Abstract
We identified a novel type of staphylococcal cassette chromosome mec (SCCmec) element carried by methicillin-resistant Staphylococcus aureus (MRSA) strain JCSC6082 isolated in Sweden. The SCCmec element was demarcated by characteristic nucleotide sequences at both ends and was integrated at the 3′ end of orfX. The element carried a novel combination of a type 5 ccr gene complex and class C1 mec gene complex. The J regions of the element were homologous to those of the SCCmercury element of S. aureus strain 85/2082, with nucleotide identity greater than 99%. However, the novel SCCmec element from JCSC6082 did not carry the mer operon nor Tn554, suggesting that evolution to SCCmec could have been from a common ancestor by acquisition of the class C1 mec gene complex. The novel SCCmec element from JCSC6082 was flanked by a novel SCC-like chromosome cassette (CC6082), which was demarcated by two direct repeats and could be excised from the chromosome independently of the SCCmec element. Our data suggest that novel SCCmec elements can be generated on the staphylococcal chromosome through the recombination between extant SCC elements and mec gene complexes.
Since the staphylococcal cassette chromosome mec (SCCmec) was first described as the carrier of the mecA gene (9, 13), at least six structural variants have been identified, namely types I, II, III, IV, V, and VI SCCmec. They are typically composed of two essential components, the mec gene complex (mec) and the cassette chromosome recombinase (ccr) gene complex (8, 10, 16, 18). In addition, they are integrated at a specific integration site sequence (ISS) of orfX on the staphylococcal chromosome (11).
Three major classes of mec gene complex have been identified in methicillin-resistant Staphylococcus aureus (MRSA) strains. The prototypic mec (class A) is composed of mecA, its regulatory genes, mecR1 and mecI, and insertion sequence IS431 downstream of mecA. Certain variants of the mec gene complex contain insertion of either IS1272 (class B) or IS431 (class C1 and C2) at the 3′ portion of mecR1. The ccr genes are responsible for the chromosomal integration and excision of the SCCmec, and there are at least five types of the ccr gene complex. The first four types contain two site-specific recombinase genes, ccrA and ccrB, with surrounding open reading frames (ORFs), while the fifth type contains one site-specific recombinase gene, ccrC, with surrounding ORFs. The classification of SCCmec is based upon the combination of the mec class and the ccr type (11).
Recently, SCCmec elements with more complex structures have been reported, which contain multiple ccr gene complexes (7, 19). In addition, mobile genetic elements that did not themselves carry ccr gene complexes have been found integrated downstream of orfX, such as the 31-kb arginine catabolic mobile element (ACME). Mobilization of these elements has been suggested to occur by the activity of the recombinases encoded by the ccrAB genes located on the adjacent SCCmec (4, 5).
In Sweden, the prevalence of MRSA is still below 1%, and MRSA isolates are genetically diverse (1). At present, the community-acquired MRSA strains are more prevalent than the nosocomial MRSA strains (Swedish Institute for Infectious Disease Control; www.smittskyddsinstitutet.se) (1). In addition, several MRSA isolates that were not carrying extant types of SCCmec elements have been identified in various regions of Sweden (C. Berglund, unpublished data).
In this work, we describe a novel type of SCCmec element from an MRSA strain isolated in Sweden.
MATERIALS AND METHODS
MRSA strain JCSC6082 (p5747/2002) (2) was isolated in 2002 from a subcutaneous abdominal wall abscess of a previously healthy 42-year-old woman. The infection was designated as community acquired; however, the patient had undergone abdominal surgery due to salpingitis 24 years earlier, and in the present abscess cavity the remnants of a suture were found.
The MRSA strain belonged to the epidemic and widespread sequence type 5 (ST5) and clonal complex 5 (CC), and carried neither the genes for Panton-Valentine leukocidin (PVL) nor exfoliative toxin A or B. The results of susceptibility testing using the agar dilution method according to Clinical and Laboratory Standards Institute guidelines showed that it was resistant to oxacillin, tetracycline, and erythromycin, with MICs of 48, 32, and 8 mg/liter, respectively (1).
Construction of fosmid libraries.
