Abstract
Previous studies have demonstrated that Staphylococcus epidermidis isolates colonizing the skin of healthy humans do not typically encode icaADBC, the genes responsible for the production of polysaccharide intercellular adhesin or biofilms. It was therefore hypothesized that the presence of icaADBC was deleterious to the successful colonization of human skin by S. epidermidis. Using a human skin competition model, it was determined that the strong biofilm-producing S. epidermidis strain 1457 was outcompeted at 1, 3, and 10 days by an isogenic icaADBC mutant (1457 ica::dhfr), suggesting a fitness cost for carriage of icaADBC.
Staphylococcus epidermidis is one of the most prevalent commensals colonizing human skin. However, it is also responsible for 37% of the reported 250,000 central venous catheter bloodstream infections in the United States each year (14, 17). Formation of biofilms is a major virulence determinant of S. epidermidis that allows the organism to bind and proliferate on a variety of biomaterials (18). An S. epidermidis biofilm consists of both proteinaceous factors as well as polysaccharide intercellular adhesion (PIA), which is produced by enzymes encoded by the icaADBC operon (10, 16). Not all S. epidermidis isolates encode icaADBC, and several studies have demonstrated that strains isolated from health care settings or infections of indwelling medical devices are more likely to encode this operon (3, 4, 6, 11, 12, 16, 19). These studies also demonstrated that strains of S. epidermidis isolated from the skin of community volunteers (considered to be commensal isolates) are less likely to carry the icaADBC genes. Recently, it was found that the icaADBC locus can be either present or absent from isolates within the same multilocus sequence typing group, suggesting that these two groups (i.e., icaADBC+ and icaADBC-deficient isolates) are not separate populations (12). These data led us to hypothesize that the icaADBC locus may confer a fitness cost to the bacterium during the colonization of skin. In support of this hypothesis, other studies have shown that the chromosomal region which contains the icaADBC locus can be readily lost within a population (1, 15). In addition, PIA production is known to undergo phenotypic variation in S. epidermidis (8, 19, 20). In the present study, we performed human skin competition studies between a strong biofilm-producing strain of S. epidermidis (1457) and its isogenic icaADBC mutant.
The clinical strain S. epidermidis 1457 (13) and its isogenic icaADBC mutant 1457 ica::dhfr (9) were grown overnight in 100 ml of tryptic soy broth (Difco, Sparks, MD) at a 5:1 flask/volume ratio and with shaking at 225 rpm at 37°C. The cells were then pelleted by centrifugation and the supernatant was removed and washed in 0.9% saline. Cells (1 × 109 CFU) of each strain were resuspended in 1 ml of 0.9% saline for skin application. Viability and confirmation of these inocula were performed by serial plating to tryptic soy agar (TSA; Difco) and Congo red agar (CRA) (5, 8). CRA plates allow for detection of PIA-producing (crusty colony phenotype) versus PIA-negative (smooth colony phenotype) colonies (8). S. epidermidis 1457 is susceptible to ampicillin, erythromycin, and trimethoprim. Due to the genetic inactivation of icaADBC by dhfr, S. epidermidis 1457 ica::dhfr is resistant to trimethoprim.
Nine healthy adult human volunteers unassociated with the health care environment were each inoculated on both forearms as follows. Briefly, each forearm was cleansed with a 70% alcohol wipe (Allegiance alcohol prep pad; Cardinal Health, McGaw Park, IL) in an area of 5 by 10 cm and allowed to air dry. This same area was then inoculated with 100 μl (1 × 108 CFU) of S. epidermidis 1457 or 1457 ica::dhfr by pipetting the cell suspension over the surface of the skin; each forearm was subsequently allowed to air dry. One forearm was then covered with sterile gauze (Allegiance gauze sponges; Cardinal Health, McGaw Park, IL), and the other was left exposed. Volunteers were instructed to go about their normal activities, including bathing, but were asked to re-cover the covered forearm with clean sterile gauze after the material was exposed to liquid (i.e., bathing). The volunteers did not receive antibiotics during at least the 2 weeks prior to or during the study period. After 1, 3, and 10 days, each forearm was then swabbed vigorously with a polyurethane foam swab (CultureSwab EZ II; Becton Dickinson and Company, Sparks, MD) for 30 s; the swabs were then placed in sterile tubes containing 1 ml of saline. The tubes were vigorously agitated in a vortex mixer for 1 min and then serially plated to CRA plates and incubated for 48 h at 37°C. After 48 h of incubation, colony counts were performed and those colonies with a crusty phenotype (i.e., 1457) were replica plated to TSA plates (without antibiotic), TSA plates containing 10 μg/ml erythromycin (Sigma, St. Louis, MO), or TSA plates containing 50 μg/ml ampicillin (Sigma). Colonies from CRA with a smooth phenotype (i.e., 1457 ica::dhfr) were replica plated to TSA plates and TSA plates containing 10 μg/ml trimethoprim (Sigma). This protocol was approved by the University of Nebraska Medical Center Institutional Review Board.
