Abstract
We have identified a developmentally essential gene, UbcB, by insertional mutagenesis. The encoded protein (UBC1) shows very high amino acid sequence identity to ubiquitin-conjugating enzymes from other organisms, suggesting that UBC1 is involved in protein ubiquitination and possibly degradation during Dictyostelium development. Consistent with the homology of the UBC1 protein to UBCs, the developmental pattern of protein ubiquitination is altered in ubcB-null cells. ubcB-null cells are blocked in the ability to properly execute the developmental transition that occurs between the induction of postaggregative gene expression during mound formation and the induction of cell-type differentiation and subsequent morphogenesis. ubcB-null cells plated on agar form mounds with normal kinetics; however, they remain at this stage for ∼10 h before forming multiple tips and fingers that then arrest. Under other conditions, some of the fingers form migrating slugs, but no culmination is observed. In ubcB-null cells, postaggregative gene transcripts accumulate to very high levels and do not decrease significantly with time as they do in wild-type cells. Expression of cell-type-specific genes is very delayed, with the level of prespore-specific gene expression being significantly reduced compared with that in wild-type cells. lacZ reporter studies using developmentally regulated and cell-type-specific promoters suggest that ubcB-null cells show an unusually elevated level of staining of lacZ reporters expressed in anterior-like cells, a regulatory cell population found scattered throughout the aggregate, and reduced staining of a prespore reporter. ubcB-null cells in a chimeric organism containing predominantly wild-type cells are able to undergo terminal differentiation but show altered spatial localization. In contrast, in chimeras containing only a small fraction of wild-type cells, the mature fruiting body is very small and composed almost exclusively of wild-type cells, with the ubcB-null cells being present as a mass of cells located in extreme posterior of the developing organism. The amino acid sequence analysis of the UbcB open reading frame (ORF) and the analysis of the developmental phenotypes suggest that tip formation and subsequent development requires specific protein ubiquitination, and possibly degradation.
INTRODUCTION
A significant amount of information has been obtained about the signal transduction pathways that control many aspects of Dictyostelium differentiation (Devreotes, 1994; Firtel, 1995). Cyclic AMP, functioning as an extracellular ligand to a seven-span/serpentine class of receptors, mediates aggregation, the developmental switch to multicellular development, and cell-type differentiation through distinct signal transduction pathways. Subsequent morphogenesis and the initiation of culmination are also believed to be regulated by cAMP functioning both as an extracellular signaling molecule and intracellularly through the activation of PKA (Williams and Morrison, 1994; Firtel, 1995). While both molecular and biochemical approaches have resulted in significant insights into these and other pathways controlling Dictyostelium differentiation, it is clear that many other regulatory networks participate in controlling differentiation of this organism.
Genetic analysis in other systems has identified ubiquitination and deubiquitination as important mechanisms controlling a diversity of cellular regulatory processes, including ubiquitin-mediated protein turnover, protein targeting, and cell fate decisions. Ubiquitin conjugating enzymes UBC/E2 are thought to be involved in substrate recognition: the multiplicity of UBC identified in a number of organisms suggests that each UBC may be important in controlling the ubiquitination of a specific subset of proteins (Hershko and Ciechanover, 1992; Varshavsky, 1992; Ciechanover, 1994). For example, a mammalian UBC/E2 protein is involved in p53 degradation; UBC6 and UBC7 are involved in degrading MATα2 in yeast; UBC2 has been implicated in degradation of the Gα protein subunit GPA1 in yeast; UBC9 is implicated in cyclin degradation essential for the progression of the cell cycle in yeast; the Huntingtin protein interacts with hE2; PAS2 is required for peroxisome biogenesis; and the Drosophila bendless gene is involved in synaptic transmission (Dohmen et al., 1991; Glotzer et al., 1991; Wiebel and Kunau, 1992; Chen et al., 1993; Muralidhar and Thomas, 1993; Chowdary et al., 1994; Ciechhanover et al., 1994; Madura and Varshavsky, 1994; Oh et al., 1994; Palombella et al., 1994; Seufert et al., 1995; Kalchman et al., 1996). In addition, ubiquitination has been shown to be a component of the pathway by which the yeast MDR transporter Pdr5, the yeast α-factor receptor, and the mammalian growth hormone receptor are targeted for endocytosis (Egner and Kuchler, 1996; Hicke and Riezman, 1996; Strous et al., 1996), while ubiquitination/deubiquitination controls some cell fate decisions during development of the Drosophila eye (Jermyn and Williams, 1995) and larval development in Caenorhabditis elegans (Zhen et al., 1996). In Dictyostelium, the structure and regulation of expression of genes encoding ubiquitin have been examined (Muller-Taubenberger et al., 1988a and 1988b; Ohmachi et al., 1989), and the in vitro ubiquitination and degradation of Dictyostelium calmodulin have been reported in a reticulocyte extract (Gregori et al., 1985). However, little is know about the role of protein ubiquitination during Dictyostelium development.
In this article, we report the characterization of the gene UbcB that, when disrupted, results in developmental arrest during the multicellular stages of development. UbcB encodes a putative ubiquitin-conjugating enzyme designated UBC1. Disruption of UbcB affects tip formation, cell-type differentiation, the ability of ubcB-null cells to participate in normal development in chimeras containing wild-type cells, and the pattern of protein ubiquitination during development. Our results suggest that protein ubiquitination at the mound and finger stage mediated by UBC1 is required for subsequent development and may be an essential component of the developmental transition between the induction of postaggregative gene expression and subsequent cell-type differentiation and morphogenesis.
MATERIALS AND METHODS
REMI mutagenesis using uracil selection, DH1 uracil auxotroph, and the pRHI30 vector, was done as previously described (Kuspa and Loomis, 1992; Dynes et al., 1994; Insall et al., 1994). Cell culture, transformation, and selection of clones expressing lacZ reporters were performed as previously described (Dynes et al., 1994; Gaskins et al., 1996). All other techniques, including the isolation and cloning of the insertion vector from the REMI mutant, construction of vectors, sequencing, analysis, and RNA, Southern, and Western blot techniques are standard and were performed as described previously (Dynes et al., 1994; Gaskins et al., 1994, 1996). Standard Southern hybridization conditions for Dictyostelium genomic DNA, which is AT-rich, are 3× SSC, 60 mM sodium phosphate (pH 6.8), 10 mM EDTA (pH 7.2), 0.2% SDS, 4× Denhardt’s solution, and 50% deioinized formamide at 37°C as described previously (Nellen et al., 1987). For lower stringency hybridizations (see text), the salt concentrations were kept constant during the hybridization; however, the salt concentrations in the wash solutions were doubled and the last wash of 5 mM EDTA, pH 7.2, and 1% SDS was replaced with 1× SSC, 1% SDS.
Western blot analysis was performed as described previously (Gaskins et al., 1994).
