Abstract
The replication of the retrovirus human T-cell leukemia virus type 1 (HTLV-1) is linked to the development of lymphoid malignancies and inflammatory diseases. Data from in vitro, ex vivo, and in vivo studies have revealed that no specific treatment can prevent or block HTLV-1 replication and therefore that there is no therapy for the prevention and/or treatment of HTLV-1-associated diseases in infected individuals. HTLV-1 and human immunodeficiency virus type 1 (HIV-1) integrases, the enzymes that specifically catalyze the integration of these retroviruses in host cell DNA, share important structural properties, suggesting that compounds that inhibit HIV-1 integration could also inhibit HTLV-1 integration. We developed quantitative assays to test, in vitro and ex vivo, the efficiencies of styrylquinolines and diketo acids, the two main classes of HIV-1 integrase inhibitors. The compounds were tested in vitro in an HTLV-1 strand-transfer reaction and ex vivo by infection of fresh peripheral blood lymphocytes with lethally irradiated HTLV-1-positive cells. In vitro, four styrylquinoline compounds and two diketo acid compounds significantly inhibited HTLV-1 integration in a dose-dependent manner. All compounds active in vitro decreased cell proliferation ex vivo, although at low concentrations; they also dramatically decreased both normalized proviral loads and the number of integration events during experimental ex vivo primary infection. Accordingly, diketo acids and styrylquinolines are the first drugs that produce a specific negative effect on HTLV-1 replication in vitro and ex vivo, suggesting their potential efficiency for the prevention and treatment of HTLV-1-associated diseases.
Human T-cell leukemia virus type 1 (HTLV-1) and human immunodeficiency virus type 1 (HIV-1) are exogenous retroviruses pathogenic for humans. Although both viruses are lymphotropic, their pathogenicities depend on strongly distinct mechanisms. Schematically, in vivo, HIV infection triggers the progressive elimination of CD4+ lymphocytes, leading to immunosuppression, whereas HTLV-1 infection is associated with the clonal expansion of infected cells, possibly leading to malignant CD4+ proliferation or to spinal cord infiltration, infection, and inflammation. Clinically, HIV-induced cellular defects are regularly linked to the development of AIDS, whereas in a minority of carriers, HTLV-1 infection causes adult T-cell leukemia/lymphoma (ATLL) and/or tropical spastic paraparesis/HTLV-1-associated myelopathy (TSP/HAM). The median length of survival for patients with AIDS receiving modern treatment, i.e., triple therapy, is currently over 8 years; in contrast, the prognosis for HTLV-1-associated diseases remains extremely poor. To date, there is no effective treatment for TSP/HAM (32), while the median overall length of survival for patients with ATLL does not exceed a few months (3).
Integration of a DNA copy of the viral RNA genome into host cellular DNA is essential and unique to the retroviral life cycle. After completion of reverse transcription, the retroviral integrase (IN) catalyzes the removal of a dinucleotide from each 3′ end of the linear viral cDNA (processing reaction) (11, 28). Newly generated 3′-OH groups are then used to attack two phosphodiester bonds in the host DNA molecule, resulting in staggered cuts in the target molecule and covalent linkage between the 3′ ends of the viral genome and the host DNA (18, 35). This strand-transfer reaction is also mediated by IN. The steps required for transformation of this intermediate into a covalently closed double strand are currently not fully understood; it is assumed that host proteins are involved (48). Together, these events result in a provirus that displays the hallmarks of integrated retroviral DNA, i.e., a lack of 2 bp in each long terminal repeat (LTR) end of the viral sequence and a short duplication of the flanking host sequences, the length of which is specific to each individual retrovirus. In addition to being involved in processing and strand-transfer reactions, IN catalyzes the so-called disintegration reaction that is actually a reversal of the in vitro strand-transfer reaction (9).
Triple therapy, commonly referred to as highly active antiretroviral therapy, has become the standard treatment for HIV infection. It consists of a protease inhibitor or a nonnucleoside reverse transcriptase inhibitor combined with two nucleoside reverse transcriptase inhibitors. Highly active antiretroviral therapy, however, is often ill-tolerated by the patients. It requires compliance, is expensive, and leads to multidrug resistance (43). Therefore, additional therapeutic approaches have been optimized. One such new approach targets the third viral enzyme, IN. Several compounds have been found to inhibit HIV IN in vitro and ex vivo, whereas recent clinical trials have demonstrated the feasibility of the use and the efficacies of IN inhibitors in humans (22).
Styrylquinolines (SQLs) and diketo acids (DKAs) are two main classes of HIV-1 IN inhibitors. They block proviral integration through distinct mechanisms: SQLs chelate the divalent metal (Mg2+ or Mn2+) in the IN catalytic core domain. DKAs are also thought to bind to the divalent metal ions in the IN active site (23) and compete with target DNA. SQLs share a quinoline substructure linked to an aryl nucleus displaying various hydroxy substitution patterns. These efficient in vitro IN inhibitors act on both 3′ processing and strand-transfer activities (6, 50), probably interfering with LTR-IN binding (42) through a competitive inhibition mechanism (16). SQLs also interfere with the accumulation of viral DNA during reverse transcription (6) and with the nuclear transport of the preintegration complex (39). DKAs compete with target DNA and therefore specifically inhibit the strand-transfer reaction without significantly interfering with 3′ processing (19, 24). These compounds, which not only abolish productive infection but also promote the accumulation of large amounts of circular DNA forms incapable of integration, consequently affect viral DNA integration. Mutations conferring resistance to SQLs or DKAs in cell culture have been identified within IN proteins, thus demonstrating the specificity of their activity (43). The effects of SQL and DKA molecules on HTLV-1 integration have not been tested. As retroviral INs share many structural properties, we hypothesized that potent HIV-1 IN inhibitors such as SQLs and DKAs could also inhibit HTLV-1 integration.
Anti-HIV reverse transcriptase and/or protease drugs have been found to be inefficient against HTLV-1 replication, and no specific anti HTLV-1 compound has been developed to date. In the study described here, we tested HIV IN inhibitors against HTLV-1 in vitro and in vivo. By using specific assays designed to monitor the effects of IN inhibitors in vitro and in vivo, we demonstrated that some HIV IN inhibitors are efficient against HTLV-1 replication, thus encouraging the use of these drugs in clinical tests.
MATERIALS AND METHODS
In vitro strand-transfer reaction.
