Abstract
To address the need for broad-spectrum antiviral activity characterization of hepatitis C virus (HCV) polymerase inhibitors, we created a panel of intergenotypic chimeric replicons containing nonstructural (NS) protein NS5B sequences from genotype 2b (GT2b), GT3a, GT4a, GT5a, and GT6a HCV isolates. Viral RNA extracted from non-GT1 HCV patient plasma was subjected to reverse transcription. The NS5B region was amplified by nested PCR and introduced into the corresponding region of the GT1b (Con-1) subgenomic reporter replicon by Splicing by Overlap Extension (SOEing) PCR. Stable cell lines were generated with replication-competent chimeras for in vitro antiviral activity determination of HCV nonnucleoside polymerase inhibitors (NNIs) that target different regions of the protein. Compounds that bind to the NNI2 (thiophene carboxylic acid) or NNI3 (benzothiadiazine) allosteric sites showed 8- to >1,280-fold reductions in antiviral activity against non-GT1 NS5B chimeric replicons compared to that against the GT1b subgenomic replicon. Smaller reductions in susceptibility, ranging from 0.2- to 33-fold, were observed for the inhibitor binding to the NNI1 (benzimidazole) site. The inhibitor binding to the NNI4 (benzofuran) site showed broad-spectrum antiviral activity against all chimeric replicons evaluated in this study. In conclusion, evaluation of HCV NNIs against intergenotypic chimeric replicons showed differences in activity spectrum for inhibitors that target different regions of the enzyme, some of which could be associated with specific residues that differ between GT1 and non-GT1 polymerases. Our study demonstrates the utility of chimeric replicons for broad-spectrum activity determination of HCV inhibitors.
Approximately 170 million people worldwide are infected with hepatitis C virus (HCV). Persistent infection with HCV is a primary cause of debilitating liver diseases, such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma (35, 43). HCV is a member of the Flaviviridae family with a positive-sense, single-stranded RNA genome of approximately 9.6 kb in length (5). The viral genome contains one open reading frame encoding a polyprotein of approximately 3,000 amino acids. At least 10 mature proteins result from the cleavage of the polyprotein by both cellular and viral proteases (14). The structural proteins, which include core, two envelope glycoproteins (E1 and E2), and p7, are cleaved by cellular signal peptidases (14) while the nonstructural (NS) proteins, NS2, NS3, NS4A, NS4B, NS5A, and NS5B, are cleaved by the viral NS2/3 or NS3/4A protease (10, 15). The HCV RNA genome is replicated by the RNA-dependent RNA polymerase, NS5B. Since NS5B is crucial for viral replication and has distinct features compared to those of human polymerases (21), it is a desirable target for the development of HCV therapies.
HCV isolates from around the world show substantial divergence in their genomic sequences (38). On the basis of these variations, HCV isolates have been classified into six genotypes (GT) (numbered 1 to 6) with nucleotide sequence divergence of as much as 35% (37, 49). Genotypes are further classified into subtypes, such as GT1a and GT1b, which have approximately 80% genetic similarity (37, 49). Substantial regional differences exist in the global distribution of HCV genotypes. GT1, -2, and -3 are found worldwide, of which GT1a and GT1b are the most common subtypes in the United States and Europe (50). GT1b is responsible for as many as two-thirds of the HCV cases in Japan (40). GT2 is commonly found in North America and Europe, along with a prevalence of GT3a infections among intravenous drug users in these regions (50). GT4 is prevalent in North Africa and the Middle East, whereas the less-common GT5 and GT6 appear to be confined to South Africa and Hong Kong, respectively (32, 49). In a study of 81,000 HCV patients in the United States, approximately 70% were infected with GT1, while 14 and 12% of patients were infected with GT2 and GT3, respectively, and the remaining 4% of patients were infected with GT4, -5, and -6 (T. E. Schutzbank, A. Perlina, T. Yashina, N. Wylie, and S. Sevall, presented at the 43rd Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 14 to 17 September 2003).
Response to the current treatment for HCV infection, pegylated interferon (IFN) and ribavirin, varies among patients infected with different genotypes. Only about 50% of patients infected with GT1 or GT4 demonstrate a sustained virologic response after treatment for 48 weeks, compared to 80 to 90% of GT2 or GT3 patients (7, 11, 29). In addition to the low response rates associated with GT1 and GT4 infections, the pegylated IFN and ribavirin combination therapy has severe side effects that often result in high discontinuation rates and low patient compliance. Therefore, there is an unmet medical need for more effective, broad-spectrum HCV therapies with favorable safety profiles.
A significant breakthrough in HCV drug discovery was the development of the GT1b Con-1 HCV replicon system (26). Since then, replicons of GT1a and GT2a have also been generated that are amenable to cell-based screening of HCV replication inhibitors (2, 19, 20, 48). Due to the lack of replicons from other genotypes, it was not possible to determine broad-spectrum activity of HCV inhibitors in cell-based assays. In addition, replication competent GT1b, -1a, and -2a replicons are derived from a single sequence within each subtype. As a result, the variability of antiviral activity among HCV patient isolates could not be readily assessed using these reagents. To allow for broad-spectrum activity determination of HCV inhibitors against various genotypes and quasispecies encountered in the clinical setting, several chimeric replicon systems have been created that utilize replication-competent GT1b replicons as a scaffold for inserting sequences derived from patient isolates of various genotypes. Chimeric replicon systems containing NS5B, either alone or in conjunction with NS5A, from GT1a and GT1b patients were established to determine the antiviral activity spectrum of HCV polymerase inhibitors in the GT1 patient population (46). An intergenotypic chimeric replicon containing the NS5B gene from a GT2b clinical isolate in the GT1b backbone was also developed to enable the determination of susceptibility of GT2b to HCV polymerase inhibitors (9). However, generation of intergenotypic chimeric replicons that contain polymerase sequences derived from GT3, -4, -5, and -6 patient isolates has not been reported.
