Abstract
This study examined the adaptive response and survival of planktonic and biofilm phenotypes of Salmonella enterica serovar Enteritidis adapted to benzalkonium chloride (BC). Planktonic cells and biofilms were continuously exposed to 1 μg ml−1 of BC for 144 h. The proportion of BC-adapted biofilm cells able to survive a lethal BC treatment (30 μg ml−1) was significantly higher (4.6-fold) than that of BC-adapted planktonic cells. Similarly, there were 18.3-fold more survivors among the BC-adapted biofilm cells than among their nonadapted (i.e., without prior BC exposure) cell counterparts at the lethal BC concentration, and this value was significantly higher than the value for BC-adapted planktonic cells versus nonadapted cells (3.2-fold). A significantly higher (P < 0.05) proportion of surviving cells was noticed among BC-adapted biofilm cells relative to BC-adapted planktonic cells following a 10-min heat shock at 55°C. Fatty acid composition was significantly influenced by phenotype (planktonic cells or biofilm) and BC adaptation. Cell surface roughness of biofilm cells was also significantly greater (P < 0.05) than that of planktonic cells. Key proteins upregulated in BC-adapted planktonic and biofilm cells included CspA, TrxA, Tsf, YjgF, and a probable peroxidase, STY0440. Nine and 17 unique proteins were upregulated in BC-adapted planktonic and biofilm cells, respectively. These results suggest that enhanced biofilm-specific upregulation of 17 unique proteins, along with the increased expression of CspA, TrxA, Tsf, YjgF, and a probable peroxidase, phenotype-specific alterations in cell surface roughness, and a shift in fatty acid composition conferred enhanced survival to the BC-adapted biofilm cell population relative to their BC-adapted planktonic cell counterparts.
Antimicrobial resistance of bacterial pathogens has been attributed either to inherited or noninherited resistance mechanisms (24). Noninherited, or adaptive, resistance to antibiotics and other chemotherapeutic agents has been widely reported in various species of bacteria; the physiological state and the physical structure of the adapted population have been attributed to this phenomenon, which is purely phenotypic in nature (23, 24). The exact biochemical mechanisms of adaptive resistance and the cross-resistance of “adapted” strains to unrelated chemotherapeutic agents and biocides, as well as the contribution of these phenomena to multidrug-resistant “superbugs,” remain largely unknown (7, 23, 25, 34, 35, 40).
Various molecular mechanisms have been reported to play roles in bacterial adaptive responses to antimicrobial compounds. These adaptive mechanisms act either singly or synergistically in order to confer antimicrobial resistance. Among them, “slow” multiplication resulting in “persisters” (2, 26), SOS response that blocks cell division during the repair of DNA damage, cold shock response, stress response, detoxification, altered permeability of the outer membranes due to changes in lipopolysaccharide (LPS) composition, outer membrane proteins, cytoplasmic membrane proteins, fatty acid composition, content of the cytoplasmic membrane, cell surface charge, hydrophobicity, active efflux of the agent, etc., are believed to be significant (3, 8, 19, 24, 27, 28, 41). It has been reported that a single microorganism may have multiple, possibly interconnected, adaptive mechanisms depending on the nature of the antimicrobial agent (9, 42). There are also recent reports suggesting that molecular mechanisms providing bacterial resistance to biocides may provide cross-protection against certain antibiotics (5, 7, 22).
Benzalkonium chloride (BC) is a well-known quaternary ammonium compound (QAC) that has been widely used as a surface disinfectant, antiseptic, and preservative. QACs are bacteriostatic in low concentrations and bactericidal in high concentrations; their antibacterial activity is associated with adsorption by the bacterial cells and subsequent leakage of cellular constituents (30). Sakagami et al. (36) reported that the ability of BC to permeate BC-resistant Pseudomonas aeruginosa was reduced because of the increases in cellular fatty acids such as phospholipids, as well as fatty and neutral lipids in the cell wall. The development of adaptive resistance to the antibiotic erythromycin and various antimicrobial agents, such as BC, chlorhexidine, and triclosan, in Salmonella enterica serovars Enteritidis, Typhimurium, and Virchow has been reported (7, 8, 28).
It has been construed that biofilm cells respond in a significantly different manner to antimicrobial agents than do their planktonic counterparts. There is evidence that proteins involved in the oxidative stress response and cell envelope synthesis, as well as synthesis of exopolymeric substances, become upregulated in biofilms, indicating that these factors might contribute to cell survival, persistence, and growth in a biofilm environment (33). Therefore, prolonged exposure to antimicrobial agents could cause significant differences in the adaptive responses of planktonic and biofilm cells. Physiologic or phenotypic adaptation resulting in “biocide tolerance” has been attributed to biofilms (13, 40). Biofilm resistance to biocides may result from slow microbial growth rates that are attributable to nutrient depletion within biofilms, binding of the biocide to the exopolymeric substances of biofilms, and neutralization or degradation of the biocide, as well as the expression of biofilm-specific phenotypes (10, 40, 42).
Planktonic and biofilm cells of Escherichia coli, P. aeruginosa, Pseudomonas pseudomallei, Staphylococcus aureus, and Streptococcus sanguis were previously shown to have some major differences in the MICs of various antibiotics to which they were susceptible (13). The lethal effects of chlorine on planktonic and biofilm cells of Salmonella spp. were compared, and it was found that biofilm cells were significantly more resistant to chlorination than planktonic cells; the biofilm matrix was believed to provide protection against this antimicrobial agent (17, 39). It is also believed that there are various contributing factors, such as the species and strain of bacteria and the type and concentration of antimicrobial agent, that determine the susceptibility of bacteria to killing or the development of adaptive resistance (30).
Bacteria employ an array of survival mechanisms to adapt to and survive the lethal effects of antimicrobial compounds. Here we report the use of proteomics, atomic force microscopy (AFM), survival assays, and fatty acid methyl ester (FAME) analyses to examine various cellular responses of Salmonella enterica serovar Enteritidis planktonic and biofilm cells that contribute to adaptive resistance to BC and survival of BC exposure in qualitative and quantitative terms, highlighting the significance of phenotype-specific responses contributing to enhanced survival.
MATERIALS AND METHODS
Media and chemicals.
Tryptic soy agar (TSA), standard plate count agar (SPCA), and Trypticase soy broth (TSB) were purchased from Becton Dickinson (Cockeysville, MD); BC [C6H5CH2N(CH3)2RCl; R = C8H17 to C18H37], magnesium chloride, phenylmethylsulfonyl fluoride, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), DNase, RNase A, bromophenol blue, dl-dithiothreitol, and iodoacetamide were purchased from Sigma Chemical Co. (St. Louis, MO); sodium chloride was from EM Science (Gibbstown, NJ); EDTA was from J. T. Baker Chemical Co. (Philipsburg, NJ); glycerol, sodium dodecyl sulfate, Tris base, and urea were purchased from Life Technologies (Grand Island, NY); immobilized pH gradient buffer (pH 4.0 to 7.0), Immobiline DryStrip gels, and a PlusOne protein silver staining kit were purchased from GE Healthcare Bio-Sciences Inc. (Baie d'Urfé, QC, Canada).
Bacteria and culture conditions.
Salmonella enterica serovar Enteritidis ATCC 4931 (hereafter referred to as Salmonella serovar Enteritidis) was cultured from a frozen stock on TSA plates overnight at 37°C. Cells in the mid-log phase of growth were obtained by transferring a loopful of colony material from overnight TSA plates to 50 ml of 10% (wt/vol) TSB in an Erlenmeyer flask and then incubating on a gyratory shaker (150 ± 5 rpm) held at room temperature (RT; 21 ± 2°C) for approximately 12 h. These cells, which were previously determined to be in the mid-log phase of growth (28), were inoculated to cultivate planktonic and biofilm cells (see below).
Determination of MIC and lethal concentration of BC.
The MIC of BC for planktonic cells of Salmonella serovar Enteritidis was determined to be 15 μg ml−1 by doubling-dilution-based nephelometry coupled with plating of mid-log-phase planktonic cells treated with increasing concentrations of BC. A sublethal concentration of 1 μg ml−1 was selected for continuous BC application. The concentration of BC for lethal challenge was determined as 500 μg ml−1, based on the preliminary observations and recommendations of regulatory agencies for surface disinfection using QACs (14, 15).
Adaptation of Salmonella serovar Enteritidis planktonic cells to BC.
