGAAAACAATGATTCCGGGGATCCGTCGACC-3′) and hyg21_KO_Revr (5′-TCCACCGGGCCCCGCGCGGACCCGCCGCCCGGCGGATCATGTAGGCTGGAGCTGCTTC-3′) (boldfacing indicates homology to the nucleotide sequence flanking hyg21, while the italicized portions of the sequences are homologous to pIJ773). Gene replacement was carried out first in cosmid 17E3 and then in the genome of S. hygroscopicus. Gene replacement in the resulting mutant strain, Δhyg21, was confirmed by PCR amplification of chromosomal DNA using the outer primers, hyg21_Out_Forw (5′-CCGCTTTCCGTCACAACGGAT-3′) and hyg21_Out_Revr (5′-CATGCGCACCTGGCTGAC-3′) and by sequencing the PCR product.
Fermentation conditions for S. hygroscopicus strains.
S. hygroscopicus NRRL 2388 and Δhyg21 mutant strains were cultivated as reported earlier (9). To obtain cell lysates, mycelium was collected from 100 ml of culture, suspended in 5 ml of methanol, and shaken on an orbital shaker at room temperature for 6 h. The suspension was then subjected to a brief sonication, and cell debris was removed by centrifugation.
Feeding of 2-3H-labeled myo-inositol.
Feeding studies using radiolabeled precursors were carried out in 5 ml of production medium inoculated with 200 μl of seed culture. [2-3H]myo-inositol was added after 24 h to a final concentration of 0.4 μCi/ml.
Disk diffusion assays to test the bioactivity of HA-P.
The bioactivity of HA-P was tested by antibiotic diffusion method using an efflux pump deficient ΔtolC E. coli strain as test organism. Fifteen milliliters of LB agar was overlaid with 1.5 ml of soft agar (0.3% agar) seeded with 2 μl of an overnight culture of ΔtolC E. coli. Filter paper disks (5 mm in diameter) were placed on the plate, to which 30 or 60 μg of purified HA-P were applied. Similar amounts of HA were used as a positive control. The plate was incubated for 15 h at 37°C and examined for the appearance of zones of inhibition. The assay was also repeated with Streptomyces lividans. Twenty five microliters of spores was spread on an ISP2 agar plate, and filter paper disks were placed as described above. The plate was examined for zones of inhibition after 15 h of incubation at 30°C.
Hygromycin sensitivity of S. hygroscopicus and the Δhyg21 strain.
The MIC95s of HA for the wild type and the Δhyg21 mutant were determined by the agar plate dilution method. Portions (30 μl) of spore suspensions of both the strains (2,000 CFU/ml) were plated on 3-ml ISP2 agar plates containing 200, 300, or 400 μg of the antibiotic/ml. The plates were incubated at 30°C for 48 h. The MIC95 was the lowest concentration of HA that prevented visible growth of 95% or more of the CFU on the plate.
RESULTS
Sequence analysis of Hyg21.
The hyg21 ORF is 552 nucleotides long, with a putative translation product of 183 amino acids and a theoretical molecular mass of 19.87 kDa. A search of the NCBI protein sequence database using the web-based BLASTP program revealed that the deduced amino acid sequence has highest similarity of 73% to the Ard2 protein from Saccharothrix mutabilis subsp. capreolus, the producer of antibiotic A201A (compound 7 in Fig. 1). A201A is structurally homologous to HA, and cell extracts from S. capreolus wild type and S. lividans harboring a recombinant Ard2 have been shown to phosphorylate A201A at the C-2 hydroxyl of the hexafuranose moiety, leading to loss of antibiotic activity (2). There have been no published reports of purification or characterization of Ard2. No significant overall similarity of Hyg21 to any other known antibiotic modifying kinases could be detected. A BLAST/Conserved Domain search instead retrieves the gluconate kinase domain proteins, indicating that Hyg21 and Ard2 may represent a new class of antibiotic-modifying kinases. A search of PROSITE protein domain database (12) indicated the presence of a ATP/GTP binding site motif, also known as Walker A motif or P-loop, [(A/G)XXXXGK(S/T)], near the N terminus of Hyg21 within residues 24 to 31 (GIPGSGKS). This glycine-rich motif is often found in nucleotide-binding proteins and is known to form a flexible loop that interacts with ATP or GTP (18, 23). On the basis of these observations we hypothesized that the hyg21 gene encodes a kinase involved in the O phosphorylation of HA.