HCV polymerase has been the target of active research and development efforts on novel anti-HCV therapies. As is the case with other polymerases, the three-dimensional structure of the HCV polymerase resembles a right hand, complete with finger, palm, and thumb domains (1, 3, 25, 27). Several nucleoside analogues that bind to the catalytic site of the polymerase, such as NM283, R-1626, and R-7128, have been shown to be potential anti-HCV therapies (4, 39). Nonnucleoside polymerase inhibitors (NNIs) represent a more diverse class of inhibitors that bind to distinct allosteric sites in the palm, thumb, and finger domains (6, 17, 22, 23). The NNI1 (benzimidazole) and NNI2 (thiophene carboxylic acid) binding sites reside in the thumb domain, while the NNI3 (benzothiadiazine) and NNI4 (benzofuran) sites are in the palm domain of the polymerase. Since the amino acid sequences of these binding sites are more variable across genotypes than the active site, understanding of the broad-spectrum activity would be a critical step in the development of NNIs as effective HCV therapies.
In the present study, we describe the construction and characterization of an intergenotypic HCV chimeric replicon panel bearing non-GT1 polymerase sequences in a GT1b replicon backbone. The NS5B polymerase sequences were isolated from patient samples infected with GT2b, -3a, -4a, -5a, or -6a and used to replace the corresponding sequence in a highly active GT1b Con-1 reporter replicon system (12). Due to a significant reduction in the replication fitness of these chimeric replicons in transient assays, stable cell lines were created from the replication-competent intergenotypic chimeric replicons for use in susceptibility assays. All replicons showed similar susceptibilities to IFN-α and an NS3 protease inhibitor but had various levels of susceptibility to compounds that target different allosteric binding sites (NNI1 through NNI4) on NS5B. The effect of amino acid substitutions at residues 419 and 482 of the polymerase on the antiviral activity of inhibitors binding to the thumb domain (NNI1 and NNI2 sites) was also examined. Our results showed differences in activity spectrum for inhibitors that target different regions of the enzyme and indicate that the intergenotypic chimeric replicons can serve as useful tools for the discovery and development of HCV polymerase inhibitors with broad-spectrum activity.
MATERIALS AND METHODS
Cell lines and plasmids.
The Huh7.5 cell line was obtained from Apath LLC (St. Louis, MO). Cells were propagated in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 50 IU/ml penicillin, and 50 μg/ml streptomycin sulfate (Invitrogen) at 37°C with 5% CO2. All stable replicon lines were cultured using the same conditions plus the addition of 250 μg/ml G418 (Invitrogen). The GT1b Con-1 reporter replicon construct, pBB7M4 hRLuc, was constructed as described previously (12). The pBS.KS plasmid was purchased from Stratagene (La Jolla, CA).
Compounds.
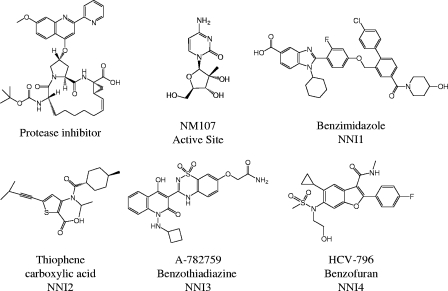
The NS3 protease inhibitor (8) and polymerase inhibitors methylcytidine (NM107) (34), benzimidazole (13), thiophene carboxylic acid (47), benzothiadiazine (A-782759) (31), and benzofuran (HCV-796) (18) were synthesized by external vendors according to literature procedures. The structures are shown in Fig. 1.
FIG. 1.
Structures of HCV protease and polymerase inhibitors used in this study.
Extraction, amplification, and sequence analysis of patient NS5B sequences.
Plasma of HCV patients infected with GT2b, -3a, -4a, -5a, and -6a were purchased from Cliniqa (San Diego, CA) and Teragenix (Fort Lauderdale, FL) corporations. Viral RNA was extracted from 540 μl of each sample using the viral RNA mini kit (Qiagen, Valencia, CA), following the manufacturer's protocol. Reverse transcription (RT) was performed using random hexamers (IDT, Coralville, OH) and Thermoscript reverse transcriptase (Invitrogen), following the manufacturer's protocol. Two rounds of PCR were performed on the resulting cDNA using the AccumPrime Taq HiFi kit (Invitrogen) and 35 cycles of the following conditions: 94°C for 45 s, 55°C for 1 min, and 72°C for 3 min. The first round of amplification was carried out in two reaction mixtures, each containing a cocktail of primers complementary to the end of the NS5A gene and the variable region of the 3′ untranslated region (UTR) (see supplemental material for primer sequences). These primers (A through D; var 1 and var 2) were designed for each genotype based on the consensus sequences generated from 2 to 14 of the published full-length polymerase sequences available in GenBank and the Los Alamos databases. Forward primers A through D were designed from the consensus sequence for each genotype. Two reverse primers were designed from the variable region sequence for each genotype (var 1 and var 2) to cover the sequence immediately downstream of the stop codon. Since sequence information for the GT6a variable region was not available, var 1 and var 2 from GT3 and GT5 were utilized in the PCRs for GT6a. The PCR products of the first-round reaction mixtures were combined and subjected to nested PCR using the two most internal primers, D and var 1. The final product was cloned into the pGEM vector by TA cloning using the pGEM-T Easy II kit (Promega, Madison, WI). Sequence analysis was performed on 10 clones for each patient, and the clone with the highest sequence homology to the patient consensus sequence was utilized for chimeric replicon construction. The amplified sequences used in this study are available in the supplemental material.
Construction of cloning vectors and chimeric replicons.
The StuI or SnaBI restriction enzyme site was introduced into the variable region of the 3′ UTR of pBB7M4 hRLuc using the QuikChange mutagenesis protocol (Stratagene) and primer pairs StuI.for and StuI.rev or SnaBI.for and SnaBI.rev (see supplemental material for primer sequences) to create pBB7M4 hRLuc.StuI or pBB7M4 hRLuc.SnaBI, respectively. QuikChange reactions were also carried out to introduce amino acid changes L419I and I482L in the NS5B of the pBB7M4 hRLuc replicon construct using primer pairs L419I.for and L419I.rev and I482L.for and I482L.rev to generate pBB7M4 hRLuc.L419I and pBB7M4 hRLuc.I482L, respectively. A replicon cDNA containing both mutations (pBB7M4 hRLuc.L419I + I482L) was constructed through consecutive mutagenesis reactions. All mutations were confirmed by sequence analysis.