Planktonic cells were grown in an Erlenmeyer flask held on a gyratory shaker (150 ± 5 rpm) and were continuously cultured at RT for 168 h in 175 ml of 10% (wt/vol) TSB; medium was continuously added and removed at a rate of 25 ml h−1, resulting in a dilution rate of 0.14 h−1. The medium was pumped into and out of the Erlenmeyer flask via silicone tubing using two peristaltic pumps (model 202U; Watson-Marlow, Cornwall, United Kingdom). The medium in the flask was inoculated with 2 ml mid-log-phase Salmonella serovar Enteritidis cells, prepared as outlined above, concentrated or diluted to an optical density (OD) equivalent to a 0.5 McFarland standard (1.5 × 108 CFU ml−1) using a Novaspec II spectrophotometer (Biochrom Ltd., Cambridge, United Kingdom) set for measuring OD at 600 nm). After 24 h of growth, the planktonic cells were continuously exposed to a sublethal concentration (1 μg ml−1) of BC over an additional 144 h of growth. Nonadapted planktonic cells grown under identical conditions (i.e., without BC exposure) for a period of 168 h were used as a control.
Adaptation of Salmonella serovar Enteritidis biofilms to BC.
Biofilms were grown in multichannel flow cells. The flow cells were constructed using polycarbonate sheets into which channels were milled, as described previously (21). Flow cell channels were sterilized by flushing with 5.25% (wt/vol) sodium hypochlorite solution for 10 min. Reservoirs of sterile nutrient medium (10% [wt/vol] TSB) were connected via silicone tubing to the flow cell channels and subsequently connected to the waste reservoir. Medium was pumped through flow cells using a Watson-Marlow peristaltic pump. In this setup, the bulk flow rate of the nutrient medium through each flow cell channel was determined to be 25 ml h−1 and the laminar flow velocity was 0.07 cm s−1. Each flow cell channel was individually inoculated with 0.5 ml mid-log-phase Salmonella serovar Enteritidis cells (0.5 McFarland standard) (see above). The inoculum was retained in flow cell channels for 15 min while nutrient flow was turned off to facilitate bacterial attachment, and then the flow was turned on. Established (24-h) biofilms were continuously exposed to a sublethal concentration of BC (1 μg ml−1) over an additional 144 h of growth. Experiments conducted using nonadapted biofilms (i.e., without BC exposure) grown under identical conditions for a period of 168 h were also performed to serve as controls.
Survival of BC-adapted and nonadapted planktonic and biofilm cells with different BC concentrations.
Biofilm cells were scraped off from glass coverslips of the flow cells, and progress was regularly monitored using a light microscope. BC-adapted and nonadapted planktonic and biofilm cells were collected and centrifuged at 4,000 rpm (model 5810 R with swinging-bucket rotor A-4-81; Eppendorf, Hamburg, Germany) for 3 min in order to pellet the bacterial cells. The supernatant was completely removed, the pellet was resuspended in phosphate-buffered saline (PBS; pH 7.2) and thoroughly vortexed, and the cell density was adjusted to an OD equivalent to a 0.5 McFarland standard. The bacterial suspension after serial dilution in PBS was then used as the inoculum for the survival assay. The serially diluted suspensions of BC-adapted and nonadapted planktonic and biofilm cells were immediately spread plated on SPCA containing increasing concentrations (0, 10, 20, 25, and 30 μg ml−1) of BC. The time required to process cells (from pellet to the inoculation of SPCA plates) was ∼10 min. The plates were then incubated at 37°C for 24 h. The proportion of survivors was determined by counting the colonies recovered on the SPCA plates containing increasing concentrations of BC and expressed in CFU ml−1.
Treatment of BC-adapted and nonadapted planktonic and biofilm cells with a lethal concentration of BC.
Ten-milliliter suspensions of BC-adapted and nonadapted planktonic and biofilm cells were prepared in PBS as described above, supplemented with 500 μg ml−1 BC (final concentration), and incubated at RT for a total of 10 min. Aliquots (0.1 ml) were then collected at 2-min intervals until the end of the 10-min incubation period, serially diluted using PBS at RT, and spread plated on SPCA. The plates were then incubated at 37°C for a 24-h period. The proportion of survivors on SPCA was expressed as CFU ml−1.
Heat shock treatment of BC-adapted and nonadapted planktonic and biofilm cells.
Suspensions of BC-adapted and nonadapted planktonic and biofilm cells were prepared in PBS as described above for the BC survival assay and then incubated on a water bath set at 55°C for a total of 10 min. Aliquots (0.1 ml) were then collected at 2-min intervals until the end of the 10-min incubation period, serially diluted using PBS at RT, and spread plated on SPCA. After incubation on SPCA for 24 h at 37°C, the number of survivors was determined and expressed as CFU ml−1.
Analysis of cell surface roughness using AFM.
Changes in cell surface roughness of BC-adapted (for 144 h) and nonadapted planktonic and biofilm cells were analyzed using AFM. The cell surface changes of BC-adapted and nonadapted planktonic and biofilm cells following exposure to lethal BC concentrations (30, 50, 100, and 500 μg ml−1 for 10 min) were compared. Planktonic cells were allowed to air dry on a coverslip for ∼15 min, washed with PBS, and then rinsed with sterile reverse osmosis (RO) water. Biofilm cells were prepared by removing pieces of coverslips from the flow cell channels and affixing the pieces onto a mounting coverslip using silicon adhesive. Subsequent washing was performed as for planktonic cells. Cells were then lethally treated for 10 min using BC solutions (see above) prepared in RO water, followed by washing with PBS and RO water. The samples were allowed to air dry for ∼15 min and placed in a humidifying chamber (RT and ∼80% relative humidity) until the AFM was performed (11, 12).
AFM was performed using a PicoSPM I with a PicoScan 2100 controller and equipped with a piezo scanner (Molecular Imaging Corp., Tempe, AZ). Imaging was carried out using oxide-sharpened silicon nitride AFM probes with a tip radius of ca. 10 nm (cantilever nominal spring constant = 0.12 N m−1; resonant frequency, 14 to 26 kHz) (model DNP-S; Veeco Probes, Camarillo, CA) in the contact mode. The scanning force constant was ca. 3 nN, and scan rates were ∼1 Hz. Image areas of 5 μm by 5 μm were collected from the same location in height and deflection modes simultaneously (11, 32, 43). Quantitative data were collected from the height mode images. Images were collected from samples derived from three independently replicated experiments.
This study employed the analytical method of Cross et al. (11) to determine bacterial cell surface roughness. The stored images were analyzed using PicoScan software, version 5.3.1a (Molecular Imaging Corp.), to determine cell surface roughness. An area of 0.25 μm by 0.25 μm was selected from the middle region of each cell image and then analyzed to determine average cell surface roughness. The data obtained from 60 randomly selected cells (i.e., 20 cells each from three independently replicated experiments) were statistically analyzed to obtain cell surface roughness pertaining to the treatment and expressed in Å.
Cell membrane fatty acid profile analyses.
FAME analyses were conducted as described previously (1, 38) using ∼50 mg of washed BC-adapted and nonadapted planktonic and biofilm cell pellets. A 5890 series 2 gas-liquid chromatograph (Hewlett-Packard Co., Avondale, PA) equipped with a flame ionization detector and a 25-m by 0.22-mm methyl phenyl silicone-fused silica capillary column (Ultra 2; Hewlett-Packard; catalog no. 19091B-102) was used for identification of FAMEs. Results were automatically integrated by Hewlett-Packard 3365 series II ChemStation software, version A.03.21, and FAMEs were identified with the MIDI microbial identification software (Sherlock TSBA Library, version 4.1; Microbial ID, Inc., Newark, DE).
Protein expression analyses.
BC-adapted and nonadapted planktonic and biofilm cells were collected after 168 h of growth and pelleted by centrifuging at 4,000 rpm (model 5810 R with swinging-bucket rotor A-4-81; Eppendorf, Hamburg, Germany) for 5 min. The cells were then disrupted, and the protein was extracted. Total cellular proteins from biofilm and planktonic cells were electrophoretically separated by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), as detailed previously (28). Differentially expressed proteins were detected and quantified from the stored images of gels using Phoretix 2D, version 2004, analysis software (Nonlinear Dynamics Ltd., Newcastle upon Tyne, United Kingdom). An increase in protein spot volume of 1.5-fold or more was interpreted as upregulation, whereas a decrease in the spot volume of 1.5-fold or more was interpreted as downregulation. Differentially expressed proteins were then identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (28). The data from this analysis were processed using ProteinLynx software (Waters-Micromass) and searched against the NCBInr, MSDB, and Swiss-Prot/TrEMBL protein databases using Mascot Search (Matrix Science Ltd., London, United Kingdom). The biological function(s) of each protein identified was determined from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk) and PUMA2 (http://compbio.mcs.anl.gov/puma2/cgi-bin/index.cgi) databases.
