Abstract
The spread of antibiotic resistance in pathogens is primarily a consequence of the indiscriminate use of antibiotics, but there is concern that food-borne lactic acid bacteria may act as reservoirs of antibiotic resistance genes when distributed in large doses to the gastrointestinal tract. Lactobacillus reuteri ATCC 55730 is a commercially available probiotic strain which has been found to harbor potentially transferable resistance genes. The aims of this study were to define the location and nature of β-lactam, tetracycline, and lincosamide resistance determinants and, if they were found to be acquired, attempt to remove them from the strain by methods that do not genetically modify the organism before subsequently testing whether the probiotic characteristics were retained. No known β-lactam resistance genes was found, but penicillin-binding proteins from ATCC 55730, two additional resistant strains, and three sensitive strains of L. reuteri were sequenced and comparatively analyzed. The β-lactam resistance in ATCC 55730 is probably caused by a number of alterations in the corresponding genes and can be regarded as not transferable. The strain was found to harbor two plasmids carrying tet(W) tetracycline and lnu(A) lincosamide resistance genes, respectively. A new daughter strain, L. reuteri DSM 17938, was derived from ATCC 55730 by removal of the two plasmids, and it was shown to have lost the resistances associated with them. Direct comparison of the parent and daughter strains for a series of in vitro properties and in a human clinical trial confirmed the retained probiotic properties of the daughter strain.
The use of probiotic bacteria, generally members of the genera Lactobacillus and Bifidobacterium, in foods and supplements that promote health or prevent disease, has followed a long history of safe use of these organisms (8). The spread of antibiotic resistance determinants in the environment is undoubtedly primarily a consequence of indiscriminate use of antibiotics in both animals and humans. However, this concern has led to the idea that food-borne lactic acid bacteria may act as reservoirs of antibiotic resistance genes when purposely distributed in large doses to the microbiota of the gastrointestinal tract (17, 38). The outcome of the EU PROSAFE project was to recommend that all future probiotics should not contain known antibiotic resistance traits (40), and currently the European Food Safety Authority qualified presumption of safety proposals seem to be following this recommendation (6).
Bacterial antibiotic resistance can be either intrinsic or acquired (16). Intrinsic resistance is a natural resistance present in all strains of a bacterial species, while acquired resistance is often identifiable as a resistance found in only a certain number of members of a particular species. Although intrinsic resistance is generally accepted to be nontransferable, acquired resistance may be more easily transferred to other bacterial species, particularly if the resistance trait is located on a plasmid in the carrier strain (37).
Lactobacillus reuteri ATCC 55730, a commercially available, well-documented probiotic (health-promoting) bacterium (1, 23, 27, 34, 46-48), has been shown to possess a series of intrinsic antibiotic resistances common to this particular species (18, 25, 26). It does, however, carry specific, unusual resistances to tetracycline and lincosamides, as well as a β-lactam resistance which appears in approximately half of the members of this species (18, 24). With a draft genome sequence of L. reuteri ATCC 55730 determined in our laboratory (7), the aim of this study was to define the origin, localization, and character of these unusual resistances. Subsequent localization of tet(W) and lnu(A) on plasmids led to the removal of these plasmids and validation of the properties retained by the daughter strain generated.
MATERIALS AND METHODS
Bacteria, culturing, DNA preparation, and primers.
The bacterial strains and primers used in this study are listed in Tables 1 and 2, respectively. The primers were obtained from Invitrogen (Carlsbad, CA). L. reuteri strains were grown in MRS broth or on MRS agar (Oxoid, Basingstoke, United Kingdom). Plates were incubated in an anaerobic atmosphere (GasPak; BD, Sparks, MD). For antibiotic supplementation, 100 μg ml−1 tetracycline (Sigma, St. Louis, MO; MRS-Tet) or 8 μg ml−1 lincomycin (Sigma; MRS-Lin) was used unless otherwise stated. Bacterial DNA was prepared with the DNeasy tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
TABLE 1.
L. reuteri strains used in this study
| Strain | Description | Origin or reference |
|---|---|---|
| ATCC 55730 | Alternative name, SD2112; Tetr Linr Ampr; human milk isolate | Biogaia AB |
| ATCC 55730Tets | ATCC 55730 cured of plasmid pLR581; Tets Linr Ampr | This study |
| DSM 17938 | ATCC 55730 cured of plasmids pLR581 and pLR585; Tets Lins Ampr | This study |
| DSM 20016 | Type strain; Tets Lins Amps; human intestinal isolate | DSMZa |
| DSM 20015 | Amps; cow manure isolate | DSMZ |
| ATCC 55148 | Amps; chicken intestinal isolate | Biogaia AB |
| ATCC 55149 | Ampr; turkey intestinal isolate | Biogaia AB |
| CF48-3A | Ampr; human fecal isolate | Biogaia AB |
DSMZ, German Collection of Microorganisms and Cell Cultures.