RESULTS
Isolation of UbcB and Identification of Transcripts
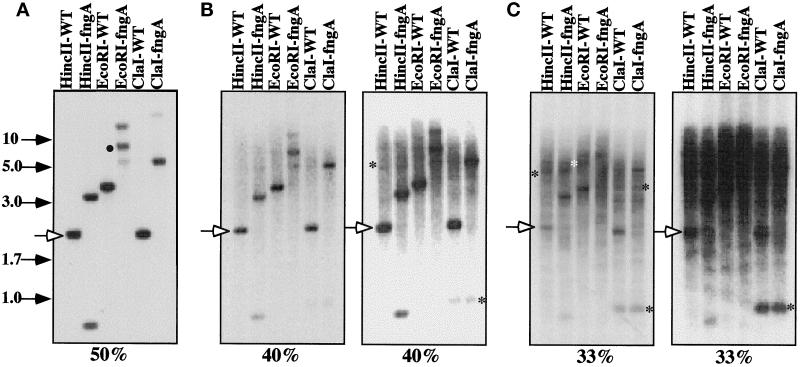
We identified UbcB (see below) as a developmentally essential gene in a REMI mutagenesis (Kuspa and Loomis, 1992) screen for genes required for multicellular development. REMI mutagenesis was performed using an insertion vector (pRHI30) carrying the Dictyostelium Pyr5–6 gene (uracil prototroph) transformed into the uracil auxotroph DH1, selecting for uracil prototrophs (Kuspa and Loomis, 1992; Insall et al., 1994). The insertion vector was mapped to a single site in the Dictyostelium genome by Southern blot analysis (Figure 1A; our unpublished observations). There was an apparent shift in the mobility of the endogenous UbcB band in the mutant strain, consistent with a UbcB gene disruption. The vector and ∼1.7 kb of flanking DNA were excised by digestion with the restriction enzyme EcoRI, circularized by ligation, and cloned into Escherichia coli (see MATERIALS AND METHODS). Restriction maps of the genomic and cDNA clones (see below) are shown in Figure 2, A and B. To confirm that the developmental phenotype resulted from insertion at this locus, we retransformed the isolated DNA into Dictyostelium after linearizing it by digestion with EcoRI. Approximately 60% of the clones had the same developmental phenotype as the original REMI mutation, while the remainder were wild type. Southern blot analysis showed a 1:1 correlation with the phenotype of the original REMI strain (see below) and insertion of the DNA at the same site (our unpublished observations).
Figure 1.
Southern blot of wild-type and ubcB-null cells. Genomic DNA isolated from wild-type cells and ubcB-null cells was digested with the restriction enzymes shown and analyzed by Southern blot hybridization as described previously at different hybridization stringencies (Nellen et al., 1987). (A, 50% formamide) The solid circle marks the proper UbcB gene band for this digest. (B, 40% formamide) The two panels are different exposures of the same filter. (C, 33% formamide) The same batch of restriction enzyme-digested DNA was used for each hybridization. The two 40% and 33% formamide panels are different exposures of the same filter. The positions of size markers (in kb) are shown. See MATERIALS AND METHODS for details. The open arrowhead marks the position of a common band (wild-type HincII band). The asterisks mark the positions of some bands not seen in the high stringency (50% formamide) hybridization blot but which appear in the lower stringency hybridizations.
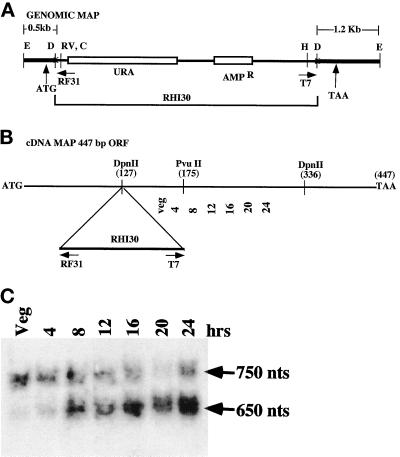
Figure 2.
Restriction maps of the UbcB genomic clone and the cDNA ORF and RNA blot hybridization. (A) The map of genomic clone shows the positions of the UbcB ORF translation initiation site (ATG) and termination site (TAA). Restriction sites for the genomic clone: E, EcoRI; D, DpnII; RV, EcoRV; C, ClaI; HP, HpaI. The transformation vector is RHI30 carrying the Pyr5–6 gene required for uracil prototrophy. The parental strain is the uracil prototroph DHI. The vector and strain were gifts from R. Insall and P. Devreotes. RF31 and T7 are sequencing primers. RF31 was obtained from W. Loomis. (B) The map of cDNA clones shows the position of the UbcB ORF translation initiation site (ATG) and termination site (TAA). The DpnII insertion site for RHI30 in the UbcB REMI mutant strain is shown. (C) Developmental RNA blot of UbcB expression. Time in development is given. Veg, vegetative cells.
DNA from both sides of the insertion site of the genomic clone hybridized to two transcripts of ∼0.65 and 0.75 kb (Figure 2C; our unpublished observations). The larger transcript was expressed during growth and development, with levels decreasing in the multicellular stages. The smaller transcript was expressed at very low levels during vegetative growth, and the levels increased during the time of aggregation. This transcript was present throughout the remainder of development. When the ubcB mutant strain is allowed to develop for 8 h (early mound formation) and RNA is isolated and hybridized to either probe, no hybridization is observed, implying that both transcripts are encoded by the same gene (our unpublished observations). From the lack of detectable transcripts in the mutant strain, we conclude this is an ubcB-null strain. Expression of the UbcB cDNA downstream from the Act15 promoter complemented the null phenotype.
Hybridization probes were then used to screen a λZap library (Schnitzler et al., 1994). Several sets of cDNA clones ranging from 0.6 to 0.8 kb were identified. The sequence of a larger and smaller clone showed that both encoded the same open reading frame (ORF) frame but had different lengths of 5′ and 3′ untranslated sequences. This suggests that differences in the two transcripts probably result from different start and/or termination sites and are not due to splicing variants within the ORF (see below). Moreover, the different temporal patterns of expression of the two RNAs suggest that UbcB transcription is regulated in a complex manner.
UbcB encodes a putative ubiquitin-conjugating enzyme
The two cDNAs of different lengths were completely sequenced in both directions and a single full-length ORF was identified (accession number U67838). Comparison of the sequence to the GenBank databases using the BLAST program showed that the UbcB ORF has very high amino acid sequence identity to E2/ubiquitin-conjugating enzymes from yeast, various metazoans, and plants (Figure 3). The extremely high amino acid sequence conservation throughout the entire protein strongly suggests that UbcB encodes a ubiquitin-conjugating enzyme. Because multiple ubiquitin-conjugating enzymes have been identified in a variety of other species, we expect that Dictyostelium also contains a complex multigene family. We have thus designated the protein encoded by UbcB as UBC1.
Figure 3.
Amino acid sequence of the UbcB ORF and comparison to UBCs from a variety of organisms. Names and accession numbers are given below. UBC1, this article; ScUBC5, Saccharomyces cerevisiae UBC5, acccession no. P15732; HsUBC2, human UBC2, accession no. P35129; CeUBC2, C. elegans UBC2, accession no. P35129; ScUBC4, S. cerevisiae UBC4, accession no. P15731; DmUBC1, Drosophila melanogaster UBC1, accession no. P25867; ScUBC5, S. cerevisiae UBC5, accession no. P15732; AtUBC1, Arabidopsis thaliana UBC1, ac. # P25865; RnUBC, rat, accession no. P23567; HsHHR6A, human UBC, accession no. P06104; ScUBC21, S. cerevisiae UBC21, accession no. P06104; ScUBC1, S. cerevisiae UBC1, accession no. P21734; ScUBC8, S. cerevisiae UBC8, accession no. P28263; Dmben, D. melanogaster Bendless (UBC), accession no. P35128.
To investigate the possibility of additional genes encoding UBC in Dictyostelium, we performed Southern blot analysis of DNA from wild-type and ubcB-null cells under different stringencies. As shown in Figure 1A, hybridization under standard conditions (see MATERIALS AND METHODS) identifies a single UbcB gene in wild-type cells. When the stringency is reduced by lowering the formamide concentration from 50% to 40% while keeping the salt concentration and hybridization and wash temperatures constant (Figure 1B), additional hybridizing bands are observed, suggesting the presence of genes that are related at the level of DNA sequence. When the stringency was reduced further (33% formamide, constant salt and temperature), some additional strong bands and a significant amount of apparently nonspecific background hybridization are observed (Figure 1C). The presence of additional bands that are observed in the 40% formamide hybridization and are stronger at 33% formamide is consistent with the possibility that there are multiple UbcB-related genes in Dictyostelium.