Experimental replication and statistical analyses.
All experimental data represent the averages of at least three independent experiments. Differential protein expression of BC-adapted planktonic and biofilm Salmonella serovar Enteritidis cells was determined from averaged spot volumes from four experimentally replicated 2D-PAGE gels. Experimental data were analyzed using SAS statistical software (version 9.1.3; SAS Institute Inc., Cary, NC), and the Fisher least significant difference method was used to test for significant (P < 0.05) differences.
RESULTS
Survival of BC-adapted and nonadapted planktonic and biofilm cells following lethal BC exposure.
Survival of BC-adapted and nonadapted planktonic and biofilm cells was determined by spread plating on SPCA containing various concentrations (0 to 30 μg ml−1) of BC. Cells belonging to all four treatments (i.e., BC-adapted and nonadapted planktonic and biofilm cells) were unable to grow on SPCA plates at BC concentrations of 35 μg ml−1 or more during some of the replications of the experiment; thus, only results from BC concentrations of 0 to 30 μg ml−1 were examined during this work (Table 1). The numbers of BC-adapted and nonadapted planktonic and biofilm cells that were able to survive on SPCA had an inverse exponential correlation with BC concentration. The most substantial difference in the number of survivors was found at the BC concentration of 30 μg ml−1. There was no significant difference (P > 0.05) in the proportions of nonadapted planktonic and biofilm cells able to survive on SPCA plates at the different BC concentrations (e.g., at 30 μg ml−1, 7.8 × 103 and 6.0 × 103 CFU ml−1 of cells survived, respectively). However, the proportion of BC-adapted biofilm cells that were able to survive 30 μg ml−1 BC (1.1 × 105 CFU ml−1) was significantly higher (P < 0.05) (4.6-fold) than that of BC-adapted planktonic cells (2.4 × 104 CFU ml−1). Furthermore, there were 18.3-fold more survivors among the BC-adapted biofilm cells than among their nonadapted (i.e., without prior BC exposure) cell counterparts at the lethal BC concentration, and this value was significantly higher than the value for BC-adapted planktonic cells versus nonadapted cells (3.2-fold). There was complete lethality of cells from each of the four treatment groups observed within 2 min of exposure to the lethal BC concentration (500 μg ml−1); no survivors were seen following extended incubation (i.e., 4, 6, 8, and 10 min) at this concentration of BC (data not shown).
TABLE 1.
Surviving population and percent survival of Salmonella serovar Enteritidis planktonic and biofilm cells in increasing concentrations of BC
| Concn of BC (μg ml−1) | Survival (CFU ml−1 [%]) of cells with:
|
|||
|---|---|---|---|---|
| Planktonic phenotype
|
Biofilm phenotype
|
|||
| Nonadapted to BCa | Adapted to BC | Nonadapted to BCa | Adapted to BC | |
| 0 | 1.5 × 108 (100)b | 1.5 × 108 (100)b | 1.5 × 108 (100)b | 1.5 × 108 (100)b |
| 10 | 5.3 × 106 (3.53) | 7.5 × 106 (5.00) | 4.3 × 106 (2.87) | 9.0 × 106 (6.00) |
| 20 | 1.2 × 105 (0.08) | 3.6 × 105 (0.24) | 1.1 × 105 (0.07) | 5.5 × 105 (0.37) |
| 25 | 5.1 × 104 (0.03) | 1.2 × 105 (0.08) | 6.1 × 104 (0.04) | 2.3 × 105 (0.15) |
| 30 | 7.8 × 103 (0.005) | 2.4 × 104 (0.016) | 6.0 × 103 (0.004) | 1.1 × 105 (0.073) |
Control for the corresponding phenotype.
Initial population unexposed to increasing concentrations of BC.
Survival of heat-shocked BC-adapted and nonadapted planktonic and biofilm cells.
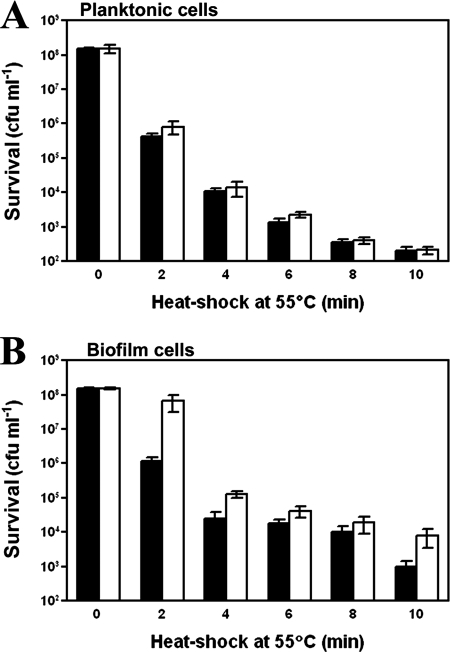
Salmonella serovar Enteritidis cells belonging to the four treatment groups were subjected to a 10-min heat shock at 55°C to determine whether BC adaptation or the biofilm phenotype resulted in acquired thermotolerance (Fig. 1). There was no significant difference (P > 0.05) in survival between BC-adapted and nonadapted planktonic cell population at different time intervals, with the exception of the 2- and 6-min time intervals (Fig. 1). The number of surviving BC-adapted biofilm cells was significantly higher (P < 0.05) than that of nonadapted biofilm cells at most time intervals (i.e., 2, 4, 6, and 10 min) of the incubation (Fig. 1B). Overall, the proportion of biofilm cells (BC adapted and nonadapted) that survived heat shock at 55°C was significantly higher (P < 0.05) than the proportion of surviving planktonic cells (BC adapted and nonadapted).
FIG. 1.
Survival of BC-adapted and nonadapted Salmonella serovar Enteritidis planktonic (A) and biofilm (B) cells following heat shock at 55°C for up to 10 min. BC adaptation of the cells was carried out by continuous exposure to 1 μg ml−1 of BC for 144 h. The measurements pertaining to nonadapted (solid bars) and BC-adapted (open bars) cells are indicated. The cells were incubated at 55°C for 10 min; samples were collected at every 2 min during the incubation period and plated on SPCA plates after serial dilution. Plates were incubated at 37°C for 24 h, and the colonies were counted in order to estimate the number of survivors in CFU ml−1. The error bars indicate the standard errors of the means.
Cell surface roughness of BC-adapted and nonadapted planktonic and biofilm cells.
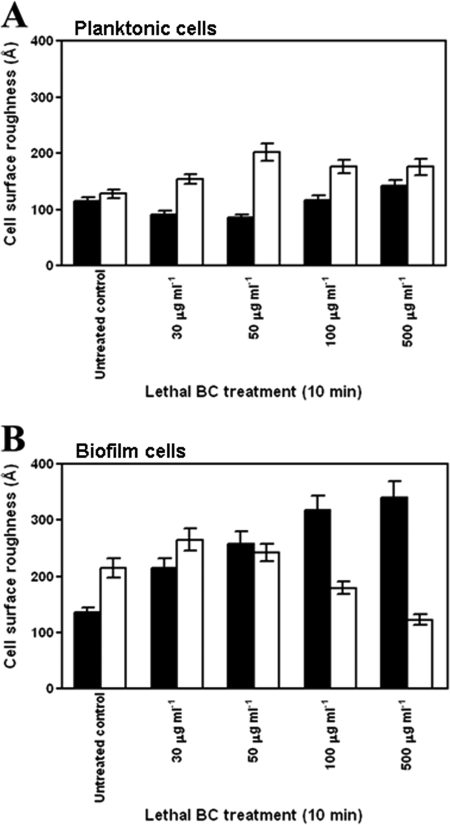
The effect of BC treatment on BC-adapted and nonadapted planktonic and biofilm cell surface roughness (in Å) was analyzed using AFM and image analysis. Levels of average cell surface roughness of BC-adapted and nonadapted planktonic cells and BC-adapted and nonadapted biofilm cells before and after exposure to lethal BC concentrations are presented in Fig. 2. The cell surface roughness of nonadapted planktonic cells was gradually reduced until exposure to a lethal BC concentration of 50 μg ml−1, at which point surface roughness values increased with exposure to higher BC concentrations. Cell surface roughness of adapted planktonic cells increased as BC concentration was increased to 50 μg ml−1 but thereafter decreased as higher BC concentrations were applied. In the case of nonadapted biofilm cells, cell surface roughness increased with increasing BC concentration. However, the roughness of BC-adapted biofilm cells increased as BC concentration was increased to 30 μg ml−1 and then decreased with application of higher BC concentrations. Overall, the roughness of biofilm cells was significantly (P < 0.05) higher than that of planktonic cells for both BC-adapted and nonadapted cells (Fig. 2). Cell surface roughness was found to be significantly (P < 0.05) influenced by the phenotype of cells (i.e., planktonic or biofilm).