Hyg21-catalyzed phosphorylation of HA.
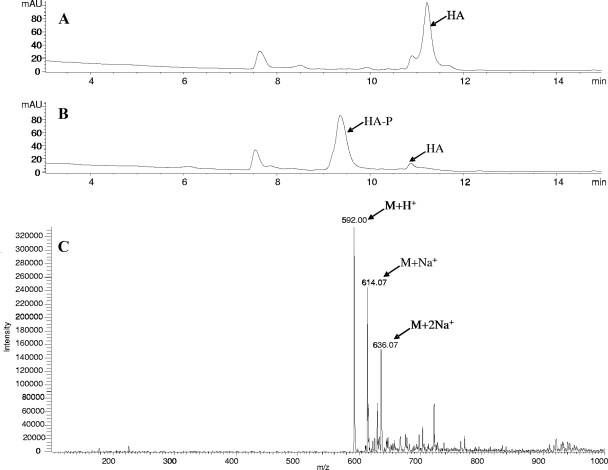
The PCR-amplified hyg21 ORF was cloned into E. coli expression vector pET15b to give pET15b-hyg21 plasmid and overexpressed in BL21-CodonPlus cells as an N-terminal His6-tagged protein. A large fraction of the expressed protein formed inclusion bodies under the given expression conditions, but it was possible to recover substantial amounts of soluble protein using a 1-liter culture and affinity chromatography purification on a Ni-NTA agarose column. SDS-PAGE analysis of the purified preparation demonstrated a prominent protein band with a relative molecular mass of 22 kDa, a finding consistent with the mass of Hyg21 (∼20 kDa) containing a His6 tag (∼2 kDa). In order to determine the biochemical function of Hyg21, the recombinant protein was incubated with HA and ATP at 30°C for 2 h. Reversed-phase HPLC analysis of the reaction mixture after incubation revealed a significant decrease in the HA peak and the appearance of a new peak with a shorter retention time (Fig. 2B). In a control incubation without Hyg21, this new peak was absent, and the levels of HA did not alter (Fig. 2A). The shorter retention time for the new peak was consistent with increased polarity of HA through the addition of a phosphoryl group. MS analysis of the reaction mixture showed that the compound under the new peak had a molecular mass of 592 ([M+H]+) (Fig. 2C). This increase of 80 atomic mass units from that of HA (512 [M+H]+) corresponds to the addition of a single phosphoryl group to the molecule. These data support the hypothesis that hyg21 encodes a phosphotransferase that carries out monophosphorylation of HA.
FIG. 2.
Reversed-phase HPLC-MS analyses of phosphorylation of HA by Hyg21. (A) HPLC chromatogram of a control reaction of HA and ATP without Hyg21. (B) HPLC chromatogram of a reaction of HA and ATP after incubation with Hyg21. The resulting HA-P product has a shorter retention time than HA under the tested HPLC conditions (see Materials and Methods). (C) MS analysis (in positive mode) of the HA-P.
The presence of 5 mM 2-mercaptoethanol in assays of Hyg21 was found to be necessary for maintaining optimal activity. The optimal pH for Hyg21 activity was measured across a range of pH values from 6 to 9. The fastest rate of reaction, determined by the amount of product formed per minute, was in the range of pH 7.5 and 8. Activity was markedly decreased at lower pH values and almost absent at pH 5. The effect of temperature on activity was also studied across a range of 23 to 50°C. In a 15-min assay, product formation was maximal at 30°C. The enzyme retained two-thirds of its activity at room temperature and 37°C but very little at 50°C.
Bioassay for antibiotic activity of HA-P.
