Abstract
An outbreak of acute hepatitis A virus in North Carolina was linked to drinking water from a contaminated shallow spring by phylogenetic analysis of hepatitis A virus (HAV) genomic sequences. Detection of HAV and fecal indicators in the water provided useful and timely information to assist with public health prevention and control measures.
Hepatitis A virus (HAV) is spread person to person by the fecal-oral route and causes acute hepatitis that can lead to jaundice and, in certain cases, liver failure. Increased use of hepatitis A vaccine during the past 10 years has led to a decline in hepatitis A incidence in the United States: only 4,488 acute cases were reported during 2005, while over 30,000 cases were reported in 1995 (20). Waterborne transmission of HAV is now rare in the United States. In 2004, waterborne and foodborne outbreaks combined accounted for only 10% of reported cases (7, 20). HAV outbreaks are commonly investigated by using serologic testing for anti-HAV immunoglobulin M to identify cases and epidemiologic analyses to assess potential risk factors (2, 5). However, using phylogenetic analysis to determine the source of exposure resulting in infection, particularly in waterborne outbreaks, is rare (4, 10).
In August 2006, the North Carolina Division of Public Health was notified of four cases of acute hepatitis A among persons who had spent time on a farm in western North Carolina. Further investigation identified 12 additional cases of acute viral hepatitis linked to the same farm. Of the 16 total cases, 9 had positive anti-HAV immunoglobulin M results and 7 were epidemiologically linked to the laboratory-confirmed cases. An epidemiologic investigation of risk factors indicated the consumption of water from the farm during the period of May to August 2006 as the likely common source of exposure associated with the outbreak. However, detailed exposure information was available for only five patients and five nonill persons. All five patients and one nonill person reported drinking water from a shallow spring that had been excavated by hand by the owner of the property. Water from the spring was fed into a ventilated prefabricated barrel reservoir and pumped to the house, a greenhouse, and a camping area downhill from the spring. The spring was located downhill from the septic tank of the farm residence, raising concerns regarding ground- and springwater contamination.
Springwater was collected from the outbreak site on 30 August 2006 in sterile containers totaling 65 liters and analyzed within 24 h for HAV and within 48 h for fecal indicators. High levels of fecal indicator bacteria were identified in the springwater, including Escherichia coli at 43 most probable number (MPN)/100 ml, enterococci at 36 MPN/100 ml, fecal coliforms at 59 MPN/100 ml, and total coliforms at 1,492 MPN/100 ml, as measured in duplicate 100-ml volumes by using a Colilert Enterolert assay (Idexx Laboratories, Westbrook, ME). F+ coliphages were not detected in a 1/3-liter volume of springwater by using U.S. Environmental Protection Agency method 1601 (12) and thus were not associated with these high levels of bacterial indicators in the grab sample. F+ coliphage occurrence is more common in sewage from populations than from individual persons or families, as demonstrated by the absence of F+ coliphages in 13 household septic tanks and their presence in 9% (2 of 22) of wastewater lift stations and 100% (14 of 14) of wastewater treatment plant influents (6). Previous studies also reported that the F+ coliphage levels were unrelated to the levels of bacterial indicators in stool (8, 14). Therefore, the presence of fecal indicator bacteria in the springwater, even without F+ coliphages, is indicative of fecal pollution.
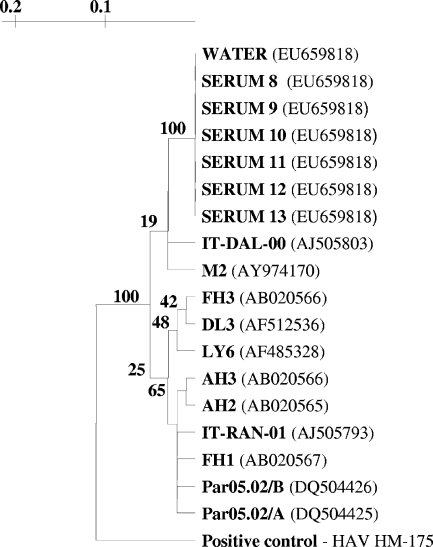
HAV was concentrated from 60 liters of springwater by using a 1MDS positively charged pleated cartridge filter (CUNO, Incorporated, Meriden, CT) as previously described (11). HAV was eluted from the filter with 550 ml of 1.5% beef extract-0.05 M glycine (pH 9.5), with the eluate adjusted to pH 7.2 before overnight 8% polyethylene glycol precipitation. The polyethylene glycol-treated eluate was then centrifuged at 6,700 × g, and the pellet was resuspended in approximately 1 ml phosphate-buffered saline (pH 7.2). The resuspended pellet was then subjected to RNA extraction (QIAamp viral RNA mini kit; Qiagen, Valencia, CA), and the RNA extract was subjected to nested reverse transcription-PCR (RT-PCR) amplification of the HAV VP1-VP3 junction (15). Standard quality assurance procedures were followed during the nested RT-PCR, and as shown in Fig. 1, the positive control was sequenced and did not match the sequence of the outbreak strain. Amplicons were visualized on 2% agarose gels, and those with appropriately sized bands were purified (Qiaquick; Qiagen). The nucleic acids were then sequenced to confirm their identities (University of North Carolina Genome Analysis Facility, Chapel Hill, NC).