FIG. 2.
Average cell surface roughness measurements, expressed in Å (n = 60), of Salmonella serovar Enteritidis planktonic (A) and biofilm (B) cells, as determined by AFM image acquisition and image analyses using PicoScan software, version 5.3.1a (Molecular Imaging Corp., Tempe, AZ). BC adaptation of the cells was carried out by continuous exposure to 1 μg ml−1 of BC for 144 h. The measurements pertaining to nonadapted (solid bars) and BC-adapted (open bars) cells are indicated. The cells were exposed to increasing lethal concentrations (30, 50, 100, and 500 μg ml−1) of BC for 10 min and then analyzed using AFM. Nonadapted cells of the respective phenotype were analyzed to serve as controls. The error bars indicate the standard errors of the means.
Fatty acid profiles of BC-adapted and nonadapted planktonic and biofilm cells.
Fatty acid profiles of BC-adapted and nonadapted planktonic and biofilm cells revealed qualitative and quantitative differences in their fatty acid compositions (Table 2). FAME profiles consisted of saturated fatty acids (SFA), branched-chain fatty acids (BCFA), unsaturated fatty acids (UFA), cyclic fatty acids (CFA), and fatty alcohols (FA). There were also three summed features (i.e., SF 2, 3, and 5) that represent structurally similar fatty acids that could not be resolved by the method used. The major differences in the FAME profiles of BC-adapted planktonic cells versus nonadapted planktonic cells was the absence of BCFA (17:1 anteiso/iso and 19:0 anteiso) and CFA (17:0 cyc); however, a significant increase (P < 0.05) in the production of SFA (12:0) and SF 3 in BC-adapted planktonic cells also occurred. Furthermore, there was a significant difference in the composition of fatty acids between BC-adapted and nonadapted biofilm cells in terms of the number of different fatty acids and their relative proportion, including the absence of SFA (17:0), BCFA (15:1 anteiso), UFA (17:1 ω9c), and CFA (19:0 cyc), as well as a significant reduction (P < 0.05) in the proportion of SFA (12:0, 14:0, 15:0, and 16:0), UFA (18:1 ω7c), CFA (17:0 cyc), and SF 2 and SF 3 in BC-adapted biofilm cells. However, there was an increase in the production of BCFA (16:0 anteiso), UFA (14:1 ω7c, 15:1 ω7c, and 16:1 ω7c), FA (16:1 ω7c alcohol), and SF 5 (18:0 anteiso/18:2 ω6, 9c) in BC-adapted biofilm cells. There were also some major differences in fatty acid composition between nonadapted planktonic and biofilm cells and between BC-adapted planktonic and biofilm cells (Table 2).
TABLE 2.
Shift in fatty acid composition of Salmonella serovar Enteritidis planktonic and biofilm cells following BC adaptationa
| Fatty acid | % of total FAME ± SD in cells with:
|
|||
|---|---|---|---|---|
| Planktonic phenotype
|
Biofilm phenotype
|
|||
| Nonadapted to BCb | Adapted to BC | Nonadapted to BCb | Adapted to BC | |
| SFA | ||||
| 12:0 | 4.0 ± 0.3 | 4.6 ± 0.2 | 2.7 ± 0.1 | 2.0 ± 0.0 |
| 14:0 | 8.3 ± 2.3 | 8.1 ± 0.4 | 7.1 ± 1.4 | 2.9 ± 0.3 |
| 15:0 | NDd | ND | 1.0 ± 0.1 | 0.7 ± 0.0 |
| 16:0 | 22.0 ± 1.9 | 24.3 ± 1.9 | 23.7 ± 2.7 | 9.7 ± 0.6 |
| 17:0 | ND | ND | 1.7 ± 2.1 | ND |
| BCFA | ||||
| 15:1 anteiso | ND | ND | 0.7 ± 0.0 | ND |
| 16:0 anteiso | ND | ND | ND | 17.9 ± 0.7 |
| 17:1 anteiso/iso | 7.4 ± 0.0 | ND | ND | ND |
| 19:0 anteiso | 3.8 ± 0.0 | ND | ND | ND |
| UFA | ||||
| 14:1 ω5c | ND | ND | ND | 5.5 ± 1.0 |
| 15:1 ω5c | ND | ND | ND | 0.7 ± 0.0 |
| 16:1 ω5c | ND | ND | ND | 19.4 ± 0.8 |
| 17:1 ω9c | ND | ND | 8.3 ± 0.0 | ND |
| 18:1 ω7c | 18.7 ± 2.2 | 20.1 ± 2.5 | 18.0 ± 3.4 | 7.4 ± 1.2 |
| CFA | ||||
| 17:0 cyc | 2.9 ± 0.1 | ND | 4.8 ± 0.5 | 0.9 ± 0.0 |
| 19:0 cyc | ND | ND | 0.7 ± 0.0 | ND |
| FA | ||||
| 16:1 ω7c alcohol | ND | ND | ND | 4.9 ± 0.0 |
| Summed featurec | ||||
| 2 (14:0 3OH/16:1 iso) | 7.5 ± 0.1 | 8.1 ± 0.5 | 7.1 ± 1.4 | 5.0 ± 0.5 |
| 3 (15:0 iso 2OH/16:1 ω7c) | 25.3 ± 3.5 | 34.8 ± 3.5 | 24.2 ± 4.7 | 11.2 ± 0.9 |
| 5 (18:0 anteiso/18:2 ω6, 9c) | ND | ND | ND | 11.9 ± 1.9 |
Data from each treatment are the means from at least three independent replications.
Control for corresponding phenotype.
Denotes structurally similar fatty acids that cannot be resolved by the methods employed.
ND, not detected.
Proteomic analyses of BC-adapted and nonadapted planktonic and biofilm cells.
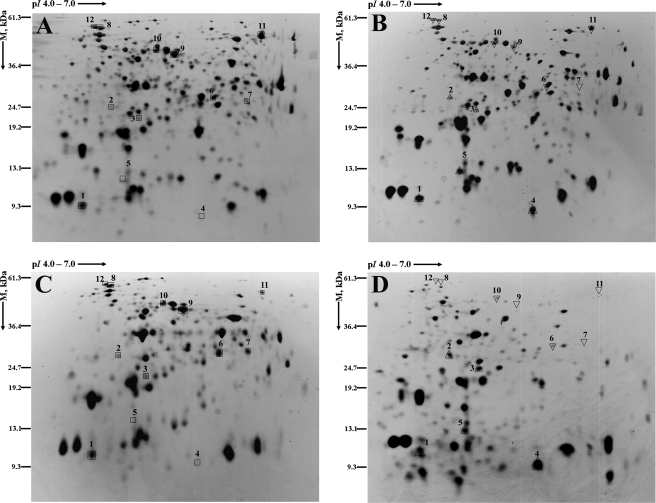
There were five proteins upregulated (Fig. 3; Table 3) and seven proteins downregulated (Fig. 3; Table 4) in both planktonic and biofilm cells following BC adaptation (relative to protein from nonadapted planktonic and biofilm cells). The upregulated proteins included TrxA (biosynthesis), Tsf (protein translation and modification), CspA (cold adaptation), YjgF (a hypothetical protein), and probable peroxidase STY0440 (putative thiol-alkyl hydroperoxide reductase) (detoxification); the downregulated proteins were GarR (degradation), GpmA (glycolysis), RpsA (ribosomal protein synthesis and modification), TufA (protein translation and modification), RfbH (cell envelope synthesis), OppA (oligopeptide-binding protein), and DnaK (chaperone protein Hsp70). Moreover, 39 proteins differentially expressed between planktonic and biofilm cells were detected following BC adaptation (Table 5). Nine unique proteins were upregulated in BC-adapted planktonic cells, and 17 unique proteins were upregulated in BC-adapted biofilm cells. It was further observed that, following BC adaptation, the majority of proteins that were upregulated in biofilm cells were either unchanged in expression or downregulated in the planktonic cell proteome, or conversely that the upregulated proteins in planktonic cells were either unchanged in expression or downregulated in the biofilm cell proteome (Table 5).