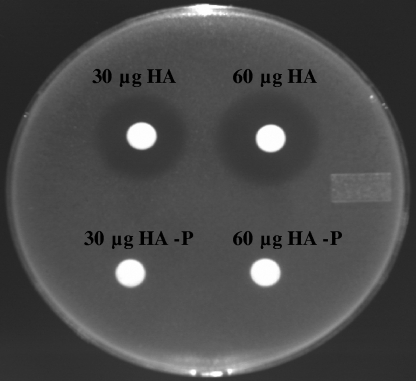
The activity of HA-P was tested in a disk diffusion assay against an HA-sensitive ΔtolC E. coli strain that has an impaired efflux mechanism due to the absence of the outer membrane component of the efflux pump (5). As seen in Fig. 3, distinct zones of inhibition were seen with HA. In contrast, no zones of inhibition were seen with the HA-P, even at 60 μg. Similar observations of antibacterial activity for HA but not HA-P were observed in disk diffusion assays against HA-sensitive S. lividans. These observations are consistent with the hypothesis that phosphorylation abolishes the antibiotic potency of HA and also indicate the likely function of hyg21 as a self-resistance determinant in S. hygroscopicus NRRL 2388.
FIG. 3.
Disk diffusion assay demonstrating that HA but not HA-P has bioactivity against the efflux pump-deficient ΔtolC E. coli strain. Filter paper disks with 30 or 60 μg of both HA and HA-P were placed on LB agar plate seeded with ΔtolC E. coli and incubated overnight at 37°C.
Site of phosphorylation in HA.
1H NMR analyses of purified HA (5 mg) and HA-P (2 mg) were carried out to determine the site of phosphorylation by Hyg21. The chemical shifts of the two samples were virtually identical, with the exception of the proton signal for the C2‴ methine (H2‴), which is shifted downfield from δ 4.36 ppm for HA to δ 4.69 ppm in HA-P. In each case the H2‴ assignment was confirmed by a 1H correlation spectroscopy experiment, which revealed a strong cross peak with the distinctive anomeric H1" resonance. The chemical shift change for the H2‴ alone is consistent with phosphorylation of the C2‴ hydroxyl substituent (Fig. 1).
Substrate specificity of Hyg21.
The substrate specificity of Hyg21 was explored by carrying out enzyme assays with various analogs of HA (compounds 1 to 6), A201A (compound 7), and puromycin (compound 8) (Fig. 1). HPLC-MS analyses revealed that, with the exception of compound 8, all of these compounds were partially or completely converted to new products with shorter retention times and with masses increased by 80 AMU (Table 1). Notable among these observations was that antibiotic A201A (compound 7), which is much larger than HA but has structurally similar fucofuranose and (E)-3-hydroxyphenyl)-2-methylacrylic acid moieties, was also a substrate for Hyg21. In addition, compound 4, which lacks the aminocyclitol and thus is significantly smaller than HA, is also a substrate. In both cases the substrate contains a fucofuranose with a C2-hydroxyl substituent. This moiety is missing in compound 8, despite its structural similarities with both HA and antibiotic A201A, and in this case phosphorylation with Hyg21 is not observed. These observations demonstrate that Hyg21 exhibits broad substrate specificity and that the fucofuranose moiety with C2-hydroxyl group is a key structural element required for phosphorylation. The latter inference is consistent with the observation that in HA, Hyg21 catalyzes phosphorylation at C2 of this moiety.
TABLE 1.
Summary of the kinetic data for Hyg21 phosphorylation
| Substrate | Compound | Mass [M+H]+
|
Mean ± SD
|
||
|---|---|---|---|---|---|
| Substrate | Product | kcat (min−1) | Km (μM) | ||
| HA | 512 | 592 | 2.23 ± 0.13 | 29.1 ± 4.4 | |
| Methoxyhygromycin A | 1 | 514 | 594 | 1.96 ± 0.13 | 29.7 ± 5.8 |
| Desmethylenehygromycin A | 5 | 500 | 580 | 1.89 ± 0.08 | 35.6 ± 3.9 |
| 5‴-Dihydrohygromycin (natural isomer) | 2a | 514 | 594 | 0.15 ± 0.03 | 8.5 ± 3.0 |
| 5‴-Dihydrohygromycin (non-natural isomer) | 2b | 514 | 594 | 1.62 ± 0.1 | 16.5 ± 2.8 |
| 5‴-Dihydromethoxyhygromycin A (natural isomer) | 3a | 516 | 596 | 0.13 ± 0.03 | 10.6 ± 2.6 |
| 5‴-Dihydromethoxyhygromycin A (non-natural isomer) | 3b | 516 | 596 | 1.31 ± 0.08 | 13.0 ± 1.7 |
| 5‴-Dihydrodesmethylenehygromycin A (natural isomer) | 6 | 502 | 582 | NDa | ND |
| 5‴-Dihydro AB subunit | 4 | 363 | 443 | ND | ND |
| A201A | 7 | 803 | 883 | ND | ND |
| ATP | 198 ± 19 | ||||
| GTP | 356 ± 62 | ||||
ND, not determined.