Developmental Phenotype of ubcB-Null Cells
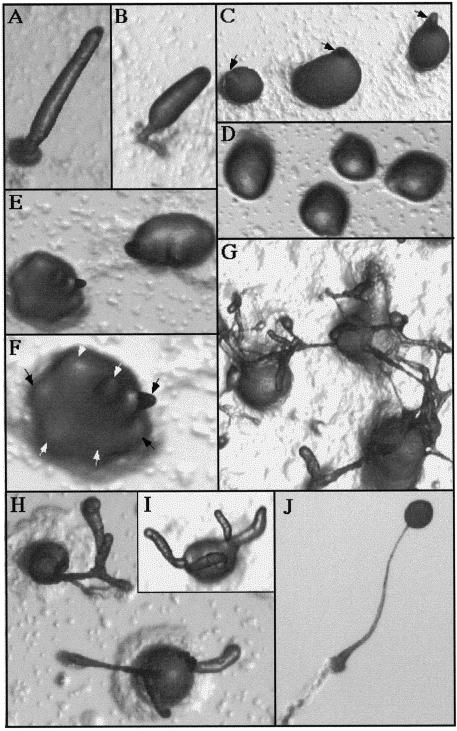
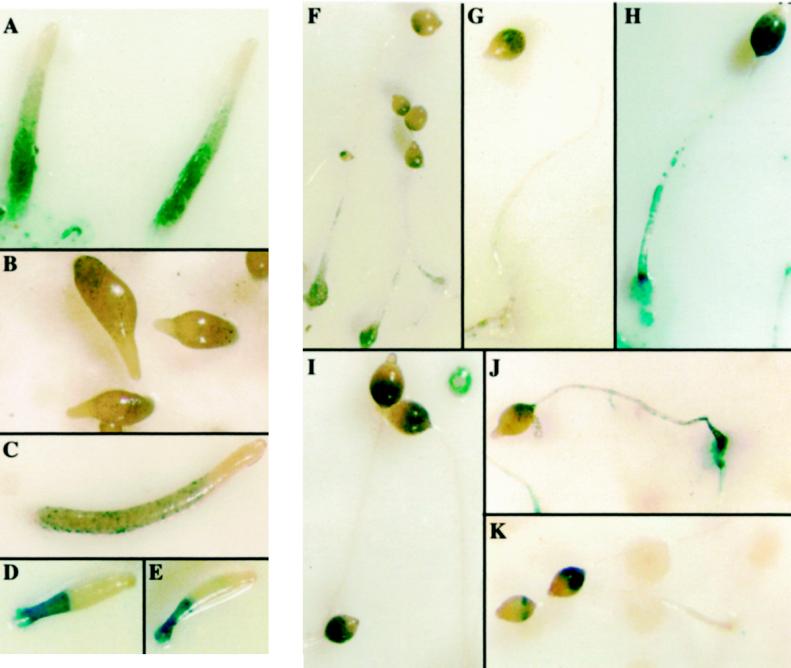
To examine the developmental morphologic phenotype of the ubcB-null mutant, cells were grown axenically, washed of nutrients, and then plated for multicellular development on sodium/postassium phosphate-containing agar plates. ubcB-null cells aggregate and form a mound at 7–8 h of development, ∼1 h earlier than the wild-type parental axenic strain. At the same plating densities, the aggregate sizes of the mutant and wild-type strains were approximately the same [our unpublished observations; Figure 4, compare panels C (wild type) and D (ubcB null)]. Wild-type mounds start to form tips at 10–11 h (Figure 4C) and begin to culminate by 18 h (Figure 4, A and B), forming a fruiting body at ∼24 h (Figure 4J). In contrast, ubcB cells arrest at the mound stage for ∼10–12 h (Figure 4D), whereupon development reinitiates with the formation of multiple tips on the same aggregate (Figure 4, E and F). These tips elongate to form fingers with a majority of the cells remaining in a mound and not participating in finger formation. Development then arrests under these conditions at ∼36 h with no migrating slugs being formed (Figure 4, H and I). At a higher cell density, not all of the cells aggregate. The mounds that form also have multiple tips and protrusions that often produce aerial tangles (Figure 4G). When cells are plated on Millipore filters, multiple fingers arise from each mound after a delay of ∼10–12 h. These fingers comprise only approximately one-half of the cells in the original mound, with the remainder found as an undifferentiated mass (see below; Figure 7B, panel B). At ∼30–36 h, some of the fingers form migrating slugs; however, none of these initiate culmination. When cells from terminal structures plated on Na/K agar or Millipore filters are disaggregated after 36 and 48 h, respectively, and assayed for heat and detergent-resistant spores, no viable cells are found. Expression of the UbcB cDNA from the Act15 promoter complements the developmental phenotypes of the ubcB-null cells, consistent with the developmental abnormalities being the result of a disruption of the UbcB gene (our unpublished observations). Overexpression of the cDNA in wild-type cells does not result in any observable morphologic defect (our unpublished observations).
Figure 4.
Developmental morphology of ubcB-null cells plated on Na/K P04 agar plates. (A–B) Wild-type cells at the second finger/early culmination stage (18 h). (C) Wild-type tight aggregates forming tips (12 h), which are indicated by arrowheads. Note that each aggregate has only one tip. (D) ubcB-null cells at 18 h. Mounds first form at ∼8 h. Notice that no tips are present. (E) ubcB-null cells at 19 h as tips are first forming. (F) Enlargement of the lower left aggregate in E. The multiple tips are indicated by arrowheads. (G) ubcB-null cells plated at a higher density at 36 h. (H–I) ubcB-null cells at 36 h. (J) Mature wild-type fruiting body (26 h).
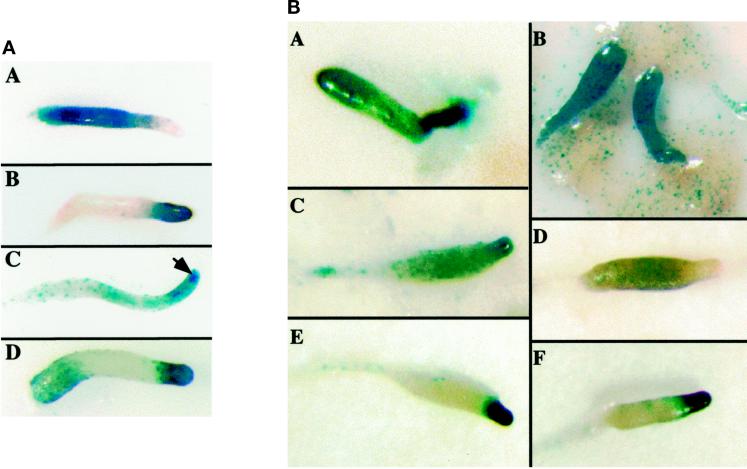
Figure 7.
Spatial patterning of cell types in wild-type and ubcB-null cells. (A) Staining of wild-type cells at the slug stage with: panel A, SP60/lacZ (prespore); panel B, ecmAO/lacZ (prestalk A, AB, and O and ALC); panel C, ecmB/lacZ (prestalk AB and ALC). Arrowhead points to the core of prestalk AB cells. Panel D, rasD/lacZ (prestalk and ALC). (B) Pattern of lacZ reporter staining of ubcB-null slugs using various reporters: panel A, ecmB; panel B, rasD; panel C, rasD; panel D, SP60; panel E, ecmAO; F, ecmAO.