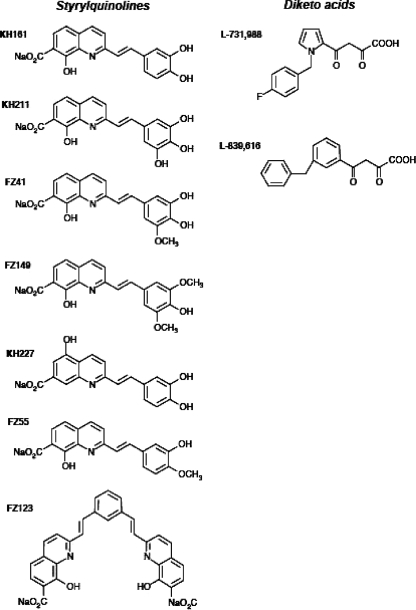
We tested the effects of seven distinct SQL derivatives and two distinct DKAs on HTLV-1 integration in vitro and ex vivo (Fig. 1). The SQLs tested were synthesized as described previously (33, 50). They corresponded to KH161, KH211, FZ41, FZ149, KH227, FZ123, and FZ55. Their effects on HIV-1 IN have been described previously (5, 10, 13, 20, 33, 40, 41). The two DKAs, a kind gift from Merck, were L-731,988 and L-839,616. Their formulas and their effects on HIV-1 IN have also been described previously (8, 15, 44). In vitro strand-transfer reactions were carried out in triplicate, as described previously (31), in the presence of increasing concentrations of the compounds and as shown in Fig. 2A. These compounds are KH161, KH211, FZ41, FZ149, KH227, FZ123, and FZ55, which correspond to the SQLs, and L-731,988 and L-839,616, which correspond to the DKAs. The same batches of each of the nine compounds were used. Their formulas and their effects on HIV-1 IN have been published elsewhere (24, 33, 50). The synthetic double-strand target sequence was similar to that of the naturally curved kinetoplast DNA containing short (5-bp) oligo(dA)-oligo(dT) tracts every 10 to 11 bp separated by the base pairs GGCC/CCGG (45) and displayed hot spots for HTLV-1 integration in vitro (31). This 50-bp target is encompassed by two 20-bp conserved primer-specific sequences. The double-strand 3′ HTLV-1 LTRs were recessed at the 3′ end and corresponded to the last 54 bp of the HTLV-1 wild-type provirus. Recombinant HTLV-1 IN was purified from Escherichia coli BL21(DE3), as described previously (1, 27, 31). Briefly, HTLV-1 IN containing plasmid pET22B, a gift from Coleen B. Jonsson, was expressed in BL21(DE3) cells, purified by using nickel nitrilotriacetate agarose (Qiagen, Chatsworth, CA), and then transferred to dialysis membrane tubes with a molecular mass cutoff of 10,000 Da (Spectra/Por; Spectrum Laboratories Inc.). The reaction mixtures were incubated at 37°C for 60 min in 15-ml reaction volumes containing the appropriate concentration of IN and 1 pmol of each oligonucleotide under the following buffer conditions: 25 mM HEPES (pH 7.4), 10 mM β-mercaptoethanol, 10% glycerol, and 7.5 mM MnCl2. The integration products were amplified by PCR with a fluorescent primer specific for the LTR combined with one of two primers complementary to the two conserved extremities of the target sequence (Fig. 2A). The amplified products were resolved on an Applied Biosystems 377 DNA sequencer with Genescan software (resolution, 0.15 bp). Approximate 50% inhibitory concentrations (IC50s) were calculated by linear regression.
FIG. 1.
SQLs and DKA used for inhibition of HTLV-1 integration.
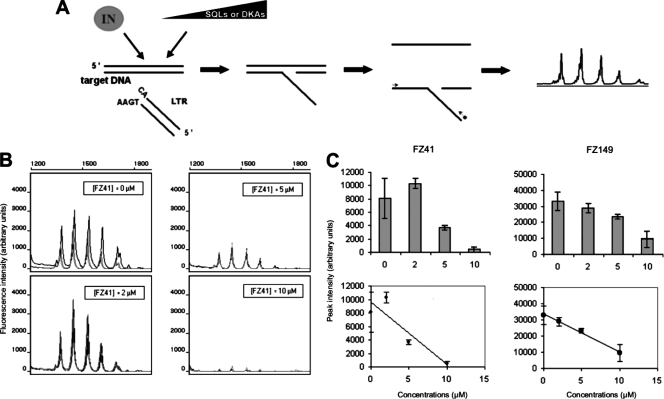
FIG. 2.
Targeting of HTLV-1 integration in vitro. (A) In vitro strand-transfer reactions catalyzed by HTLV-1 IN were performed with synthetic viral and target DNA molecules and various concentrations of SQLs or DKAs. The reaction products were quantified by fluorescent PCR. (B) Fluorescent PCR quantification of HTLV-1 strand transfer in the presence of increasing concentrations (0, 2, 5, and 10 μM) of the SQL derivative FZ41. Experiments were performed in triplicate, as shown by the electropherogram. Each peak represents a hot spot for HTLV-1 integration within the target DNA (see the text for details). (C) Concentration-dependent inhibition of the strand-transfer reaction catalyzed by recombinant HTLV-1 IN in the presence of compounds FZ41 and FZ149. The mean peak intensities from triplicate experiments were plotted as a function of the corresponding inhibitor concentrations.
Ex vivo infection with HTLV-I.
Fresh peripheral blood mononuclear cells (PBMCs) were separated from HTLV-1-negative patient blood samples by Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) density gradient centrifugation. HTLV-1 transmission was performed by coculturing the PBMCs with lethally irradiated (60 Gy) HTLV-1-positive MT2 cells at a ratio of 5:1, as described elsewhere (2, 49). MT2 is a cell line chronically infected with HTLV-1 (34). Cocultures were maintained in six-well plates in 4 ml of RPMI 1640 medium (Gibco, Paisley, United Kingdom) containing 100 U/ml of recombinant interleukin 2 in the presence or absence of increasing concentrations of SQLs or DKAs, which were dissolved in dimethyl sulfoxide.
PCRs.