The 84-nucleotide hepatitis delta virus (HDV) ribozyme sequence (42) was introduced into pBB7M4 hRLuc.StuI and pBB7M4 hRLuc.SnaBI immediately downstream of the HCV sequence so that replicon RNA transcripts with an exact 3′ terminus of the 3′ UTR could be generated by self cleavage. The incorporation of the ribozyme was accomplished with two rounds of PCRs using Herculase DNA polymerase and PCR primers rib1.for and rib1.rev for the first round and rib2.for and rib2.rev for the second round. Both rounds of amplification were carried out for 35 cycles consisting of 95°C for 60 s, 55°C for 90 s, and 72°C for 90 s. The first-round PCR generated the fusion between a sequence from a unique NcoI site in NS5B to the end of 3′ UTR and the 5′ end sequence of the ribozyme. The resulting fragment was used as a template for the second round of PCR, which fused the HDV ribozyme sequence onto the HCV 3′ UTR. The final product was cloned into pBB7M4 hRLuc at the unique NcoI and SpeI sites. These constructs are referred to as pBB7M4 hRLuc.StuI.HDV and pBB7M4 hRLuc.SnaBI.HDV. Shuttle vectors were created for precise cloning of the patient NS5B sequences by transferring the 900 bases of the 3′ end of NS5A, the entire NS5B gene, the 3′ UTR, and the HDV ribozyme from these replicons to pBS.KS at the unique EcoRI and SpeI restriction enzyme sites. These constructs are referred to as pBS.KS.StuI and pBS.KS.SnaBI.
To avoid the possible interference of the non-GT1 NS5A and 3′ UTR sequences with the replication fitness of Con-1-based chimeric replicons and their susceptibilities to polymerase inhibitors, only the NS5B region of the patient sequence was introduced into the chimeric replicons by the molecular technique of splicing by overlap extension (SOEing) PCR (16). The 900-nucleotide region encompassing the unique EcoRI site to the 3′ end of the NS5A sequence was amplified using the PCR conditions described above with the primers Eco-5A.for and Eco-5A.rev. The patient NS5B sequences were amplified from the chosen TA clone, using the same PCR conditions, with the 5B.for and 5B.rev primers specific for each genotype. The 5B.for primer is complementary to the start of the NS5B gene for the corresponding patient with an additional 20 nucleotides at the 5′ end, which are complementary to the 3′ end of the Con-1 NS5A gene. The 5B.rev primer is complementary to the 3′ end of the patient NS5B sequence with either StuI or SnaBI and a short stretch of sequences complementary to the variable region (5 or 30 bases, respectively). The two PCR products were joined by SOEing PCR with Eco-5A.for and Eco-5B.rev using the same PCR conditions. The resulting product was cloned into the appropriate pBS.KS shuttle vector at the EcoR1 and StuI (or SnaBI) sites. The chimeric sequence, along with the Con-1 3′ UTR and the HDV ribozyme, was subcloned from the pBS.KS shuttle vector to the subgenomic Con-1 replicon, pBB7M4 hRLuc, at the unique EcoR1 and SpeI restriction sites (12).
Stable cell line generation.
Subgenomic replicon RNA was transcribed in vitro from replicon constructs using the MEGAscript T7 kit (Ambion, Austin, TX), following the manufacturer's protocol. Replicon transcripts (1 to 4 μg) were electroporated into 8 × 106 Huh7.5 cells in a 0.4-cm cuvette using a Bio-Rad Gene Pulser II electroporator (Bio-Rad, Hercules, CA) at 270 V and 950 μF. Cells were resuspended in 10 ml growth medium and seeded in three 10-cm plates per construct at 1, 3, or 5 ml per plate. After 3 weeks of selection with 250 μg/ml of G418, colonies were picked and seeded in 96-well plates. Cells were transferred to wells with a greater surface area when confluence was reached, approximately every 2 to 4 days. Once cells reached confluence in T-25 flasks, the RLuc reporter activity was measured, and the cell line with the highest activity was scaled up for use in antiviral assays.
Transient and stable HCV replication assay.
Huh7.5 cells were electroporated with 10 μg of in vitro replicon transcript using the electroporation conditions described above, with the exception that 220 V was used. Electroporated cells were resuspended in Dulbecco's modified Eagle's medium (Invitrogen), seeded in 96-well plates at 6 × 104 cells per well and given a 4-h recovery period. Stable cell lines were seeded at 2e4/well and allowed to settle for 30 min. After the recovery period, compounds were added to cells and incubated for 3 days at 37°C. Compounds were tested at half-log serial dilutions over a range of concentrations with appropriate solvent controls (compound free). HCV RNA replication was monitored by the hRLuc reporter activity in the transient or stable cell line assay using the RLuc luciferase reporter kit (Promega), following the manufacturer's instructions in a Perkin Elmer 1450 MicroBeta JET (Perkin Elmer, Wellesley, MA). Fifty percent effective concentration (EC50) values were calculated as the concentration of compound that effected a decrease in viral RNA replication (as measured by hRLuc activity) in compound-treated cells to 50% of that produced in compound-free cells. The values were determined by nonlinear regression analyses.
RESULTS
Divergence of non-GT1 HCV polymerase sequences.