FIG. 1.
Phylogenetic tree of HAV strains, using the 281-bp VP1-VP3 region. Strains from water and serum samples associated with the outbreak, along with historical strains, are included, with GenBank accession numbers in parentheses. Serum samples in the figure are representative of all eight serum sequences.
RNA extraction, RT-PCR amplification, and sequencing were then performed on sera collected from eight persons with hepatitis A who were directly or indirectly linked to this outbreak. The HAV sequences from water and sera were aligned in BioEdit (13), and phylogenetic analysis was performed with TreeCon (version 1.3b) (19) by using the Jukes and Cantor correction and 100 bootstrap values. The nucleic acid sequences from sera and water were identical within a 281-bp portion of the VP1-VP3 genomic region (Fig. 1). The HAV outbreak strain was identified as being in the US-IA1 cluster of subgenotype IA (16), which is the predominant source of hepatitis A infections in North, Central, and South America (1). The outbreak strain also had 85% to 95% sequence identity with 11 similar strains selected from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/index.html) using the phylogenetic analysis methods previously described (Fig. 1).
The sample inhibition of RT-PCR amplification was determined by using a negative control sample of 60 liters of groundwater from a nearby site not linked to the outbreak. Access to the outbreak site was limited due to the sensitive nature of the investigation, so the researchers selected a nearby water source in the same aquifer as a negative control. The negative control sample was processed and RT-PCR was conducted using the same conditions and primers described previously. An HAV subgenotype IA isolate (HLD2) was spiked into the negative-control water in a dilution series of extracts beginning at 104 RT-PCR units of HAV RNA and detected by nested RT-PCR gel electrophoresis. HLD2 was selected as the model outbreak strain because it has a known infectious dose among primates, unlike cell culture-adapted laboratory strains. On the basis of the results of this experiment, the control groundwater extracts were a 10-fold dilution (1 log10) more inhibitory to RT-PCR amplification than the positive control of HLD2 diluted in Tris-EDTA buffer.
Through this investigation, we recovered HAV from a fecally contaminated drinking water source that was identical in the sequenced region to HAV isolated from the sera of persons linked to a hepatitis A outbreak. Although we were unable to prove causality of exposure through water in this outbreak, these results, in combination with those of an epidemiologic analysis of the risk factors associated with the illnesses, provide evidence that the contaminated spring was the source of this outbreak. A sanitary survey determined that the contamination might have come from a septic tank located directly uphill from the spring. These findings are similar to those for another outbreak of hepatitis A due to contamination of drinking water in which HAV traveled up to 60 m from a cesspool to neighboring wells (10). Because HAV can persist for 3 to 10 months in water (3, 18) and persistent contamination was a possibility, the local health department recommended eliminating the spring as a drinking water source and replacing it with a deep well. Environmental detection of HAV in the springwater provided useful and timely information to assist with public health prevention and control measures. Greater efforts to quantify pathogen levels in water, food, and other probable infection transmission vehicles of outbreaks are needed to better understand the quantitative risk associations between pathogen doses and human illness or infection responses.
Nucleotide sequence accession number.
The sequences described in this study have GenBank accession number EU659818.
Acknowledgments
We thank the Madison County Health Department, NC, for sample collection and epidemiologic data and Yuri Khudyakov, Division of Viral Hepatitis, National Center for Infectious Diseases, Centers for Disease Control and Prevention, for HAV strain HLD2.