Quantitative measurement of the HTLV-1 proviral loads in the PBMCs was performed by real-time quantitative PCR with DNA extracted from PBMCs as described previously by using the primers and the TaqMan probe positioned on the tax gene and albumin gene for normalization (37). TaqMan amplification was carried out in reaction volumes of 25 μl with a qPCR MasterMix (Eurogentec, Leuven, Belgium). Each sample was analyzed in triplicate with the use of 250 ng of DNA in each reaction. Thermal cycling was initiated with a 2-min incubation at 50°C, followed by a first denaturation step of 10 min at 95°C and then 45 cycles at 95°C for 15 s and 58°C for 1 min for the tax gene (or 60°C for 1 min for the albumin gene). Inverse PCR, which consists of the amplification of the 3′ extremities of the integrated proviruses with their flanking host sequences, was carried out with the extracted DNA as described previously (37). One microgram of DNA was digested with 20 U NlaIII (New England Biolabs, Montigny-Le-Bretonneux, France) in 1× NlaIII buffer for 3 h at 37°C. DNA was extracted with phenol-chloroform (1:1) and precipitated with 100% ethanol. The digested DNA was circularized for 16 h at 16°C with 20 U of T4 DNA ligase (New England Biolabs) in 600 μl of 1× T4 DNA ligase buffer and 1 mM ATP. DNA was extracted with phenol-chloroform (1:1) and precipitated with 100% ethanol. Five hundred nanograms of circularized DNA was amplified for 30 cycles with the use of 200 μM of primer pair BIO6 (5′-CTCCTGCTAGTTTATTGAGCCATA-3′; positions 8621 to 8598) and LTR1 (5′-TCGCATCTCTCCTTCACGCG-3′; positions 8657 to 8675) (the nucleotide coordinates are numbered according to the HTLV-1 ATK-1 reference sequence). Thermal cycling parameters were as follows: 96°C for 10 min and 30 cycles of 96°C for 60 s, 58°C for 60 s, and 72°C for 3 min, followed by a final elongation step of 10 min at 72°C. The length polymorphism generated by PCR amplification of the HTLV-1 flanking sequences was analyzed by the linear PCR amplification of both the 3′ extremity of the provirus and its flanking sequence (runoff). Two microliters of the amplified product was submitted to 10 cycles of linear PCR with 2 μM of primer BIO5 (5′-TGGCTCGGAGCCAGCGACAGCCCAT-3′; positions 8995 to 9020) radiolabeled with 32P at the 5′ end, 1 U of the Stoffel fragment of Taq DNA polymerase (Perkin-Elmer Applied Biosystems, Courtaboeuf, France), and 200 μM of each deoxynucleoside triphosphate in a final volume of 20 μl. The thermal cycling parameters were as follows: 95°C for 10 min and 10 cycles of 95°C for 60 s, 58°C for 60 s, and 72°C for 3 min, followed by a final elongation step of 10 min at 72°C. After the mixture was boiled, 2 μl of the runoff products was analyzed on a 6% sequencing gel.
RESULTS
Targeting HTLV-1 integration in vitro.
The effects of the SQLs and DKAs on HTLV-1 integration were first tested in vitro. As DKAs mainly interfere with strand transfer rather than with 3′ processing (24) and in order to subsequently compare the IN-inhibitory efficiencies of the SQLs and DKAs, we tested the effects of the SQLs and DKAs on the strand-transfer reaction, i.e., on the integration of 3′-recessed double-strand 3′ HTLV-1 LTRs (54 bp) within a double-strand target DNA sequence. Strand-transfer reactions were carried out as detailed above (Fig. 2A). The compounds did not interfere with the fluorescent PCR rates (data not shown); they were used at various concentrations, including those known to inhibit HIV-1 strand transfer.
Effects of IN inhibitors on the strand-transfer reaction catalyzed by the HTLV-1-encoded IN.
Typical results for HTLV-1 strand-transfer inhibition by SQL derivatives FZ41 and FZ149 are shown in Fig. 2B and C. The approximate IC50s of FZ41 and FZ149 for HIV-1 3′ processing are 0.7 μM and 4.9 μM, respectively (50). We performed HTLV-1 strand-transfer reactions with FZ41 or FZ149 at 0, 2, 5, and 10 μM (three experiments per concentration); Fig. 2C shows that both FZ41 and FZ149 displayed clear concentration-dependent strand-transfer-inhibitory activities in vitro (Fig. 2B and C). Both compounds significantly reduced the overall strand-transfer efficiency without interfering with the position of HTLV-1 3′ LTR integration along the 50-bp target DNA sequence in vitro (Fig. 2B). Data from triplicate experiments permitted a regression line to be drawn (Fig. 2C) and, thereby, the calculation of the 50% strand-transfer-inhibitory concentrations (the approximate IC50s; Fig. 2C), which were 5.2 μM and 7.2 μM for FZ41 and FZ149, respectively. The same procedure was repeated with the remaining five SQL derivatives and the two DKAs. KH161 and KH211 were tested in triplicate at the same three concentrations of FZ149 and FZ41 used (Fig. 2B). KH227, FZ123, and FZ55 were tested in triplicate at 10, 20, and 50 μM. The two DKAs were tested in triplicate at 20, 50, and 100 nM. Table 1 summarizes the approximate IC50s obtained for all nine compounds tested. None interfered with the integration position of synthetic HTLV-1 double-strand 3′ LTRs along the target double-strand DNA sequence. The mean approximate IC50 of the seven SQL compounds was 20 ± 7 μM, and the IC50 range was 4.9 to 52.5 μM. Four compounds (KH161, KH211, FZ41, and FZ149) inhibited the strand-transfer reaction at low concentrations (mean approximate IC50, 6.2 μM; range, 4.9 μM [for KH161] to 7.4 μM [for KH211]), whereas the remaining three compounds (KH227, FZ123, and FZ55) had IC50s greater than approximately 10 μM and were therefore considered to possess no significant inhibitory effect on the HTLV-1 IN.
TABLE 1.
Inhibitory concentrations of SQLs and DKAs on HTLV-1 strand-transfer reaction
| Compound | Approximate IC50 (μM) |
|---|---|
| SQLs | |
| KH161 | 4.9 |
| KH211 | 7.4 |
| FZ41 | 5.2 |
| FZ149 | 7.2 |
| KH227 | 31 |
| FZ123 | 31.8 |
| FZ55 | 52.5 |
| DKAs | |
| L-731,988 | 0.051 |
| L-839,616 | 0.069 |
Possible structure-function relationship of IN inhibitor for blocking HTLV-1 strand-transfer reaction.