For each genotype, two to five patient plasma samples were subjected to RT-PCR, and one to three products were successfully amplified for further analysis (Table 1). Despite the limited number of sequences available and the lack of information for the GT6a variable region, we successfully amplified NS5B sequences from all five genotypes. To examine the quasispecies population of each non-GT1 patient NS5B amplified, the RT-PCR products were cloned into a TA vector, and 10 clones were sequenced for each patient. The percent divergence between clones of the same patient ranged from 0.2 to 3.5% at the nucleotide level and from 0 to 3.3% at the amino acid level, irrespective of the genotypes examined. The variation among the consensus sequences of the three GT2b patients ranged from 5 to 7% at the nucleotide level and 2 to 3.5% at the amino acid level, while variation between the two GT4a patients sequenced was 7% at the nucleotide level and 5% at the amino acid level. Variation between the two GT3a or GT5a sequences was only 0.2% at both the nucleotide and amino acid levels. These results indicate that the NS5B sequences from GT3a and GT5a patient samples utilized in this study were less diverse than those from the GT2b and GT4a patients.
TABLE 1.
Amplification and analysis of NS5B sequences from patient plasma samplesa
| GT | No. of samples subjected to RT-PCR | No. of samples amplified by RT-PCR | Divergence of the NS5B sequences from Con-1b
|
|
|---|---|---|---|---|
| % Nucleotide divergence | % Amino acid divergence | |||
| 2b | 5 | 3 | 31 | 26 |
| 3a | 2 | 2 | 28 | 24 |
| 4a | 5 | 2 | 29 | 23 |
| 5a | 2 | 2 | 28 | 21 |
| 6a | 3 | 1 | 27 | 21 |
RNA was extracted from patient plasma samples using the Qiagen viral RNA mini kit. RT-PCR was performed on each sample to amplify the NS5B gene of HCV. The number of samples successfully amplified varies by genotype.
Sequence comparison was performed on the patient NS5B sequences used to construct the replication-competent chimeric replicons that produced antiviral results shown in this study (except for GT6a).
A consensus sequence was derived from 10 clones for each patient, and the one closest to the consensus sequence derived from published sequences for that subtype was chosen for chimeric replicon construction. This method was used to most closely represent the patient sequence and to afford the chimera the best possible replication advantage. Figure 2 shows the alignment of the non-GT1 HCV NS5B amino acid sequences utilized in this study. Compared to Con-1, the divergence at the nucleotide and amino acid levels of the patient NS5B sequences used in this study ranged between 27 and 31% and 21 and 26%, respectively (Table 1).
FIG. 2.
Alignment of the HCV polymerase sequences utilized in this study. Amino acid residues 419 and 482, located in the thumb domain of the polymerase (25), are highlighted in gray boxes. The key amino acid residues that are present in the active and NNI-binding sites are also highlighted.
Replication fitness of intergenotypic NS5B chimeric replicons.
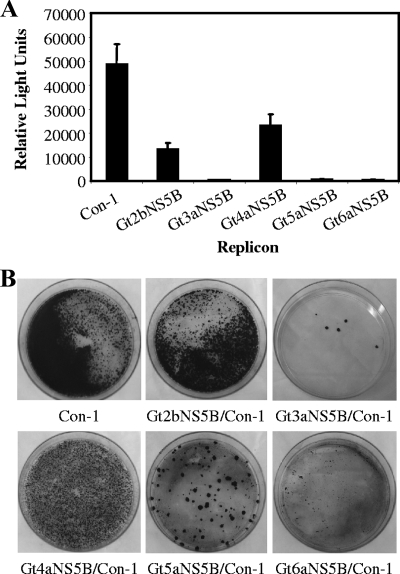
To examine the replication fitness of the chimeric replicons, in vitro-transcribed RNA was electroporated into Huh7.5 cells, and the replicon RLuc reporter activity was used as an indicator of replication activity (Fig. 3A). The results showed a 60 to 80% reduction in replication activity for GT2b and GT4a chimeras, compared to Con-1, and a nearly background level of replication activity for GT3a, -5a and -6a. Similar levels of reduction in replication fitness were also observed in a colony formation assay of Huh7.5 cells electroporated with the intergenotypic chimeras (Fig. 3B). The number of colonies produced by the GT2b and GT4a NS5B chimeras was reduced by approximately 50% and 30%, respectively, compared to the Con-1 replicon. Both GT3a and GT5a formed stable colonies despite the nearly background levels of replication in the transient assay, but the total number of colonies was dramatically reduced compared to those for Con-1 by approximately 80 and 98%, respectively. A few colonies were observed for the GT6a chimeric replicon, but none survived to become stable cell lines. The significant reduction in the replication fitness of chimeric replicons containing intergenotypic polymerase sequences was consistent with previous reports (9, 28).
FIG. 3.
Replication fitness of intergenotypic chimeric replicons in transient replication (A) and colony formation (B) assays. (A) HCV reporter replicon constructs were electroporated into Huh7.5 cells, seeded in 96-well plates at 6 × 104 cells/well, and cultured for 3 days before RLuc activity was measured. Results represent the averages of three to five individual experiments, and error bars represent standard deviations. (B) 8 × 106 Huh7.5 cells were electroporated with 1 μg reporter replicon RNA and seeded in 10-cm dishes. Colonies were selected for 3 weeks in 250 μg/ml of G418 and stained with crystal violet.
Generation of stable cell lines.
On account of the low level of replication observed for the intergenotypic chimeric replicons in the transient replication assay, stable cell lines were isolated and scaled up for use in susceptibility assays. Between 5 and 10 individual lines were expanded for each genotype, and the line with the highest RLuc activity and optimal growth kinetics was used for antiviral activity determinations of known HCV inhibitors. With the exception of GT6a, stable cell lines with high reporter activities (>120,000 relative light units) were obtained for all intergenotypic chimeric replicons. To identify any amino acid changes that may have allowed for cell culture adaptation and increases in the replication efficiency of the chimeric replicons, the NS5B region was amplified and sequenced from the total cellular RNA of the stable lines. No nucleotide sequence changes that resulted in amino acid changes were observed in the NS5B region of any of the chimeric replicon cell lines. It is possible, however, that sequence changes may have occurred outside of the NS5B region. Our attempt in generating a stable cell line from the GT6a NS5B chimeric replicon was not successful. Another chimera was generated with a different NS5B sequence from the same patient, but the alternative sequence did not result in increased replication efficiency or the successful generation of a stable cell line.