TABLE 2.
Primers used in this study
| Primer | Use | Sequence |
|---|---|---|
| 1989f | Detection of lr1989a on pLR580 | 5′-ATT TTC CAC CCG CAT ATT CA-3′ |
| 1989r | Detection of lr1989a on pLR580 | 5′-TGC ATC ACG AAT CAA ACC AT-3′ |
| 2004f | Detection of lr2004 on pLR581 | 5′-AGG TGA AGC ATT TCG AGC AT-3′ |
| 2004r | Detection of lr2004 on pLR581 | 5′-GGC TTT CCG TCA TCA TCA GT-3′ |
| 2084f | Detection of lr2084b on pLR584 | 5′-TTT GGC TGG CAA AAT CAT TC-3′ |
| 2084r | Detection of lr2084b on pLR584 | 5′-TTT TTG CAG CAT TGA AAA CG-3′ |
| 2102f | Detection of lr2102 on pLR585 | 5′-GAA CGG AAG CAA CAA CGA AT-3′ |
| 2102r | Detection of lr2102 on pLR585 | 5′-CGT TTG GTT GGA GAA GTG GT-3′ |
| 1996f | Detection of tet(W) | 5′-TTC GCT GGG ATA CTT GAA CC-3′ |
| 1996r | Detection of tet(W) | 5′-TTT TTA CCT GGA CCG TTT CG-3′ |
| 2105f | Detection of lnu(A) | 5′-TGG AAA ACA ACA AAG AGA ACA CA-3′ |
| 2105r | Detection of lnu(A | 5′-CCA GAA TGA AAA AGA AGT TGA GC-3′ |
| 545(1)f | Sequencing of pbp1a | 5′-ATC AGT CAA CGC GTA GTG AGC-3′ |
| 545(1)r | Sequencing of pbp1a | 5′-GCA TCG TCC CAA TCA GAT G-3′ |
| 545(2)f | Sequencing of pbp1a | 5′-AAC GTA AAG CCC AAG AAG CA-3′ |
| 545(2)r | Sequencing of pbp1a | 5′-TGC AGC AGA AAC TTG GAG TG-3′ |
| 889(1)f | Sequencing of pbp2b | 5′-CAC GTA CCA AAA AGC GTC AA-3′ |
| 889(1)r | Sequencing of pbp2b | 5′-AAC CCT TGG GAC TCT GAA CA-3′ |
| 889(2)f | Sequencing of pbp2b | 5′-GCA GCA ATG AGT GGA GCA TA-3′ |
| 889(2)r | Sequencing of pbp2b | 5′-GTT TCA CCA GGC AAG TCG AT-3′ |
| 1005(1)f | Sequencing of pbp2a | 5′-TCA GAT TTG AAA GAG CGA ATC A-3′ |
| 1005(1)r | Sequencing of pbp2a | 5′-ATC GAA CAT TGC TCC ACC AT-3′ |
| 1005(2)f | Sequencing of pbp2a | 5′-GCG GTT GAG GTA GAA AAC CA-3′ |
| 1005(2)r | Sequencing of pbp2a | 5′-CAA AGA CTG CAT AGG CAC GA-3′ |
| 1631(1)f | Sequencing of pbp2x | 5′-AGC CAC AAC GAA CGA AAA AT-3′ |
| 1631(1)r | Sequencing of pbp2x | 5′-TTG AAC GGT TAA AGT TGT TCC A-3′ |
| 1631(2)f | Sequencing of pbp2x | 5′-AAG GCG AAT TTG CTT CTC AA-3′ |
| 1631(2)r | Sequencing of pbp2x | 5′-CAG TTG TGG TCT TTC CAG CA-3′ |
| 1752f | Sequencing of pbpX | 5′-TTT TTG ACG CTT TGT TCC TTT-3′ |
| 1752r | Sequencing of pbpX | 5′-AAT TAT GGA GAA TAT CAT CCG AAG C-3′ |
Identification of plasmids and putative resistance genes.