It appears from Fig. 1 that some structural characteristics distinguished the three inefficient SQL derivatives from their efficient counterparts. Indeed, the unique characteristic of KH227 was the presence of a carboxy radical at position 5 of the quinoline moiety. FZ123 was the sole SQL compound that harbored two quinoline nuclei. Finally, FZ55 displayed the highest approximate IC50 and was characterized by the presence of a hydroxyl radical and a methoxy radical at positions 4 and 3 of the aryl nucleus, respectively. The four SQL derivatives active against HTLV-1 IN in vitro have been found to significantly inhibit the HIV-1 IN, with a mean approximate IC50 of 1.725 μM (50). Interestingly, FZ55, which does not inhibit HTLV-1 strand transfer, possesses an approximate IC50 of 2.8 μM for the HIV-1 IN in vitro (33, 50). To our knowledge, there are no published data on the activities of KH227 and FZ123 against HIV-1 IN. For the DKAs, the approximate IC50s of L-731,988 and L-839,616 were 0.051 μM and 0.069 μM, respectively. As reported above for the effective SQLs, the active DKAs did not modify the integration position of the HTLV-1 3′ LTR along the target DNA double-strand sequence. For HTLV-1 integration in vitro, the mean approximate IC50 of the effective DKAs was 100 times lower than that of the four effective SQLs. For HIV-1 IN inhibition, the approximate IC50s of L-731,988 and L-839,616 are 0.05 μM and 0.17 μM, respectively (24). Therefore, in contrast to the active SQLs in vitro, which exhibit approximate IC50s more than three times higher for HTLV than for HIV, the active DKAs (compound L-839,616) may be more efficient against the HTLV IN than against the HIV-1 enzyme in vitro. Together, these experiments demonstrate for the first time that some SQLs and DKAs are potent inhibitors of HTLV-1 integration in vitro. This led us to test the potential inhibitory effects of these products in vivo, i.e., on the intercellular transmission of HTLV-1 and on its cell-associated expansion and amplification.
Testing of IN inhibitors on HTLV-1 replication ex vivo.
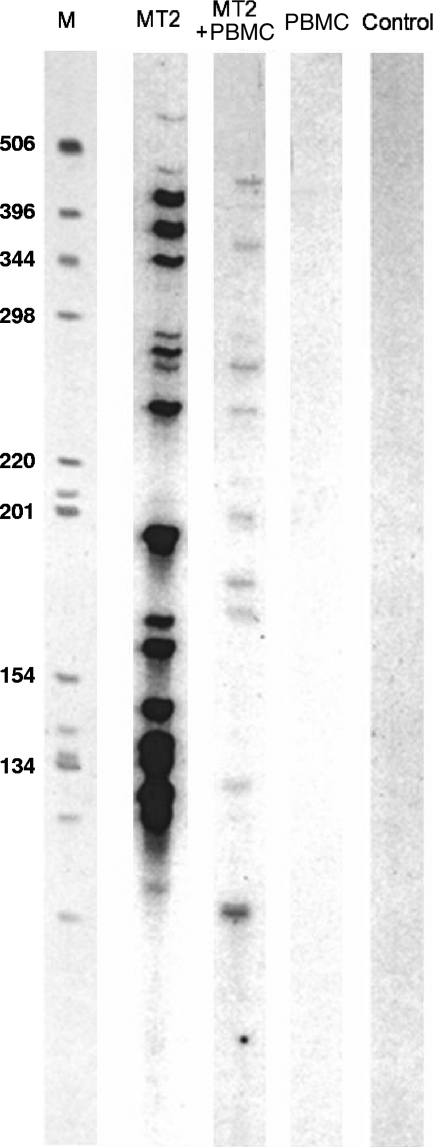
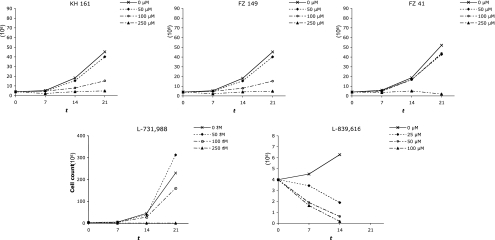
In contrast to HIV replication, HTLV-1 intercellular transmission depends on a close intercellular contact that facilitates the transmission of the virus through the recently described virological synapse (26). After viral entry, reverse transcription, and integration, the second hallmark of HTLV-1 replication is the cell-associated expansion and amplification of the HTLV provirus (36). This clonal expansion of HTLV-1-infected cells is in strong contrast to the cellular depletion associated with HIV replication ex vivo. Coculture of irradiated HTLV-1-positive cells with uninfected lymphocytes is the most appropriate model of HTLV-1 infection in vivo: it incorporates both HTLV-1 intercellular transmission and the subsequent clonal expansion of newly infected cells (30). Accordingly, HTLV-1 transmission was performed in the present study by coculturing PBMCs from healthy adult donors who were seronegative for HTLV-1 and -2, HIV, hepatitis B virus, and hepatitis C virus with lethally irradiated MT2 cells, as detailed in the Materials and Methods section. Each MT2 cell was found to harbor 18 integrated proviruses (Fig. 3), and at day 7 of coculture of fresh PBMCs with irradiated MT2 cells, inverse PCR failed to detect any MT2-specific HTLV-1 integration site; at this time point, the proviral copies detected corresponded to those from newly infected cells (Fig. 3). Five compounds were selected for use in the in vivo assays. Selection was based on a combination of low toxicity (50% cytotoxic concentration [CC50; >100 μM] and high efficiency in vitro (approximate IC50, <10 μM). The CC50s were assessed with uninfected PBMCs. Accordingly, KH227, FZ123, and FZ55 were excluded on the basis of their low inhibitory effects in vitro (Table 1), whereas KH211, although it displayed a low approximate IC50, was not tested on the basis of its low CC50 (10.7 μM) (33). The toxicities of L-731,988 and L-839,616 were assessed by cultivating fresh PBMCs with various concentrations of each IN inhibitor. Cellular viability was assessed at 250, 500, and 1,000 μM (three experiments per concentration). The CC50s of L-731,988 and L-839,616 were 520 and 220 μM, respectively. Therefore, KH161, FZ41, FZ149, L-731,988, and L-839,616 combined low cellular toxicity with significant antiviral effects in vitro and were tested for their antiviral effects ex vivo. To this end, selected SQLs and DKAs were added at final concentrations of 0, 50, 100, and 250 μM at the onset of the culture and then at half the initial concentrations 4, 7, and 10 days postinfection to ensure the presence of constant drug levels in the culture medium in the early phase of infection. We assessed the effects of the IN inhibitors on cell growth kinetics by measuring cell counts for each compound concentration on days 7, 14, and 21 of coculture. Figure 4 shows that each compound inhibited cell growth in a concentration-dependent manner.
FIG. 3.