Changes in the Pattern of Protein Ubiquitination in ubcB-Null Cells
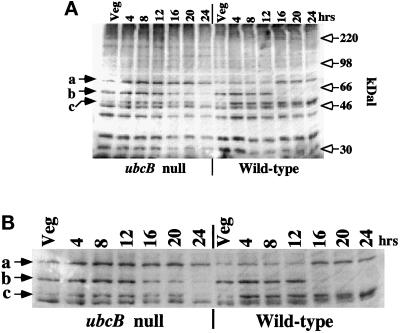
Because UbcB encodes a putative UBC, we have investigated potential changes in the pattern of ubiquitinated proteins during development on Western blots using an anti-ubiquitin antibody. While most of the pattern of ubiquitinated proteins is similar between wild-type and ubcB-null cells (Figure 5), three proteins that react strongly with the antibody show significant differences between the two strains. In wild-type cells, a protein of ca. 73 kDa (Figure 5, band a) from its mobility reacts weakly with the antibody through 12 h of development, the time at which the ubcB phenotype is first observed. Afterward, the intensity of a band of the same mobility (and thus possibly the same protein) increases, suggesting that this protein undergoes either a reduction in the rate of degradation and/or an increase in the level of ubiquitination and/or expression. Our observations cannot exclude the possibility that band a represents two proteins of the same electrophoretic mobility but with different developmental kinetics. In contrast with observations in wild-type cells, in the mutant strain, this band does not decrease in intensity at 12 h but shows strong staining throughout development. Another protein (band b, ca. 59 kDa) shows a reciprocal staining pattern in wild-type cells: it is detected through 12 h of development cells, after which the level of the protein or the level of ubiquitination of the protein rapidly declines. In ubcB-null cells, the band continues to stain strongly for a longer time in development. A third protein (band c, ca. 51 kDa) increases in staining intensity at 4 h and remains high throughout the remainder of development in wild-type cells. The staining is generally weaker in ubcB-null cells and the level of staining decreases after the 12-h time point. The level of ubiquitination of other proteins may also be affected but is not detectable on our blots. While these data do not directly address whether any of these proteins are direct substrates of UBC1, they indicate that ubcB-null cells show observable changes in the quantitative and/or qualitative pattern of protein ubiquitination as compared with wild-type cells (see DISCUSSION).
Figure 5.
Anti-ubiquitin Western blot. (A) Cells were taken at the times in development indicated, lysed in PAGE-SDS gel buffer, sized on a 10% gel, blotted to Immobilon-P membrane (Millipore), and probed with a commercial antiubiquitin antibody (Sigma, St. Louis, MO). The position of the size markers are shown. Band a, ca. ∼73 kDa; band b, ca. ∼59 kDa; band c, ca. ∼51 kDa. (B) Enlargement of the section of the blot containing bands a–c.
Control of Gene Expression in ubcB-Null Organisms
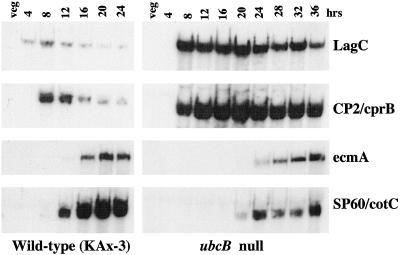
To better understand regulatory changes that might be occurring in the ubcB-null cells, we examined the developmental regulation of genes preferentially expressed during multicellular development. The expression of LagC and CP2/cprB was used as a molecular marker for postaggregate gene regulation during mound formation (Pears et al., 1985; Datta et al., 1986; Dynes et al., 1994). In wild-type cells, both genes are induced by a high, constant level of extracellular cAMP that is thought to occur as the mound forms (Abe and Yanagisawa, 1983; Firtel, 1995). CP2/cprB encodes a cysteine protease and has been used as a marker for postaggregate gene expression (Pears et al., 1985; Datta et al., 1986). LagC, which is similarly regulated, encodes a putative transmembrane protein that is essential for development past the mound stage and is required for prestalk and prespore cell differentiation (Dynes et al., 1994). In wild-type cells, CP2/cprB and LagC transcripts start to accumulate late in aggregation as the mound forms. Transcript levels remain high for 4–6 h through the tipped aggregate and first finger stages (∼11–13 h), after which levels of the transcripts decrease significantly (Figure 6). In ubcB-null cells, both genes are induced during mound formation, but in contrast to observations in wild-type cells, the level of transcript accumulation is >10-fold higher and remains high for ∼10 h, until the time ubcB-null cells start to form tips, before decreasing only moderately. This extended expression of CP2/cprB and LagC is consistent with the developmental delay of the mutant at the mound stage and its subsequent arrest at the finger stage. The high, extended level of expression suggests that the regulatory mechanism involved in down-regulating these genes during finger formation is blocked in ubcB-null cells.
Figure 6.
RNA blot hybridization of reporter genes during development. Expression of developmentally regulated genes in wild-type and ubcB-null strains: LagC and CP2/cprB, postaggregative genes; ecmA, prestalk-specific; SP60/cotC, prespore-specific. The filters assaying the expression level of the same gene were probed together.
Analysis of the expression of the endogenous prestalk-specific gene ecmA demonstrated that the gene was induced ∼12 h later in ubcB-null cells than wild-type cells and showed a similar level of expression in both wild-type and ubcB-null strains (Figure 6). The time of ecmA induction corresponds approximately to the time of tip formation in the mutant strain. Expression of the prespore-specific gene SP60/cotC shows a similar delay in induction, indicating that the onset of expression of both prestalk and prespore genes is equally affected. However, in contrast to ecmA expression, SP60/cotC mRNA accumulation in ubcB-null cells is significantly lower than in wild-type cells.
Spatial Patterning of Prestalk and Prespore Cells in ubcB-Null Cells and ubcB-Null/Wild-Type Cell Chimeras
The effect of the ubcB-null mutation on spatial patterning of prestalk and prespore cells in the multicellular organisms was examined using cell-type-specific lacZ reporters and histochemical staining. Several cell-type-specific and cell-type-enriched reporters were examined. The cloned promoter of the prestalk-specific gene ecmA has been dissected into two domains by deletion analysis using whole-mount histochemical staining of lacZ reporters to define spatial patterning into regulatory elements that are preferentially expressed in prestalk A and AB cells and in prestalk O and anterior-like cells (ALC), respectively (Early et al., 1993, 1995). The vector containing the entire promoter driving lacZ (ecmAO/lacZ) exhibits maximal expression in the anterior prestalk A and AB cells and is expressed at a lower level in prestalk O cells in the slug. It is also expressed in ALC (Early et al., 1993), a regulatory cell population comprising ∼5–10% of the cells that are scattered throughout the slug. The prestalk gene ecmB is expressed primarily in a central core of prestalk AB cells localized in the very anterior of the slug but is also expressed in the ALC population (Jermyn et al., 1989; Jermyn and Williams, 1991). rasD is a prestalk-enriched gene that is expressed predominantly in prestalk A, prestalk O, and ALC in the slug (Esch, 1991), while SP60/cotC is a prespore-specific gene expressed in the posterior 80% of the slug (Haberstroh and Firtel, 1990). Figure 7A shows the staining pattern of ecmAO/lacZ, SP60/lacZ, ecmB/lacZ, and rasD/lacZ in wild-type slugs.