This work was supported in part by funds from a cooperative agreement between the Joint Institute for Food Safety and Applied Nutrition of the University of Maryland, College Park, and the Food and Drug Administration, College Park, MD.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Arauz-Ruiz, P., L. Sundqvist, Z. Garcia, L. Taylor, K. Visona, H. Norder, and L. O. Magnius. 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J. Med. Virol. 65:449-456. [PubMed] [Google Scholar]
- 2.Bergeisen, G. H., M. W. Hinds, and J. W. Skaggs. 1985. A waterborne outbreak of hepatitis A in Meade County, Kentucky. Am. J. Public Health 75:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biziagos, E., J. Passagot, J. M. Crance, and R. Deloince. 1988. Long-term survival of hepatitis A virus and poliovirus type 1 in mineral water. Appl. Environ. Microbiol. 54:2705-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch, A. B., S. L. Stramer, J. D. Smith, H. S. Margolis, H. A. Fields, T. W. McKinley, C. P. Gerba, J. E. Maynard, and R. K. Sikes. 1990. Recovery of hepatitis A virus from a water supply responsible for a common source outbreak of hepatitis A. Am. J. Public Health 80:428-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brundage, S. C., and A. N. Fitzpatrick. 2006. Hepatitis A. Am. Fam. Physician 73:2162-2168. [PubMed] [Google Scholar]
- 6.Calci, K. R., W. Burkhardt III, W. D. Watkins, and S. R. Rippey. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 64:5027-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2006. Hepatitis surveillance report no. 61. Centers for Disease Control and Prevention, Atlanta, GA.
- 8.Cornax, R., M. A. Morinigo, F. Gonzalez-Jaen, M. C. Alonso, and J. J. Borrego. 1994. Bacteriophages presence in human faeces of healthy subjects and patients with gastrointestinal disturbances. Zentralbl. Bakteriol. 81:214-224. [DOI] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.De Serres, G., T. L. Cromeans, B. Levesque, N. Brassard, C. Barthe, M. Dionne, H. Prud'homme, D. Paradis, C. N. Shapiro, O. V. Nainan, and H. S. Margolis. 1999. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 179:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Environmental Protection Agency. 1984. Manual of methods for virology, EPA 600-4-84-013. Microbiological and Chemical Exposure Assessment Research Division, Environmental Protection Agency, Washington, DC.
- 12.Environmental Protection Agency. 2001. Method 1601: Male-specific (F+) and somatic coliphage in water by two-step enrichment procedure, EPA 821-R-01-030. Office of Water, Environmental Protection Agency, Washington, DC.
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Havelaar, A. H., K. Furuse, and W. M. Hogeboom. 1986. Bacteriophages and indicator bacteria in human and animal faeces. J. Appl. Bacteriol. 60:255-262. [DOI] [PubMed] [Google Scholar]
- 15.Hutin, Y. J., V. Pool, E. H. Cramer, O. V. Nainan, J. Weth, I. T. Williams, S. T. Goldstein, K. F. Gensheimer, B. P. Bell, C. N. Shapiro, M. J. Alter, and H. S. Margolis. 1999. A multistate, foodborne outbreak of hepatitis A. N. Engl. J. Med. 340:595-602. [DOI] [PubMed] [Google Scholar]
- 16.Nainan, V. O., G. Xia, G. Vaughan, and H. S. Margolis. 2006. Diagnosis of hepatitis A virus infection: a molecular approach. Clin. Microbiol. Rev. 19:63-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwab, K. J., F. H. Neill, R. L. Fankhauser, N. A. Daniels, S. S. Monroe, D. A. Bergmire-Sweat, M. K. Estes, and R. L. Atmar. 2000. Development of methods to detect “Norwalk-like viruses” (NLVs) and hepatitis A virus in delicatessen foods: application to a food-borne NLV outbreak. Appl. Environ. Microbiol. 66:213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobsey, M. D., P. A. Shields, F. S. Hauchman, A. L. Davis, V. A. Rullman, and A. Bosch. 1988. Survival and persistence of hepatitis A virus in environmental samples, p. 121-124. In A. J. Zuckerman (ed.), Viral hepatitis and liver disease. Alan R. Liss, New York, NY.
- 19.Van de Peer, J., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 20.Wasley, A., J. T. Miller, and L. Finelli. 2007. Surveillance for acute viral hepatitis, United States, 2005. MMWR Surveill. Summ. 56(SS-3):1-24. [PubMed] [Google Scholar]