FIG. 3.
Total proteins with pI values ranging from 4 and 7 were differentially expressed in planktonic and biofilm cells of Salmonella serovar Enteritidis adapted to BC. The images illustrate the representative 2D-PAGE gels pertaining to proteins extracted from nonadapted (control) planktonic cells (A), planktonic cells adapted to continuous exposure to BC (1 μg ml−1) (B), nonadapted (control) biofilm cells (C), and biofilm cells adapted to continuous exposure to BC (1 μg ml−1) (D). The description of the proteins and their levels of expression are illustrated in Tables 3 (upregulation) and 4 (downregulation). Boxes indicate the location of the protein spot in the controls; triangles indicate upregulation and inverted triangles indicate downregulation of the protein relative to the expression in the respective nonadapted controls. Protein spot numbering is the same for all four gels. M, molecular mass.
TABLE 3.
Proteins upregulated in planktonic and biofilm cells of Salmonella serovar Enteritidis following continuous sublethal (1-μg ml−1) BC exposure for 144 h
| Class or function(s) | Spot no. | Gene | Protein description | Ma (kDa) | pIb | Fold changec for:
|
|
|---|---|---|---|---|---|---|---|
| Planktonic cells | Biofilm cells | ||||||
| Biosynthesis of cofactors, prosthetic groups, and carriers | 1 | trxA | Thioredoxin 1 | 11.68 | 4.67 | 1.58 | 1.86 |
| Protein translation and modification | 2 | tsf | Protein chain elongation factor (EF-Ts) | 30.36 | 5.13 | 1.54 | 8.81 |
| Cell processes (detoxification) | 3 | NKd | Probable peroxidase STY0440 | 22.32 | 5.24 | 3.88 | 6.75 |
| Adaptation and atypical conditions | 4 | cspA | 7.4-kDa cold shock protein | 7.27 | 5.57 | 32.10 | 110.13 |
| Hypothetical proteins | 5 | yjgF | Conserved hypothetical protein YjgF | 13.57 | 5.36 | 6.84 | 33.81 |
M, molecular mass.
pI, isoelectric point. Values are theoretical values obtained from the Swiss-Prot or PUMA2 databases.
Ratio of protein expression in adapted cells to the protein expression in nonadapted (control) cells.
NK, not known.
TABLE 4.
Proteins downregulated in planktonic and biofilm cells of Salmonella serovar Enteritidis following continuous sublethal (1-μg ml−1) BC exposure for 144 h
| Class or function(s) | Spot no. | Gene | Protein description | Ma (kDa) | pIb | Fold changec for:
|
|
|---|---|---|---|---|---|---|---|
| Planktonic cells | Biofilm cells | ||||||
| Degradation (carbon compounds) | 6 | garR | Tartronate semialdehyde reductase | 30.73 | 5.59 | 0.58 | 0.09 |
| Energy metabolism (glycolysis) | 7 | gpmA | Phosphoglycerate mutase 1 | 28.36 | 5.78 | 0.46 | 0.02 |
| Ribosomal protein synthesis and modification | 8 | rpsA | 30S ribosomal subunit protein S1 | 61.25 | 4.89 | 0.38 | 0.08 |
| Protein translation and modification | 9 | tufA | Translation elongation factor (EF-Tu.A) | 43.40 | 5.30 | 0.27 | 0.25 |
| Cell envelope (LPS) | 10 | rfbH | LPS biosynthesis protein | 48.10 | 5.27 | 0.66 | 0.06 |
| Cell processes (binding proteins, other) | 11 | oppA | Oligopeptide-binding protein complexed with Kvk, chain A | 58.81 | 5.85 | 0.54 | 0.09 |
| Cell processes (chaperones) | 12 | dnaK | Chaperone protein Hsp70 | 69.13 | 4.83 | 0.65 | 0.17 |
M, molecular mass.
pI, isoelectric point. Values are theoretical values obtained from the Swiss-Prot or PUMA2 database.
Ratio of protein expression in adapted cells to the protein expression in nonadapted (control) cells.
TABLE 5.
Proteins differentially expressed in planktonic and biofilm cells of Salmonella serovar Enteritidis following continuous sublethal (1-μg ml−1) BC exposure for 144 h
| Class or function(s) | Gene | Protein description | Ma (kDa) | pIb | Fold changec for:
|
|
|---|---|---|---|---|---|---|
| Planktonic cells | Biofilm cells | |||||
| Degradation (carbon compounds) | pduA | Putative propanediol utilization protein A | 9.59 | 6.72 | 0.79* | 5.25 |
| pduJ | Putative propanediol utilization protein J | 9.07 | 6.50 | 1.00* | 84.76 | |
| Degradation (proteins, peptides, and glycoproteins) | degQ | Serine endoprotease | 47.28 | 6.80 | 0.92* | 0.28 |
| Energy metabolism (ATP synthesis) | atpA | ATP synthase α-subunit | 54.98 | 5.80 | 0.77* | 0.13 |
| Energy metabolism (glycolysis) | eno | Enolase | 45.47 | 5.25 | 0.32 | 36.54 |
| fbaA | Fructose 1,6-bisphosphate aldolase class II | 39.36 | 5.58 | 0.96* | 0.18 | |
| gapA | Glyceraldehyde-3-phosphate dehydrogenase A | 35.46 | 6.32 | 0.79* | 0.11 | |
| pgk | Phosphoglycerate kinase | 41.00 | 5.09 | 0.19 | 1.12* | |
| tpiA | Triose phosphate isomerase | 26.92 | 5.68 | 0.98* | 3.27 | |
| Energy metabolism (tricarboxylic acid cycle) | mdh | Malate dehydrogenase | 32.48 | 6.01 | 0.73* | 0.02 |
| Energy metabolism (nonoxidative | rpiA | Ribose-5-phosphate isomerase A | 22.90 | 5.08 | 0.97* | 0.42 |
| phase of pentose phosphate pathway) | talB | Transaldolase B | 35.04 | 5.09 | 5.99 | 0.83* |
| Energy metabolism (glyoxylate | aceA | Isocitrate lyase | 47.56 | 5.22 | 2.15 | 0.78* |
| bypass) | aceK | Isocitrate dehydrogenase kinase/phosphatase | 46.09 | 5.99 | 0.87* | 0.48 |
| Aromatic amino acid biosynthesis | wrbA | Flavoprotein WrbA | 20.74 | 5.79 | 0.42 | 8.10 |
| Ribosomal protein synthesis and modification | rplL | 50S ribosomal subunit proteins L7 and L12 | 12.17 | 4.60 | 1.00* | 1.78 |
| rplL | 50S ribosomal subunit proteins L7 and L12 | 12.17 | 4.60 | 0.89* | 1.87 | |
| rpsB | 30S ribosomal subunit protein S2 | 26.76 | 6.62 | 5.60 | 0.70* | |
| Protein translation and | tuf | Elongation factor Tu (fragment) | 29.29 | 5.30 | 0.81* | 41.83 |
| modification | dsbA | Thiol:disulfide interchange protein | 22.91 | 5.64 | 1.19* | 2.48 |
| RNA synthesis, RNA modification, and DNA transcription | rpoZ | DNA-directed RNA polymerase ω-chain | 10.24 | 4.87 | 0.55 | 4.61 |
| Cell envelope (surface structures) | fljB | Phase 2 flagellin | 52.41 | 4.75 | 0.33 | 1.02* |
| fliC | Phase 1 flagellin | 51.48 | 4.79 | 0.30 | 1.23* | |
| Cell processes (amino acid-binding proteins) | argT | Lysine-arginine-ornithine-binding periplasmic protein | 28.20 | 5.99 | 2.52 | 0.89* |
| artI | Arginine-binding periplasmic protein 1 precursor | 27.00 | 7.66 | 1.73 | 0.24 | |
| Cell processes (carbohydrate-binding proteins) | crr | Phosphotransferase system enzyme II (glucose-permease IIA component) | 18.12 | 4.73 | 4.17 | 1.08* |
| fruB | Phosphotransferase system enzyme II (fructose-specific IIA/FPr component) | 39.59 | 4.87 | 0.71* | 4.17 | |
| malE | Maltose-binding periplasmic protein | 43.18 | 6.27 | 0.66 | 0.76* | |
| mglB | d-Galactose/d-glucose-binding periplasmic protein | 35.81 | 5.81 | 0.93* | 0.57 | |
| Cell processes (binding proteins, other) | ptsH | Phosphocarrier protein HPr (histidine-containing protein) | 9.12 | 5.65 | 1.50 | 1.41* |
| Cell processes (detoxification) | sodB | Superoxide dismutase (Fe) | 21.18 | 5.58 | 0.34 | 27.13 |
| tpx | Thiol peroxidase | 17.98 | 4.75 | 1.42* | 36.82 | |
| Broad regulatory functions | hns | DNA-binding protein H-NS (histone-like protein II) | 15.41 | 5.32 | 1.85 | 0.11 |
| Atypical conditions and adaptation | grcA | Autonomous glycyl radical cofactor | 14.34 | 5.10 | 2.24 | 1.03* |
| uspA | Universal stress protein A | 15.95 | 5.12 | 0.88* | 0.18 | |
| Hypothetical proteins | gntY | Hypothetical protein GntY | 20.94 | 4.52 | 1.14* | 30.70 |
| ycbL | Hypothetical protein YcbL (putative metallo-β-lactamase) | 23.73 | 4.95 | 0.31 | 6.27 | |
| ynaF | Conserved hypothetical protein STY1416 (putative universal stress protein) | 15.70 | 5.93 | 1.13* | 2.06 | |
| Miscellaneous proteins | NKd | Putative periplasmic protein (Salmonella serovar Typhi strain CT18) | 21.44 | NKd | 0.98* | 2.86 |
M, molecular mass.