Distribution of HTLV-1 integration and clonality during experimental infection of PBMCs with irradiated HTLV-1-positive MT2 cells. Inverse PCRs were carried out with DNA extracted from MT2 cells (lane MT2), from PBMCs before coculture (lane PBMC) and after coculture with irradiated MT2 cells (lane PBMC + MT2), as detailed in the text. The DNA of peripheral blood lymphocytes obtained from an additional uninfected donor served as a negative control (lane Control). Lane M, molecular weight marker.
FIG. 4.
Altered growth kinetics of PBMCs cocultured with irradiated HTLV-1-positive MT2 cells in the presence of various concentrations (0, 50, 100, and 250 μM) of SQLs (compounds KH161, FZ149, and FZ41) or DKAs (compounds L-731,988 and L-839,616). Cell preparation and coculture were performed as detailed in the text. x axis, time (t) that has elapsed from coculture (days); y axis, cell counts (106).
Effects of SQLs and DKAs on overall HTLV-1 proviral loads ex vivo.
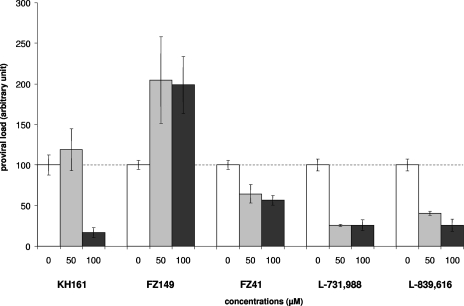
The antiviral effects of the IN inhibitors on the overall proviral loads were first evaluated after coculture of fresh PBMCs from uninfected donors with irradiated MT2 cells. Viral loads were measured on day 7 by real-time quantitative PCR (47). In the absence of inhibitor, the HTLV-1 proviral loads on day 7 of coculture ranged from 0.5 to 12 copies per cell (data not shown). It is clear from Fig. 5 that IN inhibitors displayed distinct effects on the HTLV-1 proviral loads after experimental ex vivo infection. The SQL derivative FZ149 increased the number of viral copies, but the remaining five compounds significantly decreased the proviral loads. A dose-dependent effect was clearly evidenced with the SQL KH161 and the DKA L-839,616, whereas no significant differences were noted at concentrations between 50 and 100 μM for the remaining three effective compounds. Overall, the antiviral effects of the DKAs were greater than those of the SQLs in vivo (Fig. 5), but the difference was significantly lower than that observed in vitro (Fig. 2 and Table 1).
FIG. 5.
Effects of IN inhibitors on HTLV-1 proviral loads after coculture of PBMCs with irradiated HTLV-1-positive MT2 cells in the presence of 0, 50, and 100 μM of SQLs or DKAs. DNA was extracted at 7 days of coculture, and for each concentration, real-time quantitative PCR was carried out in triplicate, as detailed in the text. In the absence of compound, proviral loads were set to 100 units in order to better compare the effects of the compounds tested.
Effects of SQLs and DKAs on HTLV-1 integration events ex vivo.
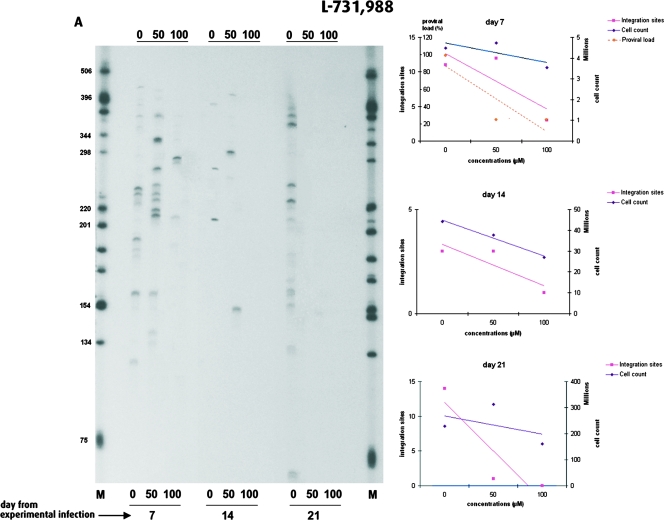
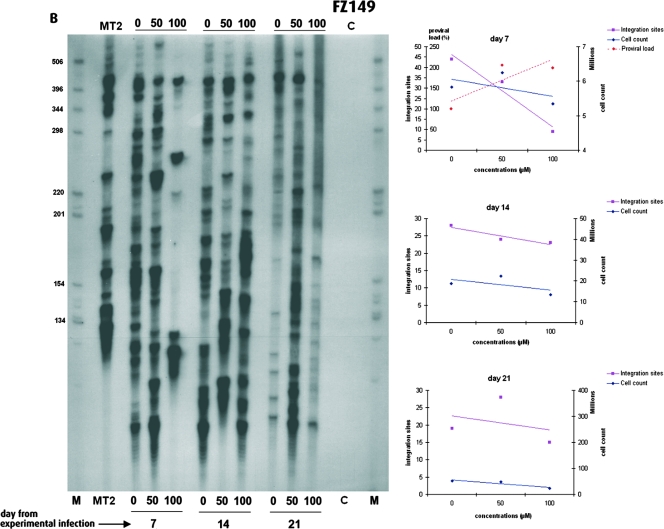
As the HTLV-1 proviral load depends on the combination of new integration events and the subsequent clonal expansion of newly infected cells, the decreased proviral loads observed with the efficient DKA or SQL derivatives might result from a decreased number of integration events specifically attributable to IN inhibitors, from the cellular toxicity of the compounds, or from a combination thereof. We therefore investigated the effects of the IN inhibitors on the number of new integration events following experimental infection. To this end, we measured the frequency of new integration events after coculture of fresh PBMCs with irradiated MT2 cells in the presence of various concentrations of an efficient IN inhibitor and an inefficient IN inhibitor, i.e., L-731,988 and FZ149, respectively. The DKA L-731,988 was the most efficient HTLV-1 IN inhibitor both in vitro (Table 1) and ex vivo (Fig. 5), while its CC50 was in the lower range (518 μM). In contrast, the SQL FZ149, which had a CC50 of 258 μM, was more than 10 times less efficient in vitro (approximate IC50, 7.2 μM) and had no antiviral effect in vivo (Fig. 5). Figure 6A represents the HTLV-1 integration patterns of experimentally infected PBMCs in the presence of various concentrations of L-731,988. As observed for the proviral loads on day 7 (Fig. 5), the number of HTLV-1 integration events decreased in a concentration-dependent manner. On day 14, no signal could be obtained at 100 μM, whereas on day 21, no signal was observed at 50 and 100 μM. At each time point the number of HTLV-1 integration sites detected decreased with a steeper slope than that reflecting the cellular toxicity of L-731,988. Therefore, after experimental infection, the significant negative effect of L-731,988 on proviral loads correlated with its strong negative effect on HTLV-1 integration. In contrast to L-731,988, the SQL FZ149 had only a little effect on HTLV-1 integration ex vivo. As can be seen from Fig. 6B, this negative effect was modest and was observed only at 100 μM, 7 days after the initiation of the experimental infection. In contrast to the observation made with L-731,988 (Fig. 6A), no significant variation in the number of subsequent HTLV-1 integration events could be observed over time (Fig. 6B). Therefore, after experimental infection, the absence of an effect of FZ149 on the proviral loads (Fig. 5) correlated with the absence of an effect of FZ149 on HTLV-1 integration. Furthermore, data from Fig. 5 and 6 suggest that FZ149 actually stimulates infection. In addition to L-731,966, the remaining three compounds tested decreased both the HTLV-1 proviral loads and the numbers of integration events. For these four efficient IN inhibitors, the correlation between the proviral loads and the number of integration events on day 7 of coculture was statistically significant (P < 0.02; R of ∼0.7, Spearman rank correlation).