In vitro antiviral activities of HCV inhibitors against intergenotypic chimeric replicons.
Susceptibility assays were carried out for stable lines containing the non-GT1 NS5B chimeric replicons in order to determine the in vitro antiviral activities of HCV inhibitors. As expected, there were no significant changes in the antiviral activity of IFN or a macrocyclic protease inhibitor (Fig. 1) against any of the intergenotypic chimeric replicons compared to that against Con-1 (Table 2). The protease inhibitor had a 112-fold reduction in activity against the GT2a JFH1 subgenomic replicon, which is consistent with previously reported results (41, 45).
TABLE 2.
In vitro antiviral activity of HCV inhibitors against intergenotypic chimeric repliconsa
| Replicon construct | RLuc count (103) | Macrocyclic protease inhibitor
|
IFN-α
|
Nucleoside active site
|
Benz-imidazole NNI1
|
Thiophene carboxylic acid NNI2
|
Benzothiadiazine NNI3
|
Benzofuran NNI4
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (μM) | FC | EC50 (IU/ml) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | EC50 (μM) | FC | ||
| GT1b, Con-1 | 490 | 0.0066 | 1.3 | 0.91 | 0.90 | 0.025 | 0.033 | 0.042 | |||||||
| GT1a, H77 | 450 | 0.020 | 3 | 1.1 | 1 | 0.53 | 1 | 0.49 | 1 | 0.02 | 1 | 0.14 | 4 | 0.038 | 1 |
| GT2a, JFH1 | 318 | 0.74 | 112 | 1 | 1 | 3.6 | 4 | 6.9 | 8 | 6.1 | 244 | >32 | >970 | 0.13 | 3 |
| GT2bNS5B, Con-1 | 370 | 0.026 | 4 | 0.48 | 0.4 | 1.3 | 1 | 30 | 33 | >32 | >1,280 | >32 | >970 | 0.055 | 1 |
| GT3aNS5B, Con-1 | 120 | 0.022 | 3 | 0.23 | 0.2 | 2.9 | 3.2 | 0.21 | 0.2 | >32 | >1,280 | >32 | >970 | 0.051 | 1 |
| GT4aNS5B, Con-1 | 500 | 0.053 | 8 | 2.5 | 2 | 1.4 | 2 | 1.5 | 2 | 2.2 | 88 | 25 | 758 | 0.069 | 2 |
| GT5aNS5B, Con-1 | 350 | 0.023 | 3 | 0.62 | 0.5 | 1.8 | 2 | 0.16 | 0.2 | 0.2 | 8 | 5 | 152 | 0.0032 | 0.1 |
The susceptibilities of the chimeric replicon cell lines to HCV inhibitors were evaluated in the reporter replicon assay. Cells were exposed to compounds for 3 days before RLuc activity was determined. Results represent the averages of duplicate values. FC, fold change in EC50 over that of the wild-type Con-1 replicon.
A set of polymerase inhibitors that bind to different regions of the enzyme (Fig. 1) was tested against the intergenotypic chimeric replicon panel in stable cell lines (Table 2) in order to understand the broad-spectrum activity of these compounds. The nucleoside polymerase inhibitor, NM107, exhibited similar antiviral activity across all genotypes evaluated (fold change values, ≤4), consistent with the notion that nucleoside inhibitors targeting the active site have broad-spectrum activities against polymerases of different genotypes. A polymerase inhibitor from the benzimidazole series that interacts with the NNI1-binding site of the enzyme displayed an 8- to 33-fold reduction in activity against GT2a and GT2b compared to Con-1, but no reduction in activity against GT3a, 4a, and 5a. Another NNI, a thiophene carboxylic acid that binds to the NNI2 pocket, showed a modest reduction in antiviral activity against GT5a (8-fold) but lost considerable activity against GT4a (88-fold), -2b, and -3a (>1,280-fold). The benzothiadiazine polymerase inhibitor that binds to the NNI3 region, A-782759, showed a significant reduction in activity against all non-GT1 chimeric replicons, with fold reductions in EC50 values ranging from 152 to >970. The benzofuran inhibitor that binds to the NNI4 pocket, HCV-796, exhibited comparable antiviral activity against all GT1 and non-GT1 replicons tested (Table 2). The evaluation of HCV polymerase inhibitors against intergenotypic chimeric replicons demonstrates differences in the antiviral activity spectrum for inhibitors that target different regions of the polymerase enzyme. It appears that inhibitors that target either the active site or the NNI4 pocket evaluated in this study may have a broader activity spectrum against HCV genotypes.
Mechanism of reduced activity of NNI2 inhibitors against non-GT1 polymerases.
The thiophene carboxylic acid compound tested in this study that binds to the thumb base site (NNI2) had reduced antiviral activity against all non-GT1 polymerase chimeric replicons with the exception of GT5. Analysis of the NNI2-binding site sequences available from GenBank and the Viral Bioinformatics Resource Center revealed that amino acid residues L419 and I482 were present in 99.4% of 792 GT1 and 100% of three GT5a NS5B sequences. L419 and I482 were substituted with I419 and L482, respectively, in 92.3% of 78 GT2 and 100% of 11 GT3, 21 GT4, and 68 GT6 NS5B sequences. Furthermore, L419 and I482 have been previously shown by crystallography studies to be among the amino acid residues that define the NNI2-binding pocket in a GT1 polymerase (27). To examine whether these residue changes could affect the susceptibilities of replicons to inhibitors that target the thumb site, L419I and I482L were introduced into the NS5B region of the GT1b Con-1 reporter replicon either individually or in combination. The mutant replicons were tested for their susceptibilities to IFN, the benzimidazole (NNI1), and the thiophene carboxylic acid (NNI2) compounds in a transient replication assay. While no change in susceptibility to IFN was observed, the NNI2 binder displayed a 221- and 150-fold loss in activity against replicons containing L419I and I482L, respectively (Table 3). Furthermore, a greater than 2,000-fold reduction in the activity of this compound was observed when both substitutions were present. These results are consistent with the observation that the GT1b subgenomic replicon and the GT5a NS5B chimeric replicon that contain L419 and I482 are more susceptible to the thiophene carboxylic acid compound than the GT2b, -3a, and -4a NS5B chimeric replicons that have I419 and L482 instead (Table 3). These results suggest that the I419 and L482 residues are largely responsible for the resistance of GT2b and GT4a chimeric replicons to this NNI2 binder. In addition, the I419 and L482 changes also resulted in a small decrease (eightfold) in susceptibility of the replicon to the benzimidazole compound, which binds to the tip of the thumb domain (NNI1) on the other side of the helix (22). In conclusion, our results show that the 419 and 482 residues of NS5B play an important role in determining the susceptibilities of HCV polymerases to certain inhibitors binding to either the NNI1 or NNI2 pocket.