When ecmAO/lacZ- and ecmB/lacZ-expressing ubcB-null cells were examined, little or no staining was observed until ∼15 h after the time of mound formation (our unpublished observations). In ubcB-null migrating slugs, the spatial pattern of ecmAO/lacZ expression was similar to that seen in wild-type cells (Figure 7B, panels E and F). ecmB expression, however, was abnormal. Staining is significantly stronger than in wild-type cells at similar copy numbers of the reporter vector and is observed throughout the developing first finger slug (Figure 6B, panel A; our unpublished observations). This may be an indication of an increase in the level of staining in each expressing cell or an increase in the number of ALC. Similarly, when rasD was examined, ubcB-null mounds produced a finger in which almost the entire finger was stained intensely (Figure 7B, panel B), instead of the anterior, prestalk zone staining that is seen in wild-type cells at this same stage (Esch and Firtel, 1991; our unpublished observations). Cells remaining in the mound that did not participate in finger formation showed only scattered staining. In the migrating slug, the very anterior of the slug stained the strongest (prestalk A domain) but ubcB-null slugs also showed strong staining through the posterior of the slug (Figure 7B, panel C). Wild-type slugs expressing rasD/lacZ showed strong staining in the prestalk A/O region but only light scattered staining of ALC in the prespore region and moderate staining of the posterior-most rearguard zone (Figure 7A, panel D; Esch and Firtel, 1991), which forms the basal disk in the mature fruiting body. A similar pattern was observed for all clonal isolates studied. As rasD/lacZ stains predominantly prestalk and ALC in wild-type slugs (Esch and Firtel, 1991), the more intense staining in the posterior may indicate a more intense staining of the ALC or an increase in the number of ubcB-derived ALC. It has been suggested that rasD is initially expressed in all cells and then the expression becomes significantly weaker in prespore cells (Jermyn and Williams, 1995). However, the observed rasD/lacZ staining pattern cannot simply be due to a continued expression of rasD in all ubcB-null cells, as there is negligible staining in the cells left in the mound.
When the prespore-specific marker SP60/cotC was examined, staining was seen in the posterior 80% of the slug, as in wild-type cells (Figure 7B, panel D). However, the staining was weaker in the mutant compared with that in wild-type cells. This pattern was also reproducible in multiple clones from independent transformations. When wild-type and ubcB-null slugs were dissociated into single cells and the number of prespore cells counted (SP60/lacZ staining cells), a similar fraction of cells stained in both strains (our unpublished observations), indicating that the number of prespore cells in the slugs is not altered significantly in the ubcB-null mutant.
To obtain a better understanding of the properties of the ubcB cells, experiments were performed in which lacZ-tagged ubcB-null and wild-type cells were mixed in a ratio of 1:3 and allowed to coaggregate, forming chimeric organisms. These chimeras were then histochemically stained for β-galactosidase activity for localization of lacZ-expressing ubcB-null cells. Such studies allow the examination of both the spatial localization of the ubcB cells and the ability of the cells to participate in different terminal structures of the fruiting body. When ecmAO/lacZ and ecmB/lacZ expression was examined in the chimeric slug, no staining was detectable (our unpublished observations). For rasD/lacZ, staining was significantly less intense compared with that seen in ubcB-null slugs. The stained cells were concentrated in the posterior of the slug, and no staining was detected in the anterior prestalk region (Figure 8B). When SP60/lacZ was examined, staining was seen in the prespore zone in migrating slugs (Figure 8C), but in second fingers at the onset of culmination, staining was restricted to the posterior half of the prespore zone (our unpublished observations). The Act15/lacZ reporter driven by the Actin 15 (Act15) promoter is expressed in all cell types (Mann and Firtel, 1991). At the slug stage, Act15/lacZ staining was found predominantly in the posterior 50% of the slug with the level of staining, and thus the proportion of ubcB-null cells, decreasing toward the anterior of the slug (Figure 8A). During early culmination, staining was seen almost exclusively in the posterior 70% of the chimera, indicating ubcB cells were being excluded from the anterior part of the prespore and prestalk zones (Figure 8, D and E).
Figure 8.
Localization of ubcB-null cells expressing lacZ reporters in chimeras of wild-type and ubcB-null cells (ratio of 3:1). Wild-type KAx-3 cells and ubcB-null cells expressing various reporters were mixed in a ratio of 3:1 and allowed to develop. Multicellular organisms were stained at the slug/finger stages (A–E) and culminants (F–K). Act15, panels A, D, E, and H; rasD, panels B and I; SP60, panels C and G. ecmAO, panel F (no staining is seen in the slug stage); ecmBΔ89, panel J; SpiA, panel K.
In the mature fruiting body the ubcB cells were, as determined from Act15/lacZ staining, found in the basal disk, lower portions of the stalk, and the lower cup and lower regions of the sorus, but were absent from the tip (Figure 8H). Much of the staining seen on the sorus represents surface staining of a layer that is probably an extension of the lower cup and derived from the ALC population. When the sorus was dissociated, only ∼8% of the spores stained (our unpublished observations). In mature fruiting bodies, ecmAO/lacZ staining is observed in the basal disk, the lower portion of the stalk, and the lower cup, but no staining was observed in the upper cup and little staining was seen in the upper portion of the stalk tube (Figure 8F). rasD, which displays a similar staining pattern in wild-type fruiting bodies to that of ecmAO, also showed a similar pattern in the ubcB chimeras as the ecmAO/lacZ reporter with no staining seen in the upper cup and tip, a strongly staining region in wild-type organisms (Figure 8I). lacZ expressed from the ecmBΔ89 stalk-cell-specific promoter (Ceccarelli et al., 1991) had a similar pattern of expression: staining cells were more concentrated in the basal disk and lower stalk than throughout the stalk (Figure 8J). Some staining was also seen in the lower cup. SP60/lacZ and lacZ expressed from the spore cell-specific promoter SpiA (Richardson et al., 1994) showed staining localized to the lower portion of the sorus in contrast to expression throughout the entire sorus, as seen in wild-type strains (Figure 8, G and K, respectively). The absence of ubcB-null cells in the upper cup or the upper part of the stalk tube may be due to the absence of ubcB-null cells in the anterior of the chimera.
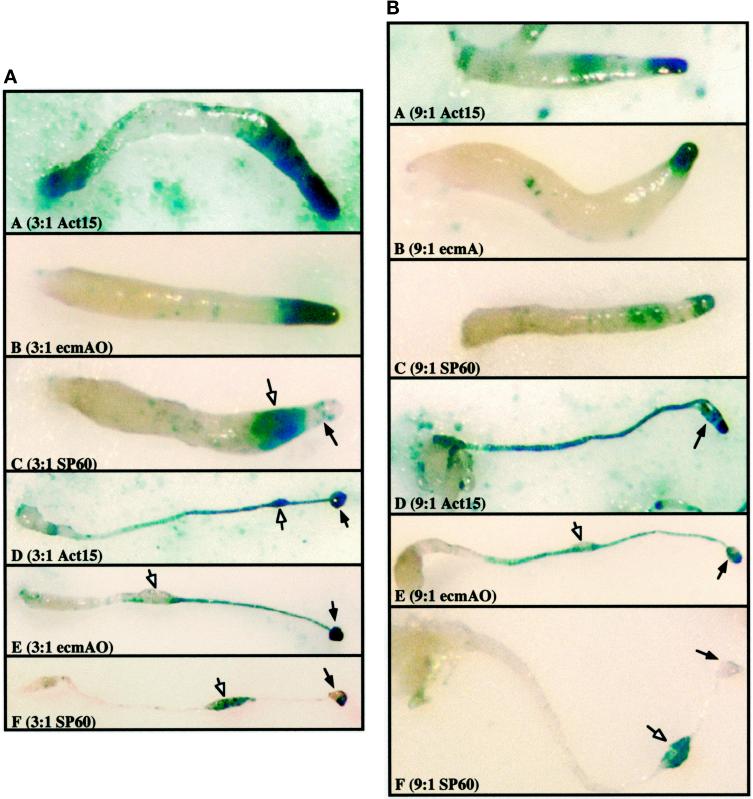
To determine whether a smaller proportion of wild-type cells is able to direct the differentiation of ubcB nulls in chimeras, we mixed ubcB-null cells with wild-type cells carrying either the ecmAO/lacZ or SP60/lacZ reporter in ratios of either 3:1 or 9:1 (ubcB:wild type). As shown in Figure 9 (A and B, panels A-C), even a small proportion (10%) of wild-type cells is able to direct tip formation in a chimeric organism. However, most, if not all, of the wild-type cells appear to be localized in the very anterior of the organism as observed in the panels in which the wild-type cells are tagged with Act15/lacZ, which marks all wild-type cells (Figure 9, A and B, panel A). This is also observed when the wild-type prespore cells are marked by SP60/lacZ. As is seen in the panel showing the 3:1 ratio mix, the SP60/lacZ expressing cells are found as a band immediately posterior to the position of the wild-type prestalk cells identified by the ecmAO/lacZ marker (Figure 9A, panel E). No ecmAO/lacZ expressing cells are observed in the posterior bulk of the chimera, which also does not show significant staining with the Act15/lacZ reporter except at the very posterior of the organism. These results indicate that the wild-type cells are localized predominantly in the anterior of the chimera (Figure 9, A and B, panel A). When the ubcB-null cells were tagged with Act15/lacZ, no ubcB-null cells were observed in the anterior of slugs that contained the wild-type cells in either the 3:1 or 9:1 ubcB-null:wild-type chimeric organisms (our unpublished observations).