Identification of plasmids in L. reuteri ATCC 55730 was achieved by analysis of the draft genome sequence of this strain (7). The genes encoding putative penicillin resistance proteins and penicillin-binding proteins (Pbp) were identified by BLAST searches (3) against the genome sequence with known resistance determinants (GenBank accession numbers: Bla1, AAK53749; Bla2, AAS42360; BlaZ, CAB94802; FibA, CAB89120; FibB, CAB89121; MecA, CAG39068; MecR1, CAG39069; MurM, NP_664650; MurN, AAK07416) and Pbp (Pbp1a, NP_720910; Pbp1b, NP_722289; Pbp2a, NP_722252; Pbp2b, NP_721030; Pbp2x, NP_720898; PbpX, NP_721297). Also, the tetracycline and lincomycin resistance genes tet(W) and lnu(A) were searched for by using known resistance genes (GenBank accession numbers ABC18266 and YP_473355, respectively).
Sequencing of Pbp-encoding genes.
For all identified Pbp-encoding genes, one primer pair located close to the start and end of the gene was designed and for all but lr1752, one pair located approximately 500 bp from the first pair was also designed (Table 2). The primers were used for PCR amplification of the genes encoding the Pbp from ampicillin-sensitive L. reuteri strains DSM 20016, DSM 20015, and ATCC 55148 and ampicillin-resistant strains ATCC 55730, ATCC 55149, and CF48-3A. illustra PuReTaq Ready-To-Go PCR Beads (GE Healthcare, Uppsala, Sweden), primers (10 pmol of each), and DNA (0.5 μl) were added to the PCR mixture and amplified with the following program: 95°C for 5 min; 30 cycles of 95°C for 30 s, 53°C for 30 s, and 72°C for 2 min; and 72°C for 10 min. The PCR products were purified and thereafter sequenced with the same set of primers. After assembly of the genes (with Contig Express [Invitrogen]), the corresponding protein sequences were aligned and compared with Lactobacillus Pbp obtained from genomes available in GenBank (L. acidophilus, CP000033; L. brevis, CP00041; L. casei, CP000423; L. delbrueckii, CR954253; L. gasseri, CP000413; L. helveticus, CP000517; L. johnsonii, AE017198; L. plantarum, AL935263; L. sakei, CR936503; L. salivarius, CP000233).
Curing of plasmids.
Plasmid curing was done by protoplast formation and regeneration essentially as described by Vescovo et al. (43). An overnight culture of ATCC 55730 was diluted to an optical density at 600 nm (OD600) of 0.1 in a 10-ml MRS culture and grown at 37°C to an OD600 of 0.7 to 0.8. Cells were collected by centrifugation at 3,000 × g for 10 min, washed in 10 ml Nanopure water, recentrifuged, and resuspended in 2 ml of protoplast buffer (0.2 M sodium phosphate, 0.5 M sucrose, 20 mM MgCl2, pH 7.0). The cells were then mixed with an equal volume of protoplast buffer containing 10 mg ml−1 lysozyme (Sigma) and incubated at 37°C for 1 h. Protoplasts were harvested by centrifugation at 3,000 × g for 15 min, washed with 20 ml protoplast buffer, recentrifuged, and resuspended in 1 ml of protoplast buffer. Dilutions in protoplast buffer were then plated on MRS agar with 0.5 M sucrose for regeneration. Dilutions in Nanopure water were plated on MRS agar to assess the number of remaining whole cells. The number of CFU was determined after 1 and 2 days of anaerobic incubation at 37°C. Regenerated colonies were picked to MRS and MRS-Tet agar for identification of bacteria cured of pLR581 and to MRS and MRS-Lin agar for bacteria cured of pLR585. Plasmid-cured candidates were identified by nongrowth on MRS agar supplemented with antibiotics.
PCR detection of plasmids and resistance genes and repetitive-sequence-based PCR (rep-PCR) typing.
The plasmids and resistance genes of strain ATCC 55730 were detected by PCR by the same method as for Pbp-encoding genes. Primers 1989f and 1989r, 2004f and 2004r, 2084f and 2084r, and 2102f and 2102r (Table 2), detecting replication protein genes on pLR580, pLR581, pLR584, and pLR585, respectively (10 pmol of each), and primers 1996f and 1996r and primers 2105f and 2105r, detecting tet(W) and lnu(A), respectively, were used.
Bacterial isolates were fingerprinted by rep-PCR. PuReTaq Ready-To-Go PCR Beads, DNA (0.5 μl), and primer 5′-GTG GTG GTG GTG GTG-3′ (20 pmol) were mixed, and the reaction was performed according to the program described by Versalovic et al. (41): 95°C for 7 min; 30 cycles of 90°C for 30 s, 95°C for 1 min, 40°C for 1 min, and 65°C for 8 min; and 65°C for 16 min. The PCR products were separated by electrophoresis (1% agarose gel in 0.5× TBE buffer) and stained with ethidium bromide. Digitalized images were captured under UV light transillumination.