TABLE 3.
Effect of L419I and I482L substitutions on the susceptibility of the Con-1 replicon to NNIsa
| Construct | Fold change in EC50 over the wild-type Con-1 replicon
|
|||||
|---|---|---|---|---|---|---|
| NM107 nucleoside active site | Benzimidazole NNI1 | Thiophene carboxylic acid NNI2 | A-782759 benzothiadiazine NNI3 | HCV-796 benzofuran NNI4 | IFN-α | |
| L419I | 1 ± 0 | 4 ± 2 | 326 ± 150 | 0.2 ± 0.0 | 2 ± 0 | 1 ± 0 |
| I482L | 1 ± 0 | 3 ± 1 | 201 ± 72 | 0.5 ± 0.1 | 1 ± 0 | 1 ± 0 |
| L419I and I482L | 1 ± 1 | 8 ± 0 | 2,238 ± 217 | 0.2 ± 0.1 | 3 ± 0 | 2 ± 0 |
The susceptibilities of the L419I and I482L mutant replicons to HCV inhibitors were evaluated in the transient replicon assay. Replicon RNA was introduced into Huh7.5 cells and exposed to compounds for 3 days before RLuc activity was determined. Results represent the mean ± the standard deviation of two or three independent experiments.
DISCUSSION
In the present study, chimeric replicons that contained the NS5B genes from patients infected with GT2b, -3a, -4a, -5a, and -6a in the GT1b Con-1 backbone were constructed. The fitness of the chimeras was significantly impaired in comparison to the Con-1 replicon in both transient and colony formation assays. The antiviral activities of inhibitors binding to each of the four allosteric binding pockets of the polymerase (Fig. 4) were determined using stable cell lines created from each of the replication-competent non-GT1 NS5B chimeras. This evaluation revealed differences in the activity spectrum for specific inhibitors that target different regions of the polymerase enzyme (Table 2). This study further validates the utility of chimeric replicons for broad-spectrum antiviral activity determination of HCV polymerase inhibitors.
FIG. 4.
The NNI-binding sites of the HCV polymerase. The X-ray crystal structure of the 1b-BK NS5BΔ21 protein is shown (27, 36), with protein backbone in a ribbon structure and the key amino acid residues that are present in the active and NNI-binding sites indicated and highlighted in yellow. Amino acid residues L419 and I482, located in the thumb domain of the polymerase, are also indicated.
Previous studies have utilized chimeric replicons constructed using GT1b as a scaffold for inserting sequences amplified from infected patient samples. Systems expressing polymerase sequences from GT1 and GT2 patients have been previously reported (9, 24, 46). In particular, chimeric replicons with NS5B from GT1a and GT1b patients were developed to determine the antiviral activities of clinical candidates against a panel of GT1 clinical isolates (28, 30, 46). Therefore, chimeric replicons may be a useful tool for identifying broad-spectrum HCV inhibitors of non-GT1 targets that have not yet been developed as full-length replicons. One caveat to these previous studies is that the chimeric replicons often contain additional patient-derived sequences flanking NS5B, which may complicate interpretations of results for NS5B inhibitors. In addition, only a limited number of genotypes were evaluated in previous studies. To date, there have been no reports of GT4a or GT5a NS5B chimeric replicons. This study describes for the first time a more comprehensive panel of chimeric replicons that contains patient-derived NS5B sequences representing GT1 to GT5. Furthermore, the chimeric replicons generated for the current study contain exact NS5B-coding region replacements, allowing for the interrogation of NS5B inhibitors without the influence of adjacent sequences derived from patient isolates. Our expectation is that this may allow for better correlations of activity with biochemical assays. Alternatively, one limitation of the chimeric replicon system presented here or in previous studies is that any differential effects on antiviral activity of RNA-protein or protein-protein interactions that occur between the patient-derived NS5B and Con-1 RNA proteins in the context of replication complexes cannot be fully evaluated.
Clonal sequencing analysis of the NS5B region was performed on 10 clones from each patient sample. Although the sample set examined was relatively small, it is clear that the divergence between GT1 and non-GT1 NS5B amino acid sequences ranged from 25 to 31%, consistent with those previously reported in the literature. The GT6a NS5B is more closely related to the Con-1 sequence than to the other non-GT1 sequences, but the GT6a chimera was not able to maintain the level of replication needed to create a stable cell line. The GT3a and GT5a chimeras also had severely impaired fitness, as shown in the transient replication and colony formation assays. In contrast, the GT2b and GT4a chimeras reached viable levels of replication even in the transient assay and produced a large number of colonies compared to other genotypes. The GT4a chimera had the highest level of replication fitness despite the relatively higher divergence of GT4a NS5B from the Con-1 sequence. These results suggest that specific amino acid residues of the non-GT1 NS5B sequence, rather than its overall sequence similarity to Con-1, may determine the fitness of the intergenotypic chimeric replicons. Despite the low replication fitness observed in transient assays, stable cell lines that produced high levels of reporter activity were successfully generated for all chimeric replicons, except for GT6a (Table 2). Two different patient sequences were tested for stable cell line generation for both GT3a and GT5a chimeras, and only one from each genotype was replication competent. These results show that not all patient NS5B sequences are viable in the Con-1 backbone due to sequence incompatibility. The NS5B sequence from two clones of the only GT6a patient successfully amplified were used to generate chimeric replicons, but neither of these sequences supported stable cell line formation under various conditions. Future attempts to establish this chimera would include obtaining more GT6a patient samples or cloning the GT6a NS5B sequences into a different replicon backbone, such as the GT1b BK and the GT2a JFH1 subgenomic replicons (28).