Figure 9.
Localization of ubcB-null cells expressing lacZ reporters in chimeras of ubcB-null and wild-type cells at ratios of 3:1 and 9:1. (A) ubcB-null and wild-type cells a ratios of 3:1. Panel A, slug stage, Act15/lacZ; panel B, slug stage, ecmAO/lacZ; panel C, slug stage, SP60/lacZ; panel D, final structure, Act15/lacZ; panel E, final structure, ecmAO/lacZ; panel F, final structure, SP60/lacZ. (B) ubcB-null and wild-type cells at ratios of 9:1. Panel A, slug stage, Act15/lacZ; panel B, slug stage, ecmAO/lacZ; panel C, slug stage, SP60/lacZ; panel D, final structure, Act15/lacZ; panel E, final structure, ecmAO/lacZ; panel F, final structure, SP60/lacZ. In A and B, panels D-F, closed arrowheads point to enlarged region on stalk that does not stain with the prestalk/stalk marker (ecmAO/lacZ) but does stain with the prespore/spore marker (SP60/lacZ). The open arrowhead points to a small “sorus-like” structure. In A, panel C, the open arrowhead points to a wild-type prespore domain; the closed arrowhead points to a wild-type prestalk domain.
In the final structure (Figure 9, A and B, panels D-F), the wild-type cells appear to form a fruiting body with little participation of the ubcB-null cells and <1% of the mature spores derived from the ubcB-null strain. The patterns are very similar for 3:1 and 9:1 cell ratios with the patterning and morphology being more severe in the chimera containing only 10% wild-type cells. A very small anterior structure is seen, which does not appear to contain the spore cells, as it is not labeled with the SP60/lacZ reporter. These may be ALC or prestalk cells that have not entered the stalk tube, as they stain strongly with ecmAO/lacZ (Figure 9, A and B, panels E). In contrast, in many of the mature organisms, a central swelling in the stalk appears to contain SP60/lacZ-expressing cells (Figure 9, A and B, panels F). These data are consistent with a model proposing that as the fraction of wild-type cells in the chimera decrease, the wild-type cells are unable to orchestrate the differentiation of the mutant cells, suggesting that the ubcB-null phenotype is predominantly cell autonomous.
DISCUSSION
UbcB Protein Is Required for Developmental Transitions during Multicellular Development
We have identified a developmentally essential gene, UbcB, that is required at two stages during multicellular differentiation. ubcB-null cells form mounds and remain at this stage for ∼10 h before the mounds develop multiple tips and then finally arrest at the finger stage when plated on agar. When the cells are plated on Millipore filters, development proceeds slightly further, with a fraction of the fingers forming migrating slugs. In both cases, not all of the cells participate in finger and slug formation and under the conditions in which cells are plated on agar, most of the cells remain in the mound. The nature of these cells is unclear. They do not stain efficiently with rasD/lacZ or cell-type-specific lacZ reporters, suggesting that these cells arrest before or very early during cell-type differentiation. The observation that ubcB-null cells are able to form tips and fingers after an extended delay at the mound stage indicates that they are eventually able to bypass the initial block at the mound stage. The pathway required for tip formation may proceed, albeit very slowly, without the processes mediated by the UbcB protein, which we have designated UBC1. Alternately, another ubiquitin-conjugating enzyme may substitute for UBC1 function at a low efficiency at the mound stage during tip formation, but cannot substitute later in development after finger/slug formation. Low stringency hybridization suggests the presence of additional genes encoding UBCs in Dictyostelium, which would be consistent with published data showing that in organisms ranging from yeast to humans in which UBCs have been identified, multiple UBCs are found (see INTRODUCTION).
In wild-type cells, postaggregative genes such as those encoding CP2 and LagC proteins are maximally expressed at the mound stage, after which their expression decreases. The higher level and extended time of expression of these two genes in ubcB-null cells suggest that there is a regulatory mechanism in wild-type cells, leading to the down-regulation of both of these genes after the mound stage and that this is absent in the ubcB-null cells. The delay of induction of prestalk and prespore cell-type-specific genes in ubcB cells suggests that the developmental arrest at the mound stage occurs between the induction and down-regulation of the postaggregative genes and the induction of the cell-type-specific genes. UbcB function is essential for the developmental transition required for the down-regulation of postaggregative gene expression.
ubcB-null cells show normal spatial patterning of prespore cells in slugs, although the intensity of the staining is reduced, consistent with the lower level of expression of the endogenous SP60/cotC gene, indicating prespore gene expression is affected. The ecmAO/lacZ staining pattern also appears similar to that of wild-type cells; however, for ecmB/lacZ- and rasD/lacZ-expressing cells, the level of their expression is very high and/or an unusually high number of cells express these two reporters in comparison to what is observed in wild-type cells. The high level of staining with both ecmB/lacZ and rasD/lacZ suggests that the ubcB-null strain may have an unusually high proportion of ALCs. It is possible, as with LagC and CP2/cprB, that expression of rasD may continue to increase in the subset of ubcB-null cells that form the slug, resulting in a higher, more extended expression pattern. However, rasD expression is not induced in the loose mass of cells that remain at the base of the developing finger, and thus, the mechanisms by which UbcB protein controls spatial patterning are complex. The phenotypes observed in ubcB-null aggregates have not been previously described in other mutant strains.