Genomic DNA of JCSC6082 was prepared using ISOPLANT (Nippon Gene Co., Tokyo, Japan) and was subsequently used as insert DNA in the fosmid library. The fosmid libraries were created using the CopyControl fosmid library production kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's recommendation. The clones were screened by PCR with primer pairs to identify orfX, mecA, and the chromosomal region located downstream of SCCmec.
DNA manipulations.
PCRs for screening of fosmid clones and identification of specific genes, long-range PCR for amplification of DNA fragments covering the entire SCCmec, and nested PCR experiments for identification of precise excision of the novel SCCmec, SCCmec and CC6082 together, and CC6082 alone were carried out as described previously (9). The sequences and locations of the PCR primer pairs used to amplify DNA fragments covering the entire SCCmec region are shown in Fig. 1 and Table 1.
FIG. 1.
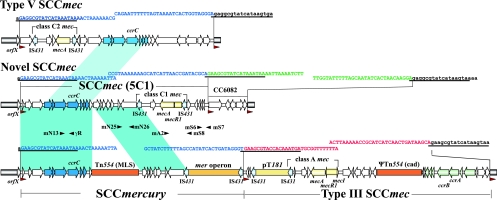
Structural comparisons of the novel SCCmec element and a chromosome cassette identified from JCSC6082 isolated in Sweden, SCCmercury, and type III SCCmec carried by 85/2082 and the type V SCCmec carried by WIS. The structures of these elements are illustrated based on the nucleotide sequence deposited in the DDBJ/EMBL/GenBank database under accession no. AB373032, AB037671, and AB121219. Conserved regions are indicated in blue: more than 99% homology is in dark blue and 19.1% to 93.6% homology is in light blue. Red arrowheads indicate integration site sequence of SCCmec (ISS) that comprises DR sequences, and black arrowheads indicate the positions and directions of primers used for detection of the novel type of SCCmec.
TABLE 1.
Oligonucleotide primers employed for PCR detection of the novel type of SCCmec
| Primer for identification of type 5C1 SCCmec | Nucleotide sequence (5′→3′) | Nucleotide positiona | Expected size of product (kbp) |
|---|---|---|---|
| mN13 | ACAACTTGCGAATTATGACGA | 6132-6152 | 1.9 |
| γR | CCTTTATAGACTGGATTATTCAAAATAT | 8071-8044 | |
| mN25 | TCCGCTTGTACAATTAATATTGAGCGT | 14310-14336 | 2.5 |
| mN26 | ACGAAACGACCATGTCTGCTCTAACA | 16848-16823 | |
| mA2 | AACGTTGTAACCACCCCAAGA | 21149-21169 | 3.5 |
| mS8 | CAAAATTGAGCGGAATCGTT | 24693-24674 | |
| mS6 | TGTTTTCGAGTATCGCTTTGA | 26080-26100 | 1.8 |
| mS7 | GAAAGTGCGTTTATATCTGCAAAA | 27870-27847 |
Nucleotide positions based on the nucleotide sequence in the DDBJ/EMBL/GenBank database (accession no. AB373032) are indicated.
Nucleotide sequencing was performed as described previously (15), and the sequences were analyzed and assembled using the BioEdit sequence alignment editor.
Analysis of ORFs.
ORFs longer than 100 bp were identified with the GenomeGambler v.1.5 software Japan (Marine Science and Technology Center and Xanagen, Inc., Kawasaki, Japan) and were compared with sequence databases at the National Center for Biotechnology Information with the basic local alignment search tool BLAST (National Library of Medicine, Bethesda, MD) for annotation and prediction of functions. The ccr genes were investigated using the ClustalW (1.83) multiple alignment program available at http://www.staphylococcus.net.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the DDBJ/EMBL/GenBank database under accession no. AB373032.
RESULTS AND DISCUSSION
Identification of a new member of the SCCmec family.
We identified an MRSA strain, JCSC6082, which carried an unknown type of SCCmec element according to PCR detection of the extant types of SCCmec (10, 14, 17). The MRSA strain JCSC6082 was considered to be community acquired, although the patient had a history of hospitalization 24 years previously. It may be that the patient had acquired the MRSA strain recently and that it was a true community-acquired infection. However, the specific location of the infection in addition to a difficult-to-treat infection 24 years earlier makes it tempting to consider the possibility of a dormant infection or at least persistence of MRSA, adherent to foreign material (e.g., a suture) and embedded in biofilm, for more than 20 years until it was reidentified by the presentation of the abscess in 2002.