Determination of MICs.
L. reuteri strains were grown in MRS broth for 16 h at 37°C. After dilution to 105 CFU ml−1, 1 μl of each strain was spotted onto MRS plates containing lincomycin or clindamycin (Sigma) at concentrations of 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, and 16 μg ml−1. After adsorption of the drops, the plates were incubated anaerobically at 37°C for 24 h. The MIC was defined as the lowest antibiotic concentration at which there was no visible bacterial growth. MICs of tetracycline were determined by Etest (AB Biodisk) on bacteria growing on MRS agar plates according to the instructions from the manufacturer.
Colony and cell morphology.
L. reuteri strains were grown for 48 h on MRS plates at 37°C in an anaerobic atmosphere before photographs of single colonies were taken. The morphology of the bacterial cells was investigated by phase-contrast microscopy (×400 magnification).
Fermentation pattern and reuterin production.
Fermentation patterns were determined with api 50 CHL (BioMérieux, Marcy l'Etoile, France) according to the instructions from the manufacturer. Reuterin production (13) was measured by the reaction between reuterin (3-hydroxy-propinoaldehyde) produced by the L. reuteri strains with 2,4-dinitrophenylhydrazine to form a red hydrazone. The bacteria were grown for 48 h on MRS plates (inoculated as streaks). The plates were then overlaid with 500 mM glycerol agar (1% agar) and incubated at 37°C for 30 min. Reuterin was detected by the addition of 5 ml 2,4-dinitrophenylhydrazine (0.1% in 2 M HCl). After 3 min incubation, the solution was poured off and 5 ml 5 M KOH was added. Red zones around the colonies demonstrated the presence of reuterin and the extent of its production.
Growth.
Growth was determined by inoculation of an overnight culture to an OD600 of 0.1 in MRS broth in tubes prewarmed to 37°C. The tubes were incubated at 37°C, and samples were taken for 8 h and the OD600 was measured. Each strain was tested in triplicate.
Binding to mucus.
Binding to ovine small intestinal mucus was performed according to Roos et al. (31). All bacterial strains were analyzed in triplicate.
Acid tolerance.
Testing of survival at pH 2 was performed according to Wall et al. (45) with a synthetic stomach juice described by Cotter et al. (15) that was modified by adding no enzymes. Duplicate samples were taken on each of two occasions. Statistical significance of differences was analyzed with Student's t test.
Bile tolerance.
Strains were grown for 16 h in MRS broth at 37°C, and the stationary-phase suspensions were diluted in phosphate-buffered saline to approximately 103 to 106 CFU ml−1. Ten microliters of each dilution was dropped onto MRS plates containing concentrations of bovine bile of up to 6% (Sigma B3883). The plates were incubated for 72 h at 37°C in an anaerobic atmosphere, after which the colonies were counted. Bacteria in exponential phase (OD600 of 0.5) were also tested in the same manner. Each bacterium was analyzed on two separate occasions.
Pathogen inhibition.
Test strains were grown in MRS broth for 16 h and plated on MRS agar. The plates were incubated anaerobically (with Anaerogen [Oxoid]) for 24 h at 37°C. Isolated single colonies were then streaked in the center of fresh MRS agar plates and incubated anaerobically for 36 h at 37°C. The target pathogens were Candida albicans ATCC 28956, Enterobacter sakazakii ATCC 51329, Salmonella enterica serovar Typhimurium ATCC 14028, and Clostridium difficile ATCC 43593.
C. albicans and E. sakazakii were grown in 10 ml of tryptic soy broth (BBL, BD) plus 1.5% Lab Lemco (Oxoid) and incubated for 48 h at 37°C (Candida) or 32°C (Enterobacter). The pathogens were then inoculated into freshly prepared sterile tryptic soy broth plus 1.5% Lab Lemco and 1.5% agar with or without the addition of glycerol (150 mM) to a target concentration of 105 CFU ml−1 (Candida) or 106 CFU ml−1 (Enterobacter). The pathogen preparations (10 ml) were overlaid on the streaked test strains growing on the MRS plates. Plates were incubated anaerobically at 37°C (Candida) or 36°C (Enterobacter) for 48 h. S. enterica serovar Typhimurium and Clostridium difficile were grown in 10 ml of brain heart infusion broth (BBL, BD) and reinforced clostridial medium (BD), respectively, and incubated at 37°C for 16 h. Each pathogen was then inoculated in brain heart infusion agar or reinforced clostridial medium plus 1.5% agar (40°C) with or without the addition of glycerol (150 mM) to get a concentration of 105 CFU ml−1. Ten milliliters of this preparation was overlaid on the test strains growing on the MRS plates. Plates were incubated anaerobically at 37°C for 48 h. Inhibition halos were measured in triplicate samples on three different experimental occasions for each strain and pathogen.