Analysis of chimeric organisms containing varying ratios of wild-type and ubcB-null cells in which the two strains are marked with lacZ reporters indicates that when there is a high enough proportion of wild-type cells, a small fraction of ubcB-null cells are able to form spores and participate in stalk formation. However, it is clear that ubcB-null cells do not effectively participate in later morphogenesis, and in chimeras containing predominantly mutant cells, the null cells are found predominantly in the posterior/basal regions and do not participate in either stalk or spore formation. The results are consistent with the inability of these cells to efficiently undergo the developmental transition that occurs at the time of cell-type differentiation. While ubcB-null cells are unable to efficiently form a tip, this alone is not responsible for the ubcB-null phenotype; the addition of a small proportion of wild-type cells leads to tip formation in chimeras but this is unable to promote the proper development of the majority of the ubcB-null cells.
ubcB-Null Cells Show an Altered Pattern of Ubiquitin-containing Proteins
The very high level of amino acid sequence identity between the UbcB ORF and UBCs from a variety of organisms strongly suggests that UbcB encodes a UBC, which we have named UBC1. Our finding that UbcB is an essential gene for progression past the finger stage suggests that protein ubiquitination as an essential step in this process. This is consistent with our observations of an altered pattern of ubiquitinated proteins in ubcB-null cells compared with wild-type cells. At least three ubiquitinated proteins exhibit changes in their developmental pattern of staining with the antiubiquitin antibody on Western blots between wild-type and ubcB-null cells. One major ubiquitinated protein stains weakly in wild-type cells during early development, and then stains more intensely starting at the time of tip formation (12 h), the same time that the ubcB-null phenotype is first observed. In ubcB-null cells, this band shows a more constant staining starting at the preaggregation stage (4 h). Another protein stains strongly through 12 h and then disappears in wild-type cells. In ubcB-null cells, this band does not disappear, although the intensity of staining decreases. A third band exhibits a constant in wild-type cells from 4 h. In ubcB-null cells, this band is significantly weaker in ubcB cells and this intensity decreases even further starting at 12 h. It is of interest that the observed changes in the developmental pattern of these three proteins between wild-type and ubcB-null cells are different from each other. We do not know if the changes in staining represent changes in the level of ubiquitination, the rate of protein degradation, the kinetics of protein expression, or combinations of the three. Nor do we know whether either of these proteins are direct substrates of the UbcB protein UBC1, whether the putative protein ubiquitination directed by UBC1 targets that protein for degradation by the proteosome pathway, or whether the ubiquitination functions in some other way. The timing of the changes is consistent with UBP1 being required either directly or indirectly. Our data do not distinguish between changes in protein ubiquitination that are a direct function of UBP1 activity or pleiotrophic changes that are indirectly the result of the ubcB-null mutation. Nonetheless, these results indicate an effect of disruption of UbcB on protein ubiquitination that correlates temporally with the ubcB-null cell phenotype.
From the role of some UBCs in ubiquitin-mediated protein degradation, one possible explanation of the developmental phenotypes exhibited by the ubcB-null mutant is that the mutant cells are unable to degrade one or more proteins through the proteosome pathway whose turnover may be essential for development to proceed. This could either be the programmed degradation of an endogenous inhibitor of development that regulates tip formation or a protein whose degradation is required to activate a developmental pathway. Ubiquitination has also been linked to other cellular functions, including protein targeting and cell fate decisions. In the case of the yeast MDR transporter Pdr5, yeast α-factor receptor, and mammalian growth hormone receptor, ubiquitination is involved in regulating turnover via targeting of the protein to the endosome (Egner and Kuchler, 1996; Hicke and Riezman, 1996; Strous et al., 1996). In Drosphila, the fat facets gene, which encodes a deubiquitinating enzyme, is required for cell fate decisions in the eye (Jermyn and Williams, 1995), opening up the possibility that ubiquitination is involved in mediating a diverse set of cellular pathways.
At present, it is not known which ubiquitination-mediated pathway may be altered in ubcB-null cells, although the possibility that UBP1 may be involved in targeting specific proteins for degradation is consistent with previous results of mutations affecting a Dictyostelium homolog of the human Tat-binding protein, DdTBPα (Cao and Firtel, 1995). Members of the TBP family are also known to be components of the 26S proteosome involved in protein degradation. The DdTBPα gene appears to be essential for both vegetative growth and multicellular development. Disruption of one of the two copies of the gene results in developmental abnormalities that can first be observed at the finger stage. Expression of an antisense construct in the partial knockout results in additional morphologic abnormalities, while DdTBPα over-expression leads to a cytokinetic defect. These over-expressing cells also show abnormal morphogenesis during the multicellular stages, with development arresting at the finger stage. These results suggest that protein degradation through the 26S proteosome pathway(s) may be important for controlling specific stages during growth and development in Dictyostelium. Whether UBP1 and DdTBPα regulate the same developmental processes or whether either is involved in protein turnover is not known.
A variety of proteins in addition to UbcB have been shown to be required for cells to proceed normally past the mound stage, form a finger, and continue through morphogenesis. These include: the transcription factor GBF; LagC, a putative cell surface signaling molecule; TagB, a subunit of an MDR with a putative serine protease domain; and the cAMP receptor cAR2 (Saxe et al., 1993; Dynes et al., 1994; Schnitzler et al., 1995; Shaulsky et al., 1995). In addition, RasD has been implicated in tip formation (Reymond et al., 1986). Cells lacking myosin II and cells in which cytosolic calcium has been depleted by expressing a constitutively active Ca2+ membrane pump also arrest or are delayed at this stage (Springer et al., 1994; A.B. Cubitt, I. Reddy, J.G. McNally, and R.A. Firtel, unpublished data). The diversity of the pathways that might be affected by these gene products suggests that multiple regulatory pathways are required to control these transitions.
ACKNOWLEDGMENTS
We thank W. Loomis, A. Kuspa, and R. Hampton for helpful discussions during the process of this work. The UbcB cDNAs were sequenced by the Dictyostelium sequencing core supported by United States Public Health Service Grant PO1HD30892. This work was supported in part by United States Public Health Service grants to R.A.F.
REFERENCES
- Abe K, Yanagisawa K. A new class of rapid developing mutants in Dictyostelium discoideum: implications for cyclic AMP metabolism and cell differentiation. Dev Biol. 1983;95:200–210. doi: 10.1016/0012-1606(83)90018-0. [DOI] [PubMed] [Google Scholar]
- Cao JG, Firtel RA. Growth and developmental functions of a human immunodeficiency virus Tat-binding protein/26S protease subunit homolog from Dictyostelium discoideum. Mol Cell Biol. 1995;15:1725–1736. doi: 10.1128/mcb.15.3.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli A, Mahbubani H, Williams JG. Positively and negatively acting signals regulating stalk cell and anterior-like cell differentiation in Dictyostelium. Cell. 1991;65:983–989. doi: 10.1016/0092-8674(91)90550-i. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATa2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chowdary D, Bermody J, Jha K, Ozer H. Accumulation of p53 in a mutant cell line defective in ubiquitin pathway. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Ciechhanover A, Shkedy D, Oren M, Bercovich M. Degradation of the tumor suppressor protein p53 by the ubiquitin-mediated proteolytic system requires a novel species of ubiquitin-carrier protein, E2. J Biol Chem. 1994;269:9582–9589. [PubMed] [Google Scholar]
- Datta S, Gomer RH, Firtel RA. Spatial and temporal regulation of a foreign gene by a prestalk-specific promotor in transformed Dictyostelium discoideum. Mol Cell Biol. 1986;6:811–820. doi: 10.1128/mcb.6.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes PN. G protein-linked signaling pathways control the developmental program of Dictyostelium. Neuron. 1994;12:235–241. doi: 10.1016/0896-6273(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynes J, Clark A, Shaulsky G, Kuspa A, Loomis W, Firtel RA. LagC is required for cell-cell interactions that are essential for cell-type differentiation in Dictyostelium. Genes Dev. 1994;8:948–958. doi: 10.1101/gad.8.8.948. [DOI] [PubMed] [Google Scholar]
- Early A, Abe T, Williams J. Evidence for positional differentiation of prestalk cells and for a morphogenetic gradient in Dictyostelium. Cell. 1995;83:91–99. doi: 10.1016/0092-8674(95)90237-6. [DOI] [PubMed] [Google Scholar]
- Early AE, Gaskell MJ, Traynor D, Williams JG. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development. 1993;118:353–362. doi: 10.1242/dev.118.2.353. [DOI] [PubMed] [Google Scholar]
- Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- Esch, R.K. (1991). Expression of the developmentally-regulated, prestalk-specific ras gene of Dictyostelium discoideum. Thesis. San Diego, CA: Univeristy of California, San Diego.