The structure of the SCCmec element from JCSC6082 was further investigated by construction of fosmid libraries and determination of the entire nucleotide sequence. The overall structure of the 33,261-bp region encompassing the SCCmec element from JCSC6082 is illustrated in Fig. 1, and the ORFs carried by this region are listed in Table 2.
TABLE 2.
ORFs of the novel SCCmec and chromosome cassette CC6082 in JCSC6082
| ORF | Value for coding sequencea
|
Gene | Description of product | Data indicating homology to ORF in SCCmec or SCCmercury of
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Locationb | Gene size (bp) | Length (aa) | 85/2082
|
WIS (JCSC3624)
|
|||||
| % Identityc | Corresponding ORF(s) (size in bp) | % Identityc | Corresponding ORF(s) (size in bp) | ||||||
| SCCmec | |||||||||
| Sw01 | 135-614 | 480 | 159 | orfX | Conserved hypothetical protein OrfX | 99.4 | CZ080 (480) | 99.4 | V001 (480) |
| Sw02 | 860-1165 | 306 | 101 | Hypothetical protein | 100 | CZ078 (306) | |||
| Sw03 | 1363-2226 | 864 | 287 | Hypothetical protein | 100 | CZ077 (870) | |||
| Sw04 | 2334-3809 | 1476 | 491 | Hypothetical protein | 100 | CZ076 (1500) | 19.1 | V011 (1989) | |
| Sw05 | 4036-5136 | 1101 | 366 | Hypothetical protein | 100 | CZ075 (1101) | 77.1 | V012 (1110) | |
| Sw06 | 5129-5500 | 372 | 123 | Hypothetical protein | 100 | CZ074 (372) | 84.4 | V013 (369) | |
| Sw07 | 5497-7140 | 1644 | 547 | Hypothetical protein | 100 | CZ073 (1644) | 82.1 | V014 (1644) | |
| Sw08 | 7365-9041 | 1677 | 558 | ccrC | Cassette chromosome recombinase C | 99.9 | CZ072 (1544) | 93.6 | V015 (1623) |
| Sw09 | 9144-9485 | 342 | 113 | Hypothetical protein | 100 | CZ070 (342) | 48.6 | V016 (339) | |
| Sw10 | 9581-9892 | 312 | 103 | Hypothetical protein | 100 | CZ069 (312) | 49.1 | V017 (327) | |
| Sw11 | 9908-10414 | 507 | 168 | Hypothetical protein | 100 | CZ068 (507) | 67.1 | V019 (504) | |
| Sw12 | 10435-10881 | 447 | 148 | radC | DNA repair protein RadC | 100 | CZ067 (318) | ||
| Sw13 | 11051-11923 | 873 | 290 | Hypothetical protein | 100 | CZ059 (873) | |||
| Sw14 | 11962-12270 | 309 | 102 | Hypothetical protein | 100 | CZ058 (309) | |||
| Sw15 | 12286-12774 | 489 | 162 | Hypothetical protein | 100 | CZ057 (489) | |||
| Sw16 | 12909-13436 | 528 | 175 | Hypothetical protein | 98.9 | CZ056 (528) | |||
| Sw17 | 13498-13941 | 444 | 147 | Membrane protein homologue | 99.3 | CZ055 (489) | |||
| Sw18 | 14505-14828 | 324 | 107 | Hypothetical protein | 100 | CZ053 (324) | 98.1 | V020 (567) | |
| Sw19 | (17134-14906) | 2229 | 742 | hsdR | Truncated type I restriction modification system endonuclease homologue | 100 | Z059 (2244) | 98.2 | V021 (3120) |
| Sw20 | (17855-17181) | 675 | 224 | tnp | Transposase of IS431 | 100, 99.6, 99.1, 99.1 | Z035 (675), Z041 (675), Z046 (675), Z058 (675) | 99.1, 97.8 | V003 (675), V008 (675) |
| Sw21 | 19157-19900 | 744 | 247 | Glycerophoshoryl diester phophodiesterase | 99.6 | CZ030 (744) | 99.6 | V004 (744) | |
| Sw22 | 19997-20425 | 429 | 142 | MaoC domain protein homologue | 100 | CZ029 (429) | 100 | V005 (429) | |
| Sw23 | (22477-20471) | 2007 | 668 | mecA | Penicillin binding protein, PBP2′ | 99.8 | Z030 (2007) | 99.7 | V006 (2007) |
| Sw24 | 22577-23575 | 999 | 332 | mecR1 | Truncated signal transducer protein MecR1 | 86.4 | CZ028 (114) | 80.9 | V007 (111) |