Four allosteric binding pockets have been identified in the HCV polymerase protein (Fig. 4), and several chemical series have been shown to interact with each of these pockets by in vitro resistance and crystallographic studies (23, 27, 33). The results in Table 2 clearly show that the NNI1 binder tested in this study has comparable activity against GT1, -3a, -4a, and -5a but reduced activity against GT2a and GT2b, consistent with results generated in enzymatic assays. No variations were observed in any of the non-GT1 chimeras tested at amino acid residue 495, which was shown to be the hot spot for resistance changes (22, 44). The thiophene carboxylic acid compound that binds to the NNI2 pocket showed significantly reduced activity against the non-GT1 polymerases tested as indicated by up to >1,280-fold changes in the EC50 values compared to Con-1. However, the NNI2 binder retained considerable activity against GT5a with only an eightfold change in EC50. An alignment of the patient sequences utilized in this study showed that GT2, -3a, and -4a encoded Ile and Leu residues at NS5B positions 419 and 482, respectively, while the Con-1 and GT5a sequences encoded Leu and Ile residues at positions 419 and 482, respectively. When the L419I and I482L substitutions were introduced into the Con-1 replicon and tested in a transient assay, shifts in antiviral activity similar to those measured for GT2, -3a, and -4a were observed. These results suggest that the I419 and L482 residues are responsible for the loss in antiviral activity of this compound against GT2, -3a, and -4a polymerase sequences. Compounds that bind to the NNI3 pocket have been shown to interact with the M414 residue by in vitro resistance studies (31, 33). Instead of M414, which was observed in GT1 and GT5a NS5B, the GT2 and GT4a NS5B sequences used in this study contain Gln and Val at amino acid position 414, respectively, which may contribute to the large fold changes in activity seen with A-782759 against the GT2b and GT4a chimeras. The GT3a NS5B has a Met at residue 414, and the basis of the >970-fold reduction in activity against this chimera with A-782759 is not clear at this time. In all cases, the resistance mutations described here are likely to be only one of the factors contributing to the changes in antiviral activity seen with the intergenotypic chimeras. HCV-796 showed broad-spectrum activity with a less than threefold decrease in antiviral activity against all genotypes tested. It has been previously reported that naturally occurring polymorphism at residue 316 could result in differential susceptibility to HCV-796 (36). In all sequences used here, the amino acid at residue 316 is a Cys. Since only one chemotype for each binding site was tested in this study, we cannot rule out that different chemical classes targeting the same binding site may behave differently.
The evaluation of HCV polymerase inhibitors against intergenotypic chimeric replicons showed differences in activity spectrum for specific inhibitors that target different regions of the polymerase enzyme. The results presented in this study suggest that evaluating the antiviral activities of HCV polymerase inhibitors against a panel of non-GT1 NS5B chimeric replicons can give insight into the relationship between sequence changes and resistance mutations for compounds that bind to the four different allosteric pockets of the polymerase protein. This study demonstrates the utility of chimeric replicons for broad-spectrum activity determination of HCV inhibitors.
Supplementary Material
Acknowledgments
We acknowledge all members of the HCV polymerase project team at Pfizer PGRD La Jolla Laboratories. We are grateful to members of the virology department, particularly Wade Blair, for their scientific input and critical review of the paper. We also thank student assistants Ashley Roth and Amy Chan for their dedication and hard work.
Footnotes
Published ahead of print on 11 August 2008.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll, S. S., and D. B. Olsen. 2006. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets 6:17-29. [DOI] [PubMed] [Google Scholar]
- 5.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Marco, S., C. Volpari, L. Tomei, S. Altamura, S. Harper, F. Narjes, U. Koch, M. Rowley, R. De Francesco, G. Migliaccio, and A. Carfi. 2005. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. J. Biol. Chem. 280:29765-29770. [DOI] [PubMed] [Google Scholar]
- 7.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 8.Goudreau, N., C. Brochu, D. R. Cameron, J. S. Duceppe, A. M. Faucher, J. M. Ferland, C. Grand-Maitre, M. Poirier, B. Simoneau, and Y. S. Tsantrizos. 2004. Potent inhibitors of the hepatitis C virus NS3 protease: design and synthesis of macrocyclic substrate-based beta-strand mimics. J. Org. Chem. 69:6185-6201. [DOI] [PubMed] [Google Scholar]
- 9.Graham, D. J., M. Stahlhut, O. Flores, D. B. Olsen, D. J. Hazuda, R. L. Lafemina, and S. W. Ludmerer. 2006. A genotype 2b NS5B polymerase with novel substitutions supports replication of a chimeric HCV 1b:2b replicon containing a genotype 1b NS3-5A background. Antivir. Res. 69:24-30. [DOI] [PubMed] [Google Scholar]
- 10.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 12.Hao, W., K. J. Herlihy, N. J. Zhang, S. A. Fuhrman, C. Doan, A. K. Patick, and R. Duggal. 2007. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob. Agents Chemother. 51:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto, H., K. Mizutani, and A. Yoshida. July 2001. Fused-ring compounds and use thereof as drugs. WO 01/47883.
- 14.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. USA 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67:4665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, R. M. 1997. In vitro recombination and mutagenesis of DNA. SOEing together tailor-made genes. Methods Mol. Biol. 67:141-149. [DOI] [PubMed] [Google Scholar]
- 17.Howe, A. Y., H. Cheng, I. Thompson, S. K. Chunduru, S. Herrmann, J. O'Connell, A. Agarwal, R. Chopra, and A. M. Del Vecchio. 2006. Molecular mechanism of a thumb domain hepatitis C virus nonnucleoside RNA-dependent RNA polymerase inhibitor. Antimicrob. Agents Chemother. 50:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe, A. Y. M., H. Cheng, S. Johann, S. Mullen, S. K. Chunduru, D. C. Young, J. Bard, and R. Chopra. 2006. Identification and characterization of HCV replicon variants with reduced susceptibility to HCV-796, abstr. 717. Abstr. 13th Int. Meet. Hepat. C Virus Relat. Viruses.