FIG. 6.
Effects of the IN inhibitors L-731,988 (A) and FZ149 (B) on HTLV-1 integration. (Left) Clonality of HTLV-1-positive cells over time in the presence of various concentrations (0, 50, or 100 μM) of inhibitor. Inhibitor concentrations and days of analysis are given at the bottom of the gel. (Right) Temporal (day 7, 14, and 21) fluctuations of growth kinetics, proviral loads, and HTLV-1 integration frequency as a function of IN inhibitor concentrations.
Together, these results indicate that the antiviral effect of efficient IN inhibitors on HTLV-1 evidenced in vivo depends on a bona fide inhibition of HTLV-1 integration. It is of note that the antiviral effects of the IN inhibitors on HTLV-1 in vivo (Fig. 5 and 6) paralleled their inhibitory effects on strand transfer in vitro (Table 1).
DISCUSSION
Our data demonstrate that selected anti-HIV IN inhibitors also inhibit HTLV-1 integration both in vitro and ex vivo. This is the first evidence that drugs target the same mechanism in both viruses, i.e., the integration of viral cDNA in host cell DNA. Very recent work has shown that HIV-1 IN inhibitors can inhibit other retroviral/retroelement recombinases (12, 46). In vitro, HTLV-1 and HIV-1 strains have been found to be equally susceptible to nucleoside reverse transcriptase inhibitors, such as zidovudine, zalcitabine, didanosine and stavudine (21). However, using a quantitative reverse transcriptase activity assay, Garcia-Lerma et al. (21) have demonstrated that high-level resistance to lamivudine is characteristic of HTLV-1 isolates. Similarly, Balestrieri et al. have demonstrated ex vivo that lamivudine possesses an antiviral effect against HTLV-1 that does not interfere with HTLV-1 reverse transcription (2). The combination of zidovudine and interferon has been found to be effective in some ATLL cases (3, 25), but there is no evidence zidovudine has a specific effect on HTLV-1 reverse transcription (4, 14). More recently, the protease inhibitor ritonavir has been found to exert an antileukemic effect against ATLL cells ex vivo (17). Again, this effect is not based on activity against the HTLV-1 protease but, rather, is based on the ritonavir-dependent inhibition of NF-κB transcriptional activation in ATLL cells. Ex vivo, HIV IN inhibition may lead to the selection of resistant strains that harbor substitutions in the IN gene, demonstrating that IN is the relevant target of these drugs (43). Our system precludes the detection of such mutants because after experimental infection, HTLV-1-bearing cells proliferate ex vivo, without evidence of a frequent horizontal viral replicative cycle (7). However, the effects of the efficient SQL and DKA derivatives against HTLV-1 were found here to result from a dramatic decrease in the number of new integration events. Thus, one can reasonably propose that the anti-HTLV-1 activities of the SQLs or the DKAs result from an inhibition of the integration process.
Figure 6A shows that no integrated HTLV-1 proviral sequence could be detected on day 21 after experimental infection when cocultures were performed in the presence of the DKA L-731,988 at a concentration of 100 μM, suggesting that complete clearance of the virus could be obtained with IN inhibitors in the absence of significant cellular toxicity. To our knowledge, this is the first example of drugs that produce specific effects against HTLV-1. The question that arises is whether IN inhibitors can be used for the treatment of HTLV-1 infection. As mentioned above, the replicative pattern of the virus mainly involves the clonal expansion of its CD4+ and CD8+ host cells. The blocking of integration would thus be of little interest during the chronic phase of the infection. However, primary delta retroviral infection is characterized by a burst of horizontal viral replication (29, 37, 38), meaning that IN inhibitors could be an option for the treatment of early infection. The present results encourage the development of clinical assays.
Acknowledgments
This work was supported by the Agence Nationale pour la Recherche contre le Sida, the Centre Léon Bérard, the Centre National pour la Recherche Scientifique, and the Institut National de la Santé et de la Recherche Médicale. E.W. is supported by the Hospices Civils de Lyon and the Lyon I University.
We thank Marie-Dominique Reynaud for preparation of the manuscript.