Trimethoprim-resistant, CRA smooth colonies were confirmed to be S. epidermidis 1457 ica::dhfr by using the forward primer 5′- CAGTATAACAACATTCTATTGC-3′ specific for the intergenic region between icaR and icaA (bp 2334135 to 2334156 in the S. epidermidis RP62A genome sequence [NC_002976]) and reverse primer 5′-CCATTAAGCCTGACAATCG-3′ specific for the region immediately downstream of the dhfr gene (bp 5709 to 5691 in the pIP1630 sequence [AF045472]). Amplification yielded a 550-bp product which is specific for S. epidermidis 1457 ica::dhfr. Pulsed-field gel electrophoresis (PFGE) was performed on all colonies suspected of being S. epidermidis 1457 (ampicillin and erythromycin susceptible and crusty phenotype on CRA). Genomic DNA suitable for PFGE was isolated as previously described (2). PFGE was performed using a CHEF DR-III apparatus (Bio-Rad, Hercules, CA) with the following parameters: initial switch time, 5 s; final switch time, 40 s; 19 h at 200 V (6 V/cm); 14°C. PFGE restriction patterns were compared using a combination of the Gel Documentation System 2000 (Bio-Rad) and Bionumerics software (Applied Maths, Austin, TX).
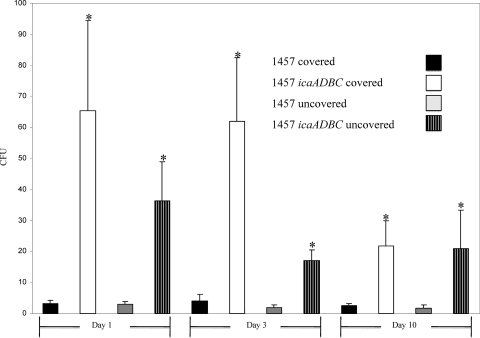
Based on previous studies demonstrating that the population of S. epidermidis isolated from the skin of healthy volunteers is primarily comprised of icaADBC-negative isolates, we hypothesized that wild-type S. epidermidis 1457 (icaADBC+) would be outcompeted by an isogenic icaADBC derivative in a human skin colonization model. Consistent with other human skin competition studies (7), there was a dramatic decrease in the number of bacteria obtained at the first skin sampling (24 h post-skin inoculation); although 108 CFU were inoculated, generally less than 102 CFU were recovered from each arm of the nine volunteers (data not shown). The total number of colonies of S. epidermidis 1457 ica::dhfr and 1457 isolated from all nine volunteers were compiled and compared (covered with gauze or uncovered) at each time point (1, 3, and 10 days). These experiments demonstrated that the mean CFU of S. epidermidis 1457 ica::dhfr isolated from the skin of healthy volunteers was significantly (P < 0.05) greater than S. epidermidis 1457 at each day regardless of whether the forearm was covered or uncovered (Fig. 1). Statistical analysis was performed using the Wilcoxon signed rank test (GraphPad Prism 2.0; San Diego, CA). All 1457 ica::dhfr and 1457 colonies were confirmed using PCR and PFGE, respectively.
FIG. 1.
CFU of S. epidermidis 1457 and 1457 ica::dhfr isolated from the human skin competition model. Bars represent mean CFU isolated from the nine healthy volunteers at days 1, 3, and 5. Error bars represent standard errors of the means, and * indicates significance (P < 0.05) compared to 1457 for the same condition and time point.