pI, isoelectric point. Values are theoretical values obtained from the Swiss-Prot or PUMA2 database.
Ratio of protein expression in adapted cells to the protein expression in nonadapted (control) cells. *, unchanged level of expression (i.e., 0.67-fold to 1.49-fold).
NK, not known.
DISCUSSION
The ability of BC-adapted and nonadapted planktonic and biofilm cell populations to survive a range of BC concentrations and heat shock treatment was examined by quantifying cell surface roughness, cell membrane fatty acid composition, and differential protein expression. Biofilm/planktonic comparison studies have been reported for different bacterial species and various antimicrobial agents (17, 39, 41), and it has been shown that biofilm cells are more tolerant to antimicrobial agents than their planktonic counterparts. In the present study, the survival of nonadapted planktonic cells and that of nonadapted biofilm cells exposed to increasing concentrations of BC were similar, whereas the survival of BC-adapted planktonic and biofilm cell populations was significantly higher (P < 0.05) than that of the controls (Table 1). The proportion of BC-adapted biofilm cells able to survive increasing BC concentrations was also significantly higher (P < 0.05) than that of BC-adapted planktonic cells. Enhanced survival of bacteria exposed to lower lethal BC concentrations was significantly influenced by previous exposure to BC and whether the cells were cultured in the planktonic or biofilm mode of growth.
Boles et al. (4) previously reported that recA-dependent random mutation events functioned to generate genetic diversity in Pseudomonas aeruginosa biofilms, thereby enhancing survival of biofilm cells under conditions of stress (as well as causing other altered traits). Notably, similar mutations were not generated when the organism was grown planktonically. In our study, the possibility that a similar genetically based mechanism enhanced the BC resistance and survival of Salmonella serovar Enteritidis biofilm cells was considered. However, control experiments using nonadapted planktonic and biofilm cells showed no significant difference (P > 0.05) in survival of lethal BC treatment (Table 1), something that would have been predicted if recA-dependent (or spontaneous) mutational events were operative. Furthermore, the sublethal BC concentration used during adaptation was very low (15-fold less than the MIC) (28); thus, a selective environment where the clonal proliferation of mutants over nonmutated cells would occur did not exist. A recent report on phenotypic adaptation of Salmonella serovar Typhimurium to three different biocides, including a QAC, did not reveal that genetic changes in the adapted strains in comparison to their wild-type parent strain had occurred (18). Consequently, while mutational events may have occurred in our model system, the evidence that they contributed to the enhanced survival of BC-exposed biofilms is not compelling. The physiological statuses of Salmonella serovar Enteritidis planktonic and biofilm cells have previously been determined to be distinct based on the proteomic analyses (29). The enhanced adaptive response and survival of BC-adapted biofilm cells, as seen in the present study, are due to the fact that a higher proportion of biofilm cells than of planktonic cells reached an advanced state of adaptation over the same adaptation period. Thus, the enhanced ability of biofilm cells to mount an early and effective physiological (phenotypic) adaptive response is likely the causative factor for enhanced survival of BC-adapted biofilm cells.
Notably, there was absence of growth of control bacteria, as well as of BC-adapted planktonic and biofilm cells that had been exposed for 10 min to 500 μg ml−1 of BC. Previous work using the same organism showed that intact BC-adapted and nonadapted biofilms exposed to the above BC treatment could regrow when BC stress was relieved for a period of 24 h (28); however, regrowth of BC-adapted biofilms was significantly higher than that of nonadapted biofilms. In a practical sense, it is clear that biofilm disruption must be achieved in conjunction with BC treatment for effective control of this organism.
Survival of Salmonella serovar Enteritidis cells after 10 min of 55°C heat treatment was significantly influenced (P < 0.05) by both biofilm phenotype and BC adaptation. Following heat treatment, the number of surviving nonadapted biofilm cells was significantly greater (P < 0.05) than that of nonadapted planktonic cells, whereas, the survival of BC-adapted biofilm cells was greater than that of nonadapted control biofilm cells. There was no significant difference (P > 0.05) in survival between BC-adapted and nonadapted planktonic cell populations. Sampathkumar et al. (38) previously reported that sublethal trisodium phosphate (TSP) treatment induced thermotolerance in Salmonella serovar Enteritidis ATCC 4931 and suggested de novo protein synthesis and altered membrane fatty acid composition as the possible mechanisms.
The likelihood that phenotypic reversion (from biofilm back to planktonic) occurred during these experiments, which involves disruption of biofilm bacteria prior to treatment, was minimized by quick processing of the cells followed by immediate plating. If phenotype reversion had occurred, there should have actually been a greater number of survivors among the BC-adapted biofilm cell population than what we observed during the survival assay and heat shock challenge, as BC-adapted planktonic cells did not show a significant cross-protection against heat treatment. With regard to survival of heat shock (55°C), nonadapted biofilm cells survived better than nonadapted planktonic cells, whereas, the survival of nonadapted planktonic cells and that of biofilm cells were more or less similar after exposure to increasing lethal BC concentrations. This is similar to the findings of Scher et al. (39), who reported that biofilm and planktonic cell phenotypes responded to various stressors (i.e., physical [heat] and chemical [hypochlorites]) in a distinct and growth phase-dependent manner. Mikkelsen et al. (31) recently reported that the proteome of P. aeruginosa biofilms more closely resembled that of exponentially growing planktonic cells than that of planktonic cells in the stationary phase. The phase of cell growth and the nature of stressor are key factors that interplay with cell phenotype to determine the outcome of a stressor's action on bacterial cells.