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Balakrishnan, M., and C. B. Jonsson. 1997. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J. Virol. 71:1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balestrieri, E., G. Forte, C. Matteucci, A. Mastino, and B. Macchi. 2002. Effect of lamivudine on transmission of human T-cell lymphotropic virus type 1 to adult peripheral blood mononuclear cells in vitro. Antimicrob. Agents Chemother. 46:3080-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bazarbachi, A., D. Ghez, Y. Lepelletier, R. Nasr, H. de The, M. E. El-Sabban, and O. Hermine. 2004. New therapeutic approaches for adult T-cell leukaemia. Lancet Oncol. 5:664-672. [DOI] [PubMed] [Google Scholar]
- 4.Bazarbachi, A., R. Nasr, M. E. El-Sabban, A. Mahe, R. Mahieux, A. Gessain, N. Darwiche, G. Dbaibo, J. Kersual, Y. Zermati, L. Dianoux, M. K. Chelbi-Alix, H. de The, and O. Hermine. 2000. Evidence against a direct cytotoxic effect of alpha interferon and zidovudine in HTLV-I associated adult T cell leukemia/lymphoma. Leukemia 14:716-721. [DOI] [PubMed] [Google Scholar]
- 5.Benard, C., F. Zouhiri, M. Normand-Bayle, M. Danet, D. Desmaele, H. Leh, J. F. Mouscadet, G. Mbemba, C. M. Thomas, S. Bonnenfant, M. Le Bret, and J. d'Angelo. 2004. Linker-modified quinoline derivatives targeting HIV-1 integrase: synthesis and biological activity. Bioorg. Med. Chem. Lett. 14:2473-2476. [DOI] [PubMed] [Google Scholar]
- 6.Bonnenfant, S., C. M. Thomas, C. Vita, F. Subra, E. Deprez, F. Zouhiri, D. Desmaele, J. D'Angelo, J. F. Mouscadet, and H. Leh. 2004. Styrylquinolines, integrase inhibitors acting prior to integration: a new mechanism of action for anti-integrase agents. J. Virol. 78:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavrois, M., S. Wain-Hobson, and E. Wattel. 1995. Stochastic events in the amplification of HTLV-I integration sites by linker-mediated PCR. Res. Virol. 146:179-184. [DOI] [PubMed] [Google Scholar]
- 8.Chi, G., V. Nair, E. Semenova, and Y. Pommier. 2007. A novel diketo phosphonic acid that exhibits specific, strand-transfer inhibition of HIV integrase and anti-HIV activity. Bioorg. Med. Chem. Lett. 17:1266-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 10.Courcot, B., D. Firley, B. Fraisse, P. Becker, J. M. Gillet, P. Pattison, D. Chernyshov, M. Sghaier, F. Zouhiri, D. Desmaele, J. d'Angelo, F. Bonhomme, S. Geiger, and N. E. Ghermani. 2007. Crystal and electronic structures of magnesium(II), copper(II), and mixed magnesium(II)-copper(II) complexes of the quinoline half of styrylquinoline-type HIV-1 integrase inhibitors. J. Phys. Chem. B 111:6042-6050. [DOI] [PubMed] [Google Scholar]
- 11.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62:829-837. [DOI] [PubMed] [Google Scholar]
- 12.Czyz, A., K. A. Stillmock, D. J. Hazuda, and W. S. Reznikoff. 2007. Dissecting Tn5 transposition using HIV-1 integrase diketoacid inhibitors. Biochemistry 46:10776-10789. [DOI] [PubMed] [Google Scholar]
- 13.d'Angelo, J., J. F. Mouscadet, D. Desmaele, F. Zouhiri, and H. Leh. 2001. HIV-1 integrase: the next target for AIDS therapy? Pathol. Biol. (Paris) 49:237-246. [DOI] [PubMed] [Google Scholar]
- 14.Datta, A., M. Bellon, U. Sinha-Datta, A. Bazarbachi, Y. Lepelletier, D. Canioni, T. A. Waldmann, O. Hermine, and C. Nicot. 2006. Persistent inhibition of telomerase reprograms adult T-cell leukemia to p53-dependent senescence. Blood 108:1021-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dayam, R., J. Deng, and N. Neamati. 2006. HIV-1 integrase inhibitors: 2003-2004 update. Med. Res. Rev. 26:271-309. [DOI] [PubMed] [Google Scholar]
- 16.Deprez, E., S. Barbe, M. Kolaski, H. Leh, F. Zouhiri, C. Auclair, J. C. Brochon, M. Le Bret, and J. F. Mouscadet. 2004. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol. Pharmacol. 65:85-98. [DOI] [PubMed] [Google Scholar]
- 17.Dewan, M. Z., J. N. Uchihara, K. Terashima, M. Honda, T. Sata, M. Ito, N. Fujii, K. Uozumi, K. Tsukasaki, M. Tomonaga, Y. Kubuki, A. Okayama, M. Toi, N. Mori, and N. Yamamoto. 2006. Efficient intervention of growth and infiltration of primary adult T-cell leukemia cells by an HIV protease inhibitor, ritonavir. Blood 107:716-724. [DOI] [PubMed] [Google Scholar]
- 18.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211-1221. [DOI] [PubMed] [Google Scholar]
- 19.Espeseth, A. S., P. Felock, A. Wolfe, M. Witmer, J. Grobler, N. Anthony, M. Egbertson, J. Y. Melamed, S. Young, T. Hamill, J. L. Cole, and D. J. Hazuda. 2000. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. USA 97:11244-11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firley, D., B. Courcot, J. M. Gillet, B. Fraisse, F. Zouhiri, D. Desmaele, J. d'Angelo, and N. E. Ghermani. 2006. Experimental/theoretical electrostatic properties of a styrylquinoline-type HIV-1 integrase inhibitor and its progenitors. J. Phys. Chem. B 110:537-547. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Lerma, J. G., S. Nidtha, and W. Heneine. 2001. Susceptibility of human T cell leukemia virus type 1 to reverse-transcriptase inhibitors: evidence for resistance to lamivudine. J. Infect. Dis. 184:507-510. [DOI] [PubMed] [Google Scholar]
- 22.Grinsztejn, B., B. Y. Nguyen, C. Katlama, J. M. Gatell, A. Lazzarin, D. Vittecoq, C. J. Gonzalez, J. Chen, C. M. Harvey, and R. D. Isaacs. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261-1269. [DOI] [PubMed] [Google Scholar]
- 23.Grobler, J. A., K. Stillmock, B. Hu, M. Witmer, P. Felock, A. S. Espeseth, A. Wolfe, M. Egbertson, M. Bourgeois, J. Melamed, J. S. Wai, S. Young, J. Vacca, and D. J. Hazuda. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA 99:6661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 25.Hermine, O., D. Bouscary, A. Gessain, P. Turlure, V. Leblond, N. Franck, A. Buzyn-Veil, B. Rio, E. Macintyre, F. Dreyfus, et al. 1995. Brief report: treatment of adult T-cell leukemia-lymphoma with zidovudine and interferon alfa. N. Engl. J. Med. 332:1749-1751. [DOI] [PubMed] [Google Scholar]