| Sw25 | (24277-23603) | 675 | 224 | tnp | Transposase of IS431 | 100, 100, 99.1, 98.6 | Z046 (675), Z058 (675), Z035 (674), Z041 (675) | 98.2, 97.8 | V003 (675), V008 (675) |
| Sw26 | 24321-24875 | 555 | 184 | Hypothetical protein, similar to glutathione synthase | |||||
| Sw27 | (25834-25403) | 432 | 143 | tnp | Truncated transposase IS1296 homologue | ||||
| Sw28 | (26236-25973) | 264 | 87 | tnp | Truncated transposase IS1296 homologue | ||||
| Sw29 | (26565-26287) | 279 | 92 | tnp | Truncated transposase homologue | ||||
| Sw30 | (26830-26534) | 297 | 98 | tnp | Truncated transposase IS3/IS911 homologue | ||||
| CC6082 | |||||||||
| Sw31 | 27446-29311 | 1866 | 621 | Hypothetical protein similar to reverse transcriptase | |||||
| Sw32 | (30451-29804) | 648 | 125 | Membrane protein homologue | |||||
| Sw33 | (31197-30472) | 726 | 241 | hsdM | Type I restriction modification system DNA methylase | 87.9 | V023 (1515) | ||
| Sw34 | 31369-32343 | 975 | 324 | Hypothetical protein | |||||
| Sw35 | 32312-32884 | 573 | 190 | Hypothetical protein | |||||
The sequenced region included three ISSs comprising directly repeated (DR) sequences typical of SCC-like cassettes, suggesting that JCSC6082 carried two SCC-like elements, including an SCCmec element and an SCC-like chromosome cassette lacking ccr recombinase genes (Fig. 1).
We then conducted nested PCR experiments in order to investigate whether these elements could be excised from the chromosome independently. DNA fragments, indicating that these elements have been excised from the chromosome, were successfully amplified by PCR, and nucleotide sequencing confirmed that excision had occurred precisely at the DR sites (data not shown).
The SCCmec element was 26,733 bp in size, i.e., as small as type IV and V SCCmec elements that are prevalent among community-acquired MRSA (3). It carried a class C1 mec complex, which was different from class C2 mec since the orientation of the IS431 inserted downstream of mecA and disrupting the mecR1 gene was opposite to that found in class C2 mec. The class C1 mec complex was first identified in methicillin-resistant Staphylococcus haemolyticus strain SH631 (12). To our knowledge, this is the first description of a class C1 mec complex in an MRSA strain. Interestingly, in the SCCmec element from JCSC6082 the deletion of ΔmecR1 was not the same as that found in the previously described C1 mec complex (12).
The SCCmec element from JCSC6082 carried a type 5 ccr gene complex, in common with the type V SCCmec and SCCmercury elements (8, 10). In this case, however, ccrC was 1,677 bp in size: i.e., larger by 133 bp than the ccrC gene carried by SCCmercury in 85/2082 (accession no. AB037671). Since the region of SCCmercury corresponding to the entire 1,677 bp of the ccrC gene had a nucleotide identity of 99.9%, it was considered that a premature stop codon had been introduced in the ccrC of SCCmercury. The ccrC genes appear to be more diversified in comparison with ccrA and ccrB, since several variants of this recombinase have been described (6, 19).
Consequently, we regarded the region between the first ISS and the second ISS in JCSC6082 as a novel type of SCCmec element, since it carried a type 5 ccr gene complex (ccrC) and a class C1 mec gene complex. No additional resistance gene other than the mecA was identified in the element.