AFM has been used to characterize surface changes associated with growth phases, mutations, and antimicrobial exposure in bacteria (6, 11, 12). Major changes associated with bacterial exposure to antimicrobials like BC are seen on their cell surfaces. BC causes concentration-dependent cell wall disruption and leakage of cellular constituents (30). Hoffmann et al. (16) reported that the surfaces of BC-resistant P. aeruginosa cells were covered by an additional layer described as an unidentified substance not present on sensitive cells. Sampathkumar et al. (37) reported that Salmonella serovar Enteritidis cells treated with TSP showed irregular cell surface topology and a wrinkled appearance, along with the disruption of the membrane system. It was thus assumed that major changes associated with BC adaptation and lethal BC treatment would include alterations in cell surface roughness mediated through altered surface composition and formation of additional structures, presumably functioning to reduce overall cell damage. The average surface roughness of nonadapted biofilm cells was significantly higher (P < 0.05) than that of nonadapted planktonic cells both before and after exposure to lethal BC concentrations. It was found that various factors such as phenotype, BC adaptation, and lethal BC exposure significantly altered bacterial cell surface roughness. Using AFM, Braga and Ricci (6) determined that cefodizime damage on E. coli cells significantly varied with the concentration and incubation time of the antibiotic treatment. Cross et al. (11) reported that cariogenicity, a virulence trait of Streptococcus mutans, was inversely proportional to cell surface roughness, as determined by AFM. Our current results also suggest an influence of phenotype (i.e., planktonic or biofilm) on bacterial cell surface roughness, with potential effects on bacterial survival following lethal BC treatments. This is the first report of alterations in bacterial cell surface roughness influenced by sublethal and lethal BC exposures. This is also the first description of biofilm cells having greater surface roughness than planktonic cells. While there was no apparent trend between BC treatment, adaptation, and cell surface roughness, the effects observed are suggestive that the cell wall is a key site of interaction with this antimicrobial compound.
Bacterial cell envelope compositional changes (e.g., membrane fatty acids and proteins) due to phenotype and BC adaptation may have influenced cell surface roughness responses and played a role in cell survival following BC and heat treatments. The fatty acid profiles of bacterial cells are influenced by both physical and chemical agents in the microenvironment, and fatty acid alterations have been suggested as a mechanism of adaptation and survival (1, 36, 38). BC-resistant P. aeruginosa cells were found to contain an increased concentration of readily extractable lipids (16), whereas the acid shock response induced the formation of cyclopropane fatty acids in Salmonella serovar Typhimurium (20). Our results show major differences in the fatty acid profiles of nonadapted planktonic and biofilm cells, as well as BC adaptation-induced shifts (Table 2). The adaptive response to BC of the planktonic phenotype was mediated mainly via SF 3, whereas in the biofilm phenotype this was mediated through BCFA (16:0 anteiso), UFA (14:1 ω5c, 15:1 ω5c, and 16:1 ω5c), FA (16:1 ω7c alcohol), and SF 5. Biosynthesis of excess UFA and reduction in CFA could be correlated with the report of cold shock response induced by the biofilm cells of Salmonella serovar Enteritidis during BC adaptation (28). TSP treatment of this organism has previously been shown to induce thermotolerance and resulted in higher SFA- and CFA-to-UFA ratios (38). Adaptation of biofilm cells to BC has resulted in a more complex fatty acid composition in terms of both number and proportion of fatty acids relative to that in planktonic cells. Similar to our findings, a significant accumulation of phospholipids, as well as fatty and neutral lipids, in the cell walls of BC-resistant P. aeruginosa cells has been reported (36). Loughlin et al. (27) reported that membrane fatty acid changes associated with BC adaptation in P. aeruginosa included an increase in the proportion of SFA (14:0 and 16:0) and a decrease in the proportion of an unknown fatty acid eluting between 2OH 10:0 and 12:0. BCFA (15:0 anteiso) was found to be associated with low-temperature growth in Listeria monocytogenes, possibly by maintaining a fluid, liquid-crystalline state in membrane lipids (1). The majority of fatty acids observed in our study are associated with cell membranes (cell envelope), where they potentially protect against the damaging effect of BC. Thus, a biofilm phenotype-specific shift in membrane fatty acids, along with a set of BC-induced fatty acid changes, occurred, and it has been suggested that these played important roles in the survival of BC-adapted biofilm cells during lethal BC exposure and heat shock.
There was a significant difference in survival between the nonadapted planktonic and biofilm cells following heat shock at 55°C, suggesting a role for cell surface alterations (i.e., increase in cell surface roughness and associated membrane changes) in biofilm cells. However, survival in increasingly lethal BC concentrations up to 30 μg ml−1 was not influenced by growth mode phenotype. It is thus suggested that phenotype-specific changes in cell surface roughness, fatty acid composition, and protein expression profile in combination with BC adaptation contributed to the enhanced survival of BC-adapted biofilm cells. Once adapted to BC, biofilm cells were more resistant to BC than planktonic cells.
We have recently identified many key proteins that were up- and downregulated during Salmonella serovar Enteritidis biofilm formation by comparison to the planktonic cell proteome (29). Proteins involved in degradation and energy metabolism, RNA and protein biosynthesis, various cell processes, and adaptation and some hypothetical proteins were upregulated, whereas some important proteins involved in biosynthesis of cell surface structures (RfbH, FljB, and FliC), detoxification (Tpx and SodB), and chaperone functions (DnaK) were downregulated in the biofilms. These findings indicated that the physiologies of planktonic and biofilm cells were distinct and that the physiologies of the phenotypes influenced the adaptive responses and survival of the two cell phenotypes, as observed in the current study.
There are few reports of the role of the proteome in the adaptive resistance of Salmonella serovar Enteritidis biofilms to BC and other agents (18, 28). In the present study, the biofilm phenotype did produce the upregulation of key proteins (CspA, TrxA, Tsf, YjgF, and probable peroxidase [putative thiol-alkyl hydroperoxide reductase] STY0440) coupled with the production of 17 unique proteins (Table 5) in the presence of BC, facilitating BC-adapted biofilm cell survival following lethal BC exposure (i.e., below the BC concentration of 30 μg ml−1). Hindered or incomplete penetration of BC into deeper layers of biofilms and detoxification processes might have influenced BC adaptation of the biofilms. There was a significant increase in the expression of two proteins involved in protection against oxidative stress (Tpx and SodB) in BC-adapted biofilm cells, in comparison to BC-adapted planktonic cells (Table 5). Tpx and SodB were relatively downregulated in nonadapted biofilms in comparison to nonadapted planktonic cells of Salmonella serovar Enteritidis (29). This scenario is a fine example to demonstrate that the biofilm adaptive response to a stressor(s) is versatile and that biofilms are highly responsive to the prevailing conditions. Bore et al. (5) recently reported the upregulation of proteins involved in protection against oxidative stress (manganese superoxide dismutase) in BC-adapted E. coli cells. The differences in expression of proteins involved in degradation (PduA and PduJ), amino acid and protein biosynthesis (DsbA, RplL, RpoZ, Tuf, and WrbA), and nutrient binding (FruB), as well as proteins with hypothetical (GntY, YnaF, and YcbL) and unknown functions (putative periplasmic protein), between BC-adapted planktonic and biofilm cells help to explain the enhanced survival of BC-adapted biofilm cells. The downregulation of RfbH (LPS biosynthesis protein) in both planktonic and biofilm cells was somewhat puzzling; however, it is possible that LPS modification (see above), rather than normal biosynthetic pathways, is involved in survival responses to BC even though the cell envelope is the primary target site of the action of this compound. Moreover, other adaptive responses might have been sufficient to provide resistance. Similar to observations made in this study, Loughlin et al. (27) reported that BC adaptation in P. aeruginosa was associated with several phenotypic changes; however, there was no mention of altered LPS composition. Recently, Braoudaki and Hilton (8) also reported that there was no significant change in the outer membrane or LPS when Salmonella spp. were exposed to sublethal BC treatment.
While it is assumed that after 6 days of sublethal BC exposure most of the planktonic and biofilm cells would have been in an advanced and uniform state of adaptation, it is possible that a small surviving subset of cells in the population were not yet adapted or contained unique and possibly relevant proteins that were undetectable due to protein concentration limitations. It was previously shown that the MIC of BC in a bacterial population could be significantly increased by extending the duration of sublethal BC exposure and gradually increasing the concentration of BC (22); thus, adaptation may be considered gradual. However, it is similarly clear that not even advanced adaptation is sufficient to provide complete protection from killing following lethal BC exposure. In this context, adaptation functions as a population response whereby a subset of the population develops a surviving phenotype.