- 19.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 21.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukolj, G., G. A. McGibbon, G. McKercher, M. Marquis, S. Lefebvre, L. Thauvette, J. Gauthier, S. Goulet, M. A. Poupart, and P. L. Beaulieu. 2005. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280:39260-39267. [DOI] [PubMed] [Google Scholar]
- 23.Lemm, J. A., M. Liu, R. E. Rose, R. Fridell, D. R. O'Boyle II, R. Colonno, and M. Gao. 2005. Replication-competent chimeric hepatitis C virus subgenomic replicons. Intervirology 48:183-191. [DOI] [PubMed] [Google Scholar]
- 24.Le Pogam, S., H. Kang, S. F. Harris, V. Leveque, A. M. Giannetti, S. Ali, W. R. Jiang, S. Rajyaguru, G. Tavares, C. Oshiro, T. Hendricks, K. Klumpp, J. Symons, M. F. Browner, N. Cammack, and I. Najera. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 30.Middleton, T., Y. He, T. Pilot-Matias, R. Tripathi, B. H. Lim, A. Roth, C. M. Chen, G. Koev, T. I. Ng, P. Krishnan, R. Pithawalla, R. Mondal, T. Dekhtyar, L. Lu, H. Mo, W. M. Kati, and A. Molla. 2007. A replicon-based shuttle vector system for assessing the phenotype of HCV NS5B polymerase genes isolated from patient populations. J. Virol. Methods 145:137-145. [DOI] [PubMed] [Google Scholar]
- 31.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, M. H., and E. B. Keeffe. 2005. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin. Gastroenterol. Hepatol. 3:S97-S101. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierra, C., A. Amador, S. Benzaria, E. Cretton-Scott, M. D'Amours, J. Mao, S. Mathieu, A. Moussa, E. G. Bridges, D. N. Standring, J. P. Sommadossi, R. Storer, and G. Gosselin. 2006. Synthesis and pharmacokinetics of valopicitabine (NM283), an efficient prodrug of the potent anti-HCV agent 2′-C-methylcytidine. J. Med. Chem. 49:6614-6620. [DOI] [PubMed] [Google Scholar]
- 35.Seeff, L. B. 1995. Natural history of viral hepatitis, type C. Semin. Gastrointest. Dis. 6:20-27. [PubMed] [Google Scholar]
- 36.Shi, S. T., K. J. Herlihy, J. P. Graham, S. A. Fuhrman, C. Doan, H. Parge, M. Hickey, J. Gao, X. Yu, F. Chau, J. Gonzalez, H. Li, C. Lewis, A. K. Patick, and R. Duggal. 2008. In vitro resistance study of AG-021541, a novel nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 52:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simmonds, P., E. C. Holmes, T. A. Cha, S. W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 38.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap, J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053-1061. [DOI] [PubMed] [Google Scholar]
- 39.Summa, V., A. Petrocchi, V. G. Matassa, M. Taliani, R. Laufer, R. De Francesco, S. Altamura, and P. Pace. 2004. HCV NS5b RNA-dependent RNA polymerase inhibitors: from α,γ-diketoacids to 4,5-dihydroxypyrimidine- or 3-methyl-5-hydroxypyrimidinonecarboxylic acids. Design and synthesis. J. Med. Chem. 47:5336-5339. [DOI] [PubMed] [Google Scholar]
- 40.Takada, N., S. Takase, A. Takada, and T. Date. 1993. Differences in the hepatitis C virus genotypes in different countries. J. Hepatol. 17:277-283. [DOI] [PubMed] [Google Scholar]
- 41.Thibeault, D., R. Maurice, L. Pilote, D. Lamarre, and A. Pause. 2001. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276:46678-46684. [DOI] [PubMed] [Google Scholar]
- 42.Thill, G., M. Blumenfeld, F. Lescure, and M. Vasseur. 1991. Self-cleavage of a 71 nucleotide-long ribozyme derived from hepatitis delta virus genomic RNA. Nucleic Acids Res. 19:6519-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas, D. L., and L. B. Seeff. 2005. Natural history of hepatitis C. Clin. Liver Dis. 9:383-398. [DOI] [PubMed] [Google Scholar]
- 44.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong, X., Z. Guo, J. Wright-Minogue, E. Xia, A. Prongay, V. Madison, P. Qiu, S. Venkatraman, F. Velazquez, F. G. Njoroge, and B. A. Malcolm. 2006. Impact of naturally occurring variants of HCV protease on the binding of different classes of protease inhibitors. Biochemistry 45:1353-1361. [DOI] [PubMed] [Google Scholar]
- 46.Tripathi, R. L., P. Krishnan, Y. He, T. Middleton, T. Pilot-Matias, C. M. Chen, D. T. Lau, S. M. Lemon, H. Mo, W. Kati, and A. Molla. 2007. Replication efficiency of chimeric replicon containing NS5A-5B genes derived from HCV-infected patient sera. Antivir. Res. 73:40-49. [DOI] [PubMed] [Google Scholar]
- 47.Wunberg, T., J. Baumeister, D. Gottschling, K. Henninger, D. Koletzki, J. Pernerstorfer, A. Urban, A. Birkmann, A. Harrenga, and M. Lobell. July 2006. Alkinyl-substituted thiophenes. WO 2006/072348.
- 48.Yi, M., and S. M. Lemon. 2004. Adaptive mutations producing efficient replication of genotype 1a hepatitis C virus RNA in normal Huh7 cells. J. Virol. 78:7904-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zein, N. N., J. Rakela, E. L. Krawitt, K. R. Reddy, T. Tominaga, D. H. Persing, et al. 1996. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann. Intern. Med. 125:634-639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.