Interestingly, the nucleotide sequence of this novel SCCmec element had 99% homology with the SCCmercury element of 85/2082 (11) but in contrast carried the mec complex instead of the mer operon and Tn554. SCCmercury has been reported to be similar to SCCcap1 carrying a type 1 capsule operon in the same manner (11). These three SCC elements might have been generated in a similar way, with incorporation of mercury resistance operon and Tn554 to generate SCCmercury, a capsule gene cluster to make SCCcap-1, or the mec gene complex to create SCCmec. We also suspect that the class C1 mec complex present in this novel SCCmec may be a composite transposon itself, since it is demarcated by two copies of IS431, which further strengthens the hypothesis that an SCC element might evolve into an SCCmec element by acquiring a mec gene complex. However, so far, there is no evidence of transposition ability of the mec gene complex.
Characteristics of chromosome cassette CC6082.
The SCC-like chromosome cassette CC6082 was 5,617 bp in length and was located between the second and third ISSs, downstream of the SCCmec element. CC6082 carried an hsdM gene encoding a type I restriction modification system DNA methylase, a gene encoding a membrane protein homologue, a gene encoding a hypothetical protein similar to a reverse transcriptase, and two additional ORFs of unknown function. Neither ccr genes nor a mec complex was identified in this region. This region may be a remnant of a previously active SCC element.
Naming of novel SCC elements.
The SCCmec element should be defined by a specific combination of the class of mec and ccr gene complex, and it should be indicated in Roman numerals. We considered that atypical elements e.g., carrying two ccr gene complexes or non-mecA-carrying SCC, at first should be analyzed by determining the entire nucleotide sequences. In the case of a composite of SCC and SCCmec, the elements should be described separately and primarily be categorized into the type of SCCmec as determined by the ccr and mec carried on the element demarcated by ISS and subsequently given the Roman numeral. Here, we designated the novel SCCmec element in JCSC6082 carrying a type 5 ccr gene complex (ccrC) and a class C1 mec gene complex as type 5C1. However, we suggest that this novel SCCmec should receive a Roman numeral as type VII. To avoid confusion on the naming of SCCmec elements, we await a decision from the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements.
Acknowledgments
We thank Tadashi Baba at the Department of Bacteriology and Infection Control Science, Juntendo University, for professional advice and assistance during the orf analysis. We also thank Li Shan Shuang at the Department of Infection Control Science, Juntendo University, Postgraduate School, for performing the nested excision PCR.
This study was supported by grants from the Sweden-Japan Foundation, the Swedish Society for Medical Research, the Swedish Institute of Biomedical Laboratory Science, and the Örebro County Council Research Committee, Sweden, and a Grant-in-Aid for 21st Century COE Research from the Ministry of Education and Science, Japan.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Berglund, C., P. Mölling, L. Sjöberg, and B. Söderquist. 2005. Multilocus sequence typing of methicillin-resistant Staphylococcus aureus from an area of low endemicity by real-time PCR. J. Clin. Microbiol. 43:4448-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund, C., P. Mölling, L. Sjöberg, and B. Söderquist. 2005. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 11:447-456. [DOI] [PubMed] [Google Scholar]
- 3.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J.-H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 5.Goering, R. V., L. K. McDougal, G. E. Fosheim, K. K. Bonnstetter, D. J. Wolter, and F. C. Tenover. 2007. Epidemiologic distribution of the arginine catabolic mobile element among selected methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates. J. Clin. Microbiol. 45:1981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanssen, A.-M., and J. U. Ericson Sollid. 2007. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 51:1671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heusser, R., M. Ender, B. Berger-Bächi, and N. McCallum. 2007. Mosaic staphylococcal cassette chromosome mec containing two recombinase loci and a new mec complex, B2. Antimicrob. Agents Chemother. 51:390-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updates 6:41-52. [DOI] [PubMed] [Google Scholar]
- 12.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma, X. X., T. Ito, P. Chongtrakool, and K. Hiramatsu. 2006. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 44:4515-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 19.Takano, T., W. Higuchi, T. Otsuka, T. Baranovich, S. Enany, K. Saito, H. Isobe, S. Dohmae, K. Ozaki, M. Takano, Y. Iwao, M. Shibuya, T. Okubo, S. Yabe, D. Shi, I. Reva, L.-J. Teng, and T. Yamamoto. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob. Agents Chemother. 52:837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]