Adaptive resistance to antimicrobial agents, including BC, and cross-resistance of adapted strains to related or unrelated antimicrobial agents have been reported among members of the family Enterobacteriaceae, which comprise many important human pathogens (7, 8, 22). In our study, there were significant differences in the pattern and degree of resistance of planktonic and biofilm Salmonella serovar Enteritidis cells to BC as well as to heat shock. We conclude that the biofilm phenotype resulted in an early, more efficient adaptive response, producing a higher proportion of adapted individuals than the planktonic phenotype over the same period. Once adapted, biofilm cells were better able to survive BC than planktonic cells. The alterations in cell surface roughness, fatty acid composition, and protein expression associated with the two phenotypes are in agreement with the ability of cells to survive. Our present findings reveal that both individual biofilm cells and individual planktonic cells (free-living cells) are susceptible to lethal exposure even after adaptation. These findings thus have significance from the clinical perspective since the cells within biofilms were resistant to lethal exposure before disruption; the state of the cells (i.e., their existence in the depth of biofilms) and BC adaptation have obvious and different roles in protecting the cells from lethal exposure, possibly in an additive or synergistic manner. These findings also have practical relevance in that the mechanical disruption of the biofilms and lethal antimicrobial treatment are both required for the successful control of biofilms (and neither technique used individually would suffice). It is noteworthy that disrupted BC-adapted biofilm cells may also have a better chance to attach, multiply, and form biofilms in BC-containing environments if the concentration is not lethal, a concern in environments such as health care facilities, the food industry, and households.
Acknowledgments
This research was supported by funding from the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi, A. A., and F. Baneyx. 1999. Stress responses as a tool to detect and characterize the mode of action of antimicrobial agents. Appl. Environ. Microbiol. 65:5023-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boles, B. R., M. Thoendel, and P. K. Singh. 2004. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. USA 101:16630-16635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bore, E., M. Hébraud, I. Chafsey, C. Chambon, C. Skjæret, B. Moen, T. Møretrø, Ø. Langsrud, K. Rudi, and S. Langsrud. 2007. Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology 153:935-946. [DOI] [PubMed] [Google Scholar]
- 6.Braga, P. C., and D. Ricci. 1998. Atomic force microscopy: application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Antimicrob. Agents Chemother. 42:18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braoudaki, M., and A. C. Hilton. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J. Clin. Microbiol. 42:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braoudaki, M., and A. C. Hilton. 2005. Mechanisms of resistance in Salmonella enterica adapted to erythromycin, benzalkonium chloride and triclosan. Int. J. Antimicrob. Agents 25:31-37. [DOI] [PubMed] [Google Scholar]
- 9.Campanac, C., L. Pineau, A. Payard, G. Baziard-Mouysset, and C. Rogues. 2002. Interactions between biocide cationic agents and bacterial biofilms. Antimicrob. Agents Chemother. 46:1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Cross, S. E., J. Kreth, L. Zhu, F. Qi, A. E. Pelling, W. Shi, and J. K. Gimzewski. 2006. Atomic force microscopy study of the structure-function relationships of the biofilm-forming bacterium Streptococcus mutans. Nanotechnology 17:S1-S7. [DOI] [PubMed] [Google Scholar]
- 12.Del Sol, R., I. Armstrong, C. Wright, and P. Dyson. 2007. Characterization of changes to the cell surface during the life cycle of Streptomyces coelicolor: atomic force microscopy of living cells. J. Bacteriol. 189:2219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Food and Agriculture Organization. 2003. Assessment and management of seafood safety and quality. FAO Fisheries technical paper 444. Food and Agriculture Organization of the United Nations, Rome, Italy.
- 15.Health Canada. 1999. Therapeutic products programme guidelines. Disinfectant drugs. Ministry of Health, Ottawa, Canada.
- 16.Hoffmann, H.-P., S. G. Geftic, J. Gelzer, H. Haymann, and F. W. Adair. 1973. Ultrastructural alterations associated with the growth resistant Pseudomonas aeruginosa in the presence of benzalkonium chloride. J. Bacteriol. 113:409-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph, B., S. K. Otta, I. Karunasagar, and I. Karunasagar. 2001. Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food Microbiol. 64:367-372. [DOI] [PubMed] [Google Scholar]
- 18.Karatzas, K. A. G., L. P. Randall, M. Webber, L. J. V. Piddock, T. J. Humphrey, M. J. Woodward, and N. G. Coldham. 2008. Phenotypic and proteomic characterization of multiply antibiotic-resistant variants of Salmonella enterica serovar Typhimurium selected following exposure to disinfectants. Appl. Environ. Microbiol. 74:1508-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlowsky, J. A., M. H. Saunders, G. A. J. Harding, D. J. Hoban, and G. G. Zhanel. 1996. In vitro characterization of aminoglycoside adaptive resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:1387-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, B. H., S. Kim, H. G. Kim, J. Lee, I. S. Lee, and Y. K. Park. 2005. The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 151:209-218. [DOI] [PubMed] [Google Scholar]
- 21.Korber, D. R., G. A. James, and J. W. Costerton. 1994. Evaluation of fleroxacin activity against established Pseudomonas fluorescens biofilms. Appl. Environ. Microbiol. 60:1663-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langsrud, S., G. Sundheim, and A. L. Holck. 2004. Cross-resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress-inducers. J. Appl. Microbiol. 96:201-208. [DOI] [PubMed] [Google Scholar]
- 23.Levin, B. R. 2004. Noninherited resistance to antibiotics. Science 305:1578-1579. [DOI] [PubMed] [Google Scholar]
- 24.Levin, B. R., and D. E. Rozen. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556-562. [DOI] [PubMed] [Google Scholar]
- 25.Levy, S. B. 2001. Antibacterial household products: cause for concern. Emerg. Infect. Dis. 7(Suppl. 3):512-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis, K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48-56. [DOI] [PubMed] [Google Scholar]
- 27.Loughlin, M. F., M. V. Jones, and P. A. Lambert. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631-639. [DOI] [PubMed] [Google Scholar]
- 28.Mangalappalli-Illathu, A. K., and D. R. Korber. 2006. Adaptive resistance and differential protein expression of Salmonella enterica serovar Enteritidis biofilms exposed to benzalkonium chloride. Antimicrob. Agents Chemother. 50:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangalappalli-Illathu, A. K., J. R. Lawrence, G. D. W. Swerhone, and D. R. Korber. 2008. Architectural adaptation and protein expression patterns of Salmonella enterica serovar Enteritidis biofilms under laminar flow conditions. Int. J. Food Microbiol. 123:109-120. [DOI] [PubMed] [Google Scholar]
- 30.Maxcy, R. B., N. P. Tiwari, and P. R. Soprey. 1971. Changes in Escherichia coli associated with acquired tolerance for quaternary ammonium compounds. Appl. Microbiol. 22:229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikkelsen, H., Z. Duck, K. S. Lilley, and M. Welch. 2007. Interrelationship between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 189:2411-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pelling, A. E., Y. Li, W. Shi, and J. K. Gimzewski. 2005. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc. Natl. Acad. Sci. USA 102:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resch, A., R. Rosenstein, C. Nerz, and F. Götz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell, A. D. 2003. Biocide use and antibiotic resistance: the relevance of laboratory findings to clinical and environmental situations. Lancet Infect. Dis. 3:794-803. [DOI] [PubMed] [Google Scholar]
- 35.Russell, A. D. 2003. Similarities and differences in the response of microorganisms to biocides. J. Antimicrob. Chemother. 52:750-763. [DOI] [PubMed] [Google Scholar]
- 36.Sakagami, Y., H. Yokoyama, H. Nishimura, Y. Ose, and T. Tashima. 1989. Mechanism of resistance to benzalkonium chloride by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 55:2036-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampathkumar, B., G. G. Khachatourians, and D. R. Korber. 2003. High pH during trisodium phosphate treatment causes membrane damage and destruction of Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 69:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampathkumar, B., G. G. Khachatourians, and D. R. Korber. 2004. Treatment of Salmonella enterica serovar Enteritidis with a sublethal concentration of trisodium phosphate or alkaline pH induces thermotolerance. Appl. Environ. Microbiol. 70:4613-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scher, K., U. Römling, and S. Yaron. 2005. Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl. Environ. Microbiol. 71:1163-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheldon, A. T. 2005. Antiseptic “resistance”: real or perceived threat? Clin. Infect. Dis. 40:1650-1656. [DOI] [PubMed] [Google Scholar]
- 41.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szomolay, B., I. Klapper, J. Dockery, and P. S. Stewart. 2005. Adaptive responses to antimicrobial agents in biofilms. Environ. Microbiol. 7:1186-1191. [DOI] [PubMed] [Google Scholar]
- 43.Touhami, A., M. H. Jericho, and T. J. Beveridge. 2004. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. 186:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]