These data suggest that S. epidermidis isolates that do not possess the icaADBC operon have the ability to outcompete isolates producing PIA. Although many studies have demonstrated that the majority of S. epidermidis isolates obtained from the skin of healthy individuals do not possess the icaADBC operon, this is the first study to address this question using an isogenic strain set and a human skin competition model. It is unknown why S. epidermidis 1457 ica::dhfr has the ability to outcompete S. epidermidis 1457 on the skin of humans. It is known, however, that the production of biofilm shunts a great deal of carbon from glycolysis to produce PIA instead of pyruvate and subsequent ATP production. Therefore, ica variants may be selected on the skin due to an increased growth rate. However, it is not known how much growth is actually occurring on the skin. For instance, less than 100 organisms are found still colonizing the skin after inoculation of 108 CFU. Therefore, it is possible that the production of PIA masks a specific adhesion factor on the surface of the bacterium that is important for adherence to epithelial surfaces. However, importantly, it has not been experimentally demonstrated that S. epidermidis produces PIA while colonizing skin. It is perplexing that although the majority of S. epidermidis organisms recovered from the skin of healthy individuals lack the icaADBC operon, most biomaterial-related infections are caused by icaADBC-positive isolates. This observation suggests that the ability to produce PIA (i.e., carrying the icaADBC locus) confers a selective advantage in certain niches, such as hospital environments. An additional, alternative interpretation of these experiments is that PIA-positive S. epidermidis is more difficult to culture from the skin when using a swab compared to PIA-negative isolates, as each may colonize unique areas of the skin structure. The use of harsh sampling techniques to further disrupt epithelial layers or other animal models where the entire skin layer is cultured could be used to address this question. Addressing these questions as well as defining mechanisms through which S. epidermidis rapidly loses icaADBC gene function (phenotypic variation) and gene loss (deletion) are a major focus of our laboratory.
Acknowledgments
This study was funded in part by Public Health Service grant AI49311 from the National Institute of Allergy and Infectious Diseases to P.D.F.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Arciola, C. R., S. Gamberini, D. Campoccia, L. Visai, P. Speziale, L. Baldassarri, and L. Montanaro. 2005. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J. Biomed. Mater. Res. A 75:408-413. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, S. H., K. Naber, J. Hacker, and W. Ziebuhr. 2002. Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int. J. Antimicrob. Agents 19:570-575. [DOI] [PubMed] [Google Scholar]
- 4.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman, D. J., F. R. Falkiner, and C. T. Keane. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42:872-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 7.Gustafsson, I., O. Cars, and D. I. Andersson. 2003. Fitness of antibiotic resistant Staphylococcus epidermidis assessed by competition on the skin of human volunteers. J. Antimicrob. Chemother. 52:258-263. [DOI] [PubMed] [Google Scholar]
- 8.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genetic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53:367-374. [DOI] [PubMed] [Google Scholar]
- 9.Handke, L. D., S. R. Slater, K. M. Conlon, S. T. O'Donnell, M. E. Olson, K. A. Bryant, M. E. Rupp, J. P. O'Gara, and P. D. Fey. 2007. σB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 53:82-91. [DOI] [PubMed] [Google Scholar]
- 10.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 11.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozitskaya, S., M. E. Olson, P. D. Fey, W. Witte, K. Ohlsen, and W. Ziebuhr. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maki, D. G., D. M. Kluger, and C. J. Crnich. 2006. The risk of bloodstream infection in adults with different intravascular devices: a systematic review of 200 published prospective studies. Mayo Clin. Proc. 81:1159-1171. [DOI] [PubMed] [Google Scholar]
- 15.Nuryastuti, T., H. C. van der Mei, H. J. Busscher, R. R. Kuijer, A. T. Aman, and B. P. Krom. 2008. recA mediated spontaneous deletions of the icaADBC operon of clinical Staphylococcus epidermidis isolates: a new mechanism of phenotypic variations. Antonie van Leeuwenhoek 94:317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohde, H., E. C. Burandt, N. Siemssen, L. Frommelt, C. Burdelski, S. Wurster, S. Scherpe, A. P. Davies, L. G. Harris, M. A. Horstkotte, J. K. Knobloch, C. Ragunath, J. B. Kaplan, and D. Mack. 2007. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28:1711-1720. [DOI] [PubMed] [Google Scholar]
- 17.Rupp, M. E. 2004. Nosocomial bloodstream infections, p. 253-265. In C. G. Mayhall (ed.), Hospital epidemiology and infection control. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 18.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]