- Esch RK, Firtel RA. cAMP and cell sorting control the spatial expression of a developmentally essential cell-type-specific ras gene in Dictyostelium. Genes Dev. 1991;5:9–21. doi: 10.1101/gad.5.1.9. [DOI] [PubMed] [Google Scholar]
- Firtel RA. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- Gaskins C, Clark AM, Aubry L, Segall JE, Firtel RA. The Dictyostelium MAP kinase ERK2 regulates multiple, independent developmental pathways. Genes Dev. 1996;10:118–128. doi: 10.1101/gad.10.1.118. [DOI] [PubMed] [Google Scholar]
- Gaskins C, Maeda M, Firtel R. Identification and functional analysis of a developmentally regulated extracellular signal-regulated kinase gene in Dictyostelium discoideum. Mol Cell Biol. 1994;14:6996–7012. doi: 10.1128/mcb.14.10.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray A, Kirschner M. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gregori L, Marriot D, West CM, Chau V. Specific recognition of calmodulin from Dictyostelium discoideum by the ATP, ubiquitin-dependent degradative pathway. J Biol Chem. 1985;260:5232–5235. [PubMed] [Google Scholar]
- Haberstroh L, Firtel RA. A spatial gradient of expression of a cAMP-regulated prespore cell type specific gene in Dictyostelium. Genes Dev. 1990;4:596–612. doi: 10.1101/gad.4.4.596. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- Insall R, Kuspa A, Lilly P, Shaulsky G, Levin L, Loomis W, Devreotes P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1994;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn K, Williams J. Comparison of the Dictyostelium rasD and ecmA genes reveals two distinct mechanisms whereby an mRNA may become enriched in prestalk cells. Differentiation. 1995;58:261–267. doi: 10.1046/j.1432-0436.1995.5840261.x. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Duffy KT, Williams JG. A new anatomy of the prestalk zone in Dictyostelium. Nature. 1989;340:144–146. doi: 10.1038/340144a0. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Williams JG. An analysis of culmination in Dictyostelium using prestalk and stalk- specific cell autonomous markers. Development. 1991;111:779–787. doi: 10.1242/dev.111.3.779. [DOI] [PubMed] [Google Scholar]
- Kalchman M, Graham R, Xia G, Koide H, Hodgson J, Graham K, Goldberg Y, Gietz R, Pickart C, Hayden M. Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem. 1996;271:19385–19394. doi: 10.1074/jbc.271.32.19385. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K, Varshavsky A. Degradation of Ga by the N-end rule pathway. Science. 1994;265:1454–1456. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech Dev. 1991;35:89–102. doi: 10.1016/0925-4773(91)90060-j. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger A, Hagmann J, Noegel A, Gerisch G. Ubiquitin gene expression in Dictyostelium is induced by heat and cold shock, cadmium, and inhibitors of protein synthesis. J Cell Sci. 1988a;90:51–58. doi: 10.1242/jcs.90.1.51. [DOI] [PubMed] [Google Scholar]
- Muller-Taubenberger A, Westphal M, Jaeger E, Noegel A, Gerisch G. Complete cDNA sequence of a Dictyostelium ubiquitin with a carboxy-terminal tail and identification of the protein using an anti-peptide antibody. FEBS Lett. 1988b;229:273–278. doi: 10.1016/0014-5793(88)81139-6. [DOI] [PubMed] [Google Scholar]
- Muralidhar MG, Thomas JB. The Drosophila bendless gene encodes a neural protein related to ubiquitin-conjugating enzymes. Neuron. 1993;11:253–266. doi: 10.1016/0896-6273(93)90182-q. [DOI] [PubMed] [Google Scholar]
- Nellen W, Datta S, Reymond C, Sivertsen A, Mann S, Crowley T, Firtel RA. Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 1987;28:67–100. doi: 10.1016/s0091-679x(08)61637-4. [DOI] [PubMed] [Google Scholar]
- Oh CE, McMahon R, Benzer S, Tanouye MA. bendless, a Drosophila gene affecting neuronal connectivity, encodes a ubiquitin-conjugating enzyme homolog. J Neurosci. 1994;14:3166–3179. doi: 10.1523/JNEUROSCI.14-05-03166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi T, Giorda R, Shaw DR, Ennis HL. Molecular organization of developmentally regulated Dictyostelium discoideum ubiquitin cDNAs. Biochemistry. 1989;28:5226–5230. doi: 10.1021/bi00438a046. [DOI] [PubMed] [Google Scholar]
- Palombella V, Rando O, Goldberg A, Maniatis T. Ubiquitin and the proteasome are required for processing the NF-κB1 precursor and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Pears CJ, Mahbubani HM, Williams JG. Characterization of two highly diverged but developmentally coregulated cysteine proteinases in Dictyostelium discoideum. Nucleic Acids Res. 1985;13:8853–8866. doi: 10.1093/nar/13.24.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond CD, Gomer RH, Nellen W, Theibert A, Devreotes P, Firtel R. Phenotypic changes induced by a mutated ras gene during the development of Dictyostelium transformants. Nature. 1986;323:340–343. doi: 10.1038/323340a0. [DOI] [PubMed] [Google Scholar]
- Richardson DL, Loomis WF, Kimmel AR. Progression of an inductive signal activates sporulation in Dictyostelium discoideum. Development. 1994;120:2891–2900. doi: 10.1242/dev.120.10.2891. [DOI] [PubMed] [Google Scholar]
- Saxe III CL, Ginsburg GT, Louis JM, Johnson R, Devreotes PN, Kimmel AR. CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum. Genes Dev. 1993;7:262–272. doi: 10.1101/gad.7.2.262. [DOI] [PubMed] [Google Scholar]
- Schnitzler G, Fischer W, Firtel R. Cloning and characterization of the G-box binding factor, an essential component of the developmental switch between early and late development in Dictyostelium. Genes Dev. 1994;8:502–514. doi: 10.1101/gad.8.4.502. [DOI] [PubMed] [Google Scholar]
- Schnitzler GR, Briscoe C, Brown JM, Firtel RA. Serpentine cAMP receptors may act through a G-protein-independent pathway to induce post-aggregative development in Dictyostelium. Cell. 1995;81:737–745. doi: 10.1016/0092-8674(95)90535-9. [DOI] [PubMed] [Google Scholar]
- Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Kuspa A, Loomis WF. A multidrug resistance transporter serine protease gene is required for prestalk specialization in Dictyostelium. Genes Dev. 1995;9:1111–1122. doi: 10.1101/gad.9.9.1111. [DOI] [PubMed] [Google Scholar]
- Springer ML, Patterson B, Spudich JA. Stage-specific requirement for myosin II during Dictyostelium development. Development. 1994;120:2651–2660. doi: 10.1242/dev.120.9.2651. [DOI] [PubMed] [Google Scholar]
- Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- Wiebel FF, Kunau WH. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugated enzymes. Nature. 1992;359:73–75. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- Williams J, Morrison A. Prestalk cell-differentiation and movement during the morphogenesis of Dictyostelium discoideum. Prog Nucleic Acid Res Mol Biol. 1994;47:1–27. doi: 10.1016/s0079-6603(08)60248-2. [DOI] [PubMed] [Google Scholar]
- Zhen M, Schein J, Baillie D, Candido E. An essential ubiquitin-conjugating enzyme with tissue and developmental specificity in the nematode Caenorhabditis elegans. EMBO J. 1996;15:3229–3237. [PMC free article] [PubMed] [Google Scholar]