HA phosphorylation was also carried out by substituting ATP with GTP, dTTP (TTP), and CTP. Besides ATP, product formation was seen only with GTP. There was no reaction with either TTP or CTP.
The rate of phosphorylation was determined by measuring the amount of product formed per minute (Table 1). The reaction was linear during the first 15 min of incubation under the given assay conditions. Under these conditions kcat values of 2.2 min−1 and Km values of approximately 30 μM were obtained for HA. The reaction rate kcat was the same with either ATP or GTP, although ATP had a lower Km. Similar but slightly slower kcat rates were observed for ATP-dependent phosphorylation of compounds 1 and 5 by Hyg21. These data and similar Km values indicated that the methylene group of HA, which is present as a methyl group in compound 1 and absent in compound 5, is not important for binding and catalysis.
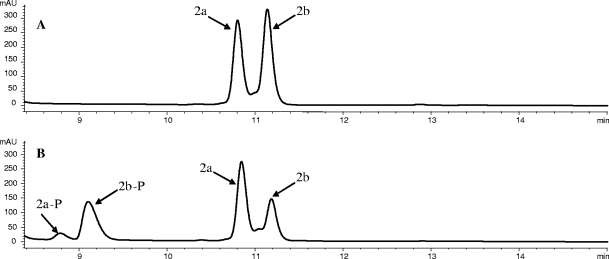
Compounds 2a (a 5‴-dihydro analog of HA) and 3a (a 5‴-dihydro analog of methoxyhygromycin A [compound 1]) have previously been identified in fermentation of the Δhyg26 S. hygroscopicus mutant (16). We produced a diastereomeric mix of compound 2a and the other 5‴-dihydroisomer (compound 2b) by chemical reduction of HA. HPLC analysis of Hyg21-catalyzed phosphorylation of this mixture (Fig. 4) revealed much faster reaction rates with compound 2b than with compound 2a. Apparent kinetic values for the reaction of each isomer in the mixture were obtained and revealed that compound 2b was processed with reaction rates (kcat = 1.68 min−1) similar to those of HA, compound 1, and compound 5. In contrast, the reaction rate is more than 10-fold slower (kcat = 0.11 min−1) with the isomer compound 2a, which appears to be a shunt metabolite in the HA biosynthetic pathway. A similar set of observations was made with a diastereomeric mixture of compounds 3a and 3b (obtained by chemical reduction of compound 1). The phosphorylation rate for the compound 3a isomer is slow (kcat = 0.11 min−1), but much faster (kcat = 1.34 min−1) for compound 3b. In contrast to the kcat values, much smaller changes in Km were observed for the various substrates. These kinetic data indicate that Hyg21 can process HA analogs in which the 5‴-keto group is reduced, but the efficiency of the process is determined by the stereochemistry at the C5‴. Kinetic studies for substrates 4, 6, and 7 were precluded because of low availability of the substrates and/or the presence of impurities. A comparison of reaction completion with HA and compound 7 carried out under standard conditions indicated that the rate of conversion of compound 7 was at least fivefold slower.
FIG. 4.
Hyg21-catalyzed phosphorylation of a diastereomeric mixture of 5‴-dihydrohygromycin A (compounds 2a and 2b). (A) HPLC chromatogram of a control reaction of dihydrohygromycin A and ATP without Hyg21. (B) HPLC chromatogram of a reaction of dihydrohygromycin A and ATP after incubation with Hyg21. Compound 2a is the naturally occurring isomer also detected in fermentation broths of the wild type and the hyg26 blocked mutant. Compound 2b is the non-natural isomer observed only in the diastereomeric mixture after chemical reduction of HA. The phosphorylated products of compounds 2a and 2b are labeled as compounds 2a-P and 2b-P, respectively.