- 26.Igakura, T., J. C. Stinchcombe, P. K. Goon, G. P. Taylor, J. N. Weber, G. M. Griffiths, Y. Tanaka, M. Osame, and C. R. Bangham. 2003. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 299:1713-1716. [DOI] [PubMed] [Google Scholar]
- 27.Jonsson, C. B., G. A. Donzella, E. Gaucan, C. M. Smith, and M. J. Roth. 1996. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J. Virol. 70:4585-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, R. A., G. Merkel, J. Kulkosky, J. Leis, and A. M. Skalka. 1990. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell 63:87-95. [DOI] [PubMed] [Google Scholar]
- 29.Kazanji, M., A. Ureta-Vidal, S. Ozden, F. Tangy, B. de Thoisy, L. Fiette, A. Talarmin, A. Gessain, and G. de The. 2000. Lymphoid organs as a major reservoir for human T-cell leukemia virus type 1 in experimentally infected squirrel monkeys (Saimiri sciureus): provirus expression, persistence, and humoral and cellular immune responses. J. Virol. 74:4860-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura, K., D. L. Rudolph, C. Goldsmith, T. M. Folks, and R. B. Lal. 1993. Isolation, characterization, and transmission of human T-lymphotropic virus types I and II in culture. Curr. Microbiol. 27:355-360. [DOI] [PubMed] [Google Scholar]
- 31.Leclercq, I., F. Mortreux, A. Gabet, C. B. Jonsson, and E. Wattel. 2000. Basis of HTLV-1 target site selection. AIDS Res. Hum. Retrovir. 16:1653-1659. [DOI] [PubMed] [Google Scholar]
- 32.Machuca, A., B. Rodes, and V. Soriano. 2001. The effect of antiretroviral therapy on HTLV infection. Virus Res. 78:93-100. [DOI] [PubMed] [Google Scholar]
- 33.Mekouar, K., J. F. Mouscadet, D. Desmaele, F. Subra, H. Leh, D. Savoure, C. Auclair, and J. d'Angelo. 1998. Styrylquinoline derivatives: a new class of potent HIV-1 integrase inhibitors that block HIV-1 replication in CEM cells. J. Med. Chem. 41:2846-2857. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi, I., I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata, and Y. Hinuma. 1981. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature 294:770-771. [DOI] [PubMed] [Google Scholar]
- 35.Mizuuchi, K. 1992. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 267:21273-21276. [PubMed] [Google Scholar]
- 36.Mortreux, F., A. S. Gabet, and E. Wattel. 2003. Molecular and cellular aspects of HTLV-1 associated leukemogenesis in vivo. Leukemia 17:26-38. [DOI] [PubMed] [Google Scholar]
- 37.Mortreux, F., M. Kazanji, A. S. Gabet, B. de Thoisy, and E. Wattel. 2001. Two-step nature of human T-cell leukemia virus type 1 replication in experimentally infected squirrel monkeys (Saimiri sciureus). J. Virol. 75:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moules, V., C. Pomier, D. Sibon, A. S. Gabet, M. Reichert, P. Kerkhofs, L. Willems, F. Mortreux, and E. Wattel. 2005. Fate of premalignant clones during the asymptomatic phase preceding lymphoid malignancy. Cancer Res. 65:1234-1243. [DOI] [PubMed] [Google Scholar]
- 39.Mousnier, A., H. Leh, J. F. Mouscadet, and C. Dargemont. 2004. Nuclear import of HIV-1 integrase is inhibited in vitro by styrylquinoline derivatives. Mol. Pharmacol. 66:783-788. [DOI] [PubMed] [Google Scholar]
- 40.Normand-Bayle, M., C. Benard, F. Zouhiri, J. F. Mouscadet, H. Leh, C. M. Thomas, G. Mbemba, D. Desmaele, and J. d'Angelo. 2005. New HIV-1 replication inhibitors of the styryquinoline class bearing aroyl/acyl groups at the C-7 position: synthesis and biological activity. Bioorg. Med. Chem. Lett. 15:4019-4022. [DOI] [PubMed] [Google Scholar]
- 41.Ouali, M., C. Laboulais, H. Leh, D. Gill, D. Desmaele, K. Mekouar, F. Zouhiri, J. d'Angelo, C. Auclair, J. F. Mouscadet, and M. Le Bret. 2000. Modeling of the inhibition of retroviral integrases by styrylquinoline derivatives. J. Med. Chem. 43:1949-1957. [DOI] [PubMed] [Google Scholar]
- 42.Ouali, M., C. Laboulais, H. Leh, D. Gill, E. Xhuvani, F. Zouhiri, D. Desmaele, J. d'Angelo, C. Auclair, J. F. Mouscadet, and M. Le Bret. 2000. Tautomers of styrylquinoline derivatives containing a methoxy substituent: computation of their population in aqueous solution and their interaction with RSV integrase catalytic core. Acta Biochim. Pol. 47:11-22. [PubMed] [Google Scholar]
- 43.Pommier, Y., A. A. Johnson, and C. Marchand. 2005. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 4:236-248. [DOI] [PubMed] [Google Scholar]
- 44.Pommier, Y., and N. Neamati. 1999. Inhibitors of human immunodeficiency virus integrase. Adv. Virus Res. 52:427-458. [DOI] [PubMed] [Google Scholar]
- 45.Pruss, D., R. Reeves, F. Bushman, and A. Wolffe. 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 40:25031-25041. [PubMed] [Google Scholar]
- 46.Shimura, K., E. Kodama, Y. Sakagami, Y. Matsuzaki, W. Watanabe, K. Yamataka, Y. Watanabe, Y. Ohata, S. Doi, M. Sato, M. Kano, S. Ikeda, and M. Matsuoka. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82:764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sibon, D., A. S. Gabet, M. Zandecki, C. Pinatel, J. Thete, M. H. Delfau-Larue, S. Rabaaoui, A. Gessain, O. Gout, S. Jacobson, F. Mortreux, and E. Wattel. 2006. HTLV-1 propels untransformed CD4 lymphocytes into the cell cycle while protecting CD8 cells from death. J. Clin. Investig. 116:974-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, J., E. Balestrieri, S. Grelli, C. Matteucci, V. Pagnini, C. D'Agostini, A. Mastino, and B. Macchi. 2001. Efficacy of 3′-azido 3′deoxythymidine (AZT) in preventing HTLV-1 transmission to human cord blood mononuclear cells. Virus Res. 78:67-78. [DOI] [PubMed] [Google Scholar]
- 50.Zouhiri, F., J. F. Mouscadet, K. Mekouar, D. Desmaele, D. Savoure, H. Leh, F. Subra, M. Le Bret, C. Auclair, and J. d'Angelo. 2000. Structure-activity relationships and binding mode of styrylquinolines as potent inhibitors of HIV-1 integrase and replication of HIV-1 in cell culture. J. Med. Chem. 43:1533-1540. [DOI] [PubMed] [Google Scholar]