Generation and analysis of a Δhyg21 S. hygroscopicus mutant.
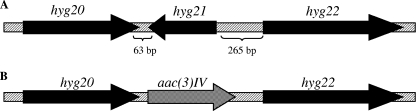
The hyg21 gene in S. hygroscopicus was replaced by the apramycin resistance gene to assess its role in HA biosynthesis and resistance. A gene replacement strategy was used because the hyg21 is well separated from its flanking genes, and its orientation relative to these indicates that it is not part of a polycistronic transcript (Fig. 5).
FIG. 5.
Schematic representation of the hyg20-22 region in the genome of S. hygroscopicus wild type (A) and Δhyg21 mutant (B). The hyg20 and hyg22 gene products are homologous to transglucosylases and acyltransferases, respectively, and have been proposed to be involved in HA biosynthesis. The hyg21 gene is replaced with the apramycin resistance gene, aac3(IV), in the Δhyg21 strain.
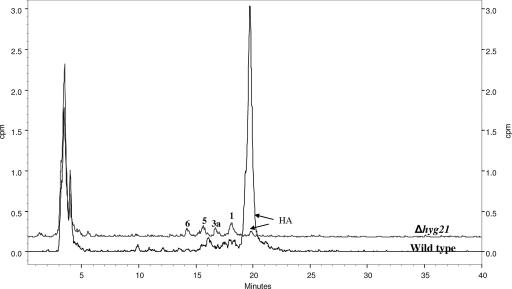
The Δhyg21 mutant strain was cultured, and the fermentation broth was analyzed for the production of HA. HA was detected, indicating that hyg21 was not essential for the biosynthetic process. Nonetheless, the yields were dramatically reduced (>90%) from the 0.8 g of HA/liter obtained in the wild-type strain. Detailed LC-MS analyses of fermentation broth of Δhyg21 also revealed the presence of compounds 1, 3a, 5, and 6. The same compounds could be detected by radioactive scintillation counting when the mutant was grown in the presence of tritiated myo-inositol (which is converted into the aminocyclitol portion of HA) (Fig. 6). Although the levels of compounds 1, 3a, 5, and 6 could not be quantitated, the analyses indicated that they were similar in both the wild type and the Δhyg21 mutant, in contrast to the observations made for the levels of HA.
FIG. 6.
Reversed-phase HPLC analyses and radiolabel detection of wild-type and Δhyg21 culture broths grown in the presence of [2-3H]myo-inositol. The production of HA was significantly decreased in the mutant. Radiolabeled analogs of HA (see Fig. 1 for structures) are present in fermentation broths of both the wild-type and the Δhyg21 strains.
The low amounts of HA in the fermentation broth of Δhyg21 raised the possibility that it is the HA-P that is exported from the S. hygroscopicus by an efflux pump and subsequently converted to HA by an extracellular phosphatase. In such a case HA might be expected to accumulate inside the cells of Δhyg21. To investigate this possibility, the mycelium of Δhyg21 was lysed by methanol treatment, and the lysate was analyzed by HPLC. However, HA could not be detected in the cell lysate, and it was estimated that nearly all of the HA made by this strain is found in the fermentation broth. Also, no HA-P could be detected in either the cell lysate or the fermentation broth of the wild-type strain.
An agar plate dilution assay was carried out to assess whether the Δhyg21 strain had increased sensitivity to HA compared to the wild-type strain. The MIC95s for both strains were found to be 400 μg/ml.
DISCUSSION
Analysis of the deduced amino acid sequence of the hyg21 gene in the HA biosynthetic gene cluster led to a hypothesis that it encodes a protein that phosphorylates HA and thus provides a resistance mechanism to the producing organism by antibiotic inactivation. The first component of this hypothesis has been unequivocally verified by in vitro assays in which Hyg21 HA-P selectively at the C2‴-OH position. Hyg21 also phosphorylates a number of HA analogs, and the site of modification in these may also be the C2‴-OH. Some of these analogs are observed at various levels in fermentations of the wild-type S. hygroscopicus (9, 16). We have also observed that these analogs, in particular methoxyhygromycin A and desmethylenehygromycin A, have some level of antibacterial activity, albeit less than that observed for HA. Presumably, resistance to these compounds, as well as to HA, can be provided in part by the Hyg21-catalyzed phosphorylation.
The enzyme-catalyzed process was most efficient with HA, a finding consistent with phosphorylation occurring with the final biosynthetic product, rather than with an intermediate. Nonetheless, the rate of reaction with HA was relatively slow (kcat of ∼2 min−1). Analyses of S. hygroscopicus fermentations reveal that the majority of the HA product is found in fermentation media, indicating an efficient export process. We see no evidence for any HA-P in the fermentation broth or in the cell lysate, and our analyses of the HA biosynthetic gene cluster to date have not revealed a candidate phosphatase that would convert HA-P to HA (the hyg25 gene product appears to be involved in the conversion of myo-inositol-1-phosphate to myo-inositol, a step in the biosynthesis of the aminocyclitol moiety of HA). Thus, all evidence indicates that it is HA that is exported and that HA-P may be generated from residual HA in the cell. In such a case, a moderately slow rate of phosphorylation by Hyg21 would allow most of the HA biosynthetic product to be exported rather than become trapped intracellularly as HA-P.
The results from these studies do not provide unequivocal evidence for our hypothesis that phosphorylation of HA is a resistance mechanism in the producing organism. We do observe that HA and not HA-P has antibacterial activity against other bacteria. However, it remains to be determined whether the loss of antibacterial activity is associated with poorer penetration into the organisms or inhibition of binding to the ribosomal target. In addition, the Δhyg21 mutant retains significant resistance to HA. This observation likely reflects the presence of other operational mechanisms of resistance. The most likely of these are methylation of the rRNA, catalyzed by the hyg29 gene product, and an efficient HA export process, catalyzed by the hyg19 and hyg28 gene products. Characterization of the remaining putative resistance determinants in the HA biosynthetic gene cluster is currently under way. Finally, the observation that insertional inactivation of hyg21 reduces the production of HA to almost negligible amounts is also curious. A polar effect from replacement of the gene cannot be ruled out but seems unlikely given the strategy used and the organization of genes surrounding hyg21 (Fig. 5). This result was consistently observed in all of the four mutant colonies that were examined. A significant decrease in HA production would be predicted in this mutant if the Hyg21-catalyzed phosphorylation process occurred at an earlier step in the biosynthetic process, with the later stages occurring with phosphorylated intermediates. Early modification would ensure that the resistance mechanism is in operation prior to assembly of the final product and has precedent in the biosynthesis of streptomycin (15) and paromomycin (17). The slow turnover of HA might also indicate that a pathway intermediate, and not the final product, may be the preferred substrate for Hyg21. However, we do not have any genetic evidence for a phosphatase that would be responsible for conversion of the HA-P to HA, we detect only HA in fermentations of the wild-type strain (see above), and HA was the preferred substrate for Hyg21 in our kinetic studies. These observations argue against Hyg21-catalyzed phosphorylation of a pathway intermediate as a step in the biosynthetic process. In the case of tylosin biosynthesis it has been shown that disruption of any of the four genes involved in the biosynthesis of the mycaminose sugar, which is the first sugar that gets attached to tylactone (the polyketide aglycone), abolishes not just tylosin production but also the accumulation of tylactone (3). In a similar way, HA-P may be required for stimulating the HA biosynthetic pathway by positive feedback regulation, accounting for the decreased production in the Δhyg21 mutant.
In conclusion, we have shown that Hyg21 catalyzes phosphorylation of HA and related compounds, leading to the loss of antibacterial activity. Thus, hyg21 is the first of the several possible resistance determinants in S. hygroscopicus NRRL 2388 that has been functionally characterized. The roles of Hyg6 and Hyg29 (methyl transferases) and of Hyg19 and Hyg28 (antibiotic efflux proteins) as additional resistance determinants are currently being investigated.
Acknowledgments
We thank A. Jimenez from Centro de Biologia Molecular Severo Ochoa, Madrid, Spain, for providing us with antibiotic A201A.
Footnotes
Published ahead of print on 21 July 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barrasa, M. I., J. A. Tercero, and A. Jimenez. 1997. The aminonucleoside antibiotic A201A is inactivated by a phosphotransferase activity from Streptomyces capreolus NRRL 3817, the producing organism: isolation and molecular characterization of the relevant encoding gene and its DNA flanking regions. Eur. J. Biochem. 245:54-63. [DOI] [PubMed] [Google Scholar]
- 3.Butler, A. R., S. A. Flint, and E. Cundliffe. 2001. Feedback control of polyketide metabolism during tylosin production. Microbiology 147:795-801. [DOI] [PubMed] [Google Scholar]
- 4.Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. [DOI] [PubMed] [Google Scholar]
- 5.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gourmelen, A., M. H. Blondelet-Rouault, and J. L. Pernodet. 1998. Characterization of a glycosyl transferase inactivating macrolides, encoded by gimA from Streptomyces ambofaciens. Antimicrob. Agents Chemother. 42:2612-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero, M. D., and J. Modolell. 1980. Hygromycin A, a novel inhibitor of ribosomal peptidyltransferase. Eur. J. Biochem. 107:409-414. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habib el, S. E., J. N. Scarsdale, and K. A. Reynolds. 2003. Biosynthetic origin of hygromycin A. Antimicrob. Agents Chemother. 47:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkey, P. M. 1998. The origins and molecular basis of antibiotic resistance. BMJ 317:657-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hopwood, D. A. 2007. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 63:937-940. [DOI] [PubMed] [Google Scholar]
- 12.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansouri, K., and W. Piepersberg. 1991. Genetics of streptomycin production in Streptomyces griseus: nucleotide sequence of five genes, strFGHIK, including a phosphatase gene. Mol. Gen. Genet. 228:459-469. [DOI] [PubMed] [Google Scholar]
- 14.Martin, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 15.Miller, A. L., and J. B. Walker. 1970. Accumulation of streptomycin-phosphate in cultures of streptomycin producers grown on a high-phosphate medium. J. Bacteriol. 104:8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palaniappan, N., S. Ayers, S. Gupta, S. Habib el, and K. A. Reynolds. 2006. Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster. Chem. Biol. 13:753-764. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Gonzalez, J. A., M. Lopez-Cabrera, J. M. Pardo, and A. Jimenez. 1989. Biochemical characterization of two cloned resistance determinants encoding a paromomycin acetyltransferase and a paromomycin phosphotransferase from Streptomyces rimosus forma paromomycinus. J. Bacteriol. 171:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraste, M., P. R. Sibbald, and A. Wittinghofer. 1990. The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15:430-434. [DOI] [PubMed] [Google Scholar]
- 19.Smith, C. A., and E. N. Baker. 2002. Aminoglycoside antibiotic resistance by enzymatic deactivation. Curr. Drug Targets Infect. Disord. 2:143-160. [DOI] [PubMed] [Google Scholar]
- 20.Thompson, C. J., R. H. Skinner, J. Thompson, J. M. Ward, D. A. Hopwood, and E. Cundliffe. 1982. Biochemical characterization of resistance determinants cloned from antibiotic-producing streptomycetes. J. Bacteriol. 151:678-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilches, C., C. Hernandez, C. Mendez, and J. A. Salas. 1992. Role of glycosylation and deglycosylation in biosynthesis of and resistance to oleandomycin in the producer organism, Streptomyces antibioticus. J. Bacteriol. 174:161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker, J. B., and M. S. Walker. 1967. Streptomycin biosynthesis. Enzymatic synthesis of O-phosphorylstreptidine from streptidine and adenosinetriphosphate. Biochim. Biophys. Acta 148:335-341. [DOI] [PubMed] [Google Scholar]
- 23.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright, G. D., and P. R. Thompson. 1999. Aminoglycoside phosphotransferases: proteins, structure, and mechanism. Front. Biosci. 4:D9-D21. [DOI] [PubMed] [Google Scholar]
- 25.Zhao, L., N. J. Beyer, S. A. Borisova, and H. W. Liu. 2003. Beta-glucosylation as a part of self-resistance mechanism in the methymycin/pikromycin producing strain Streptomyces venezuelae. Biochemistry 42:14794-14804. [DOI] [PubMed] [Google Scholar]