Gastrointestinal passage and safety in humans.
Sixteen subjects (10 female) aged 27 ± 7 (mean ± standard deviation) years were recruited after written informed consent was obtained at the Gastroenterology Unit, Department of Internal Medicine, Lund University Hospital, Lund, Sweden. The Scientific Ethical Committee of Lund University Hospital approved this study prior to its start. Inclusion criteria were an age of 18 to 65 years, written informed consent, stated availability throughout the study period, a lack of any major illnesses (allergy symptoms were acceptable), and the mental ability to understand and fulfill the protocol. Exclusion criteria were significant disease, pregnancy, use of oral antibiotics in the 2 weeks prior to ingestion of the study product, and participation in other clinical trials. Subjects were randomly assigned to receive a placebo (four subjects), an ATCC 55730 standard dose (8 × 108 CFU/day; three subjects), a DSM 17938 standard dose (8 × 108 CFU/day; four subjects), or a DSM 17938 high dose (6.5 × 1010 CFU/day; five subjects). The study was double blind with neither the subjects nor the principal investigator aware of the contents of the study product in each group.
Subjects were then asked to take a study product each day for 28 days. The study product (in sealed one-dose sachets) consisted of freeze-dried powder of the strain mixed with 1.5 g maltodextrin. The placebo contained only maltodextrin and was identical in appearance to the active products but had no detectable bacteria. Sachets were kept refrigerated at all times up to the point of consumption, and L. reuteri levels remained stable in parallel stability testing (data not shown). The subjects were instructed in writing to open the sachet immediately before consumption, pour the powder into a glass containing cold water (at least 100 ml), stir well for 15 s, and swallow directly.
Fecal samples (≥5 g) were collected at baseline and on days 7, 14, and 28 and also on days 42 and 56 (after a 2- to 4-week washout period). The samples were stored for maximally 1 h at room temperature and maximally 1 day in the refrigerator (not frozen) until they could be collected and delivered to the laboratory, where they were frozen (−20°C) upon arrival. For analysis of L. reuteri content, samples were thawed, weighed, diluted in MRS broth (1:10), and mixed thoroughly. Aliquots (1 ml) were serially diluted and spread on MRS-vancomycin agar plates (50 μg ml−1 vancomycin) and incubated anaerobically at 37°C for 48 to 72 h. L. reuteri colonies were confirmed by analysis of reuterin production with the overlay assay described above. Overlay-positive clones were picked from copied plates and analyzed for specific plasmid content to identify them as L. reuteri ATCC 55730-like or L. reuteri DSM 17938 strains.
Fasting blood samples were collected at baseline and after 28 days and analyzed by standard procedures at the Clinical Chemistry Laboratory of Lund University Hospital for Fe, total iron-binding capacity, hemoglobin, erythrocyte particle concentration (erythrocytes), blood corpuscle volume, white blood cell count, blood differential cell counts, low-density lipoprotein cholesterol, high-density lipoprotein cholesterol, triglycerides, albumin, glucose, calcium, sodium, potassium, phosphorus, total bilirubin, alanine aminotransferase, aspartate aminotransferase, glutamyl transferase, alkaline phosphatase, creatinine, urea, urate, and C-reactive protein, and even in the absence of overt fever or illness, a general bacterial analysis was done to detect the possible translocation of any bacteria into the blood (only on day 28 after last study product dose was taken).
RESULTS
Identification of plasmids and resistance genes.
Analysis of the genome sequence of ampicillin-resistant strain L. reuteri ATCC 55730 identified no known β-lactam resistance gene, but five genes encoding putative Pbp were found (Table 3). The corresponding genes from two other penicillin-resistant and three penicillin-sensitive L. reuteri strains were PCR amplified and sequenced for comparison. Alignment of the deduced protein sequences showed few differences, but there were amino acid differences at four positions that correlated with the expression of penicillin resistance. Two of those were located in Pbp1a, where aspartic acid (Asp, D) in the sensitive strains was changed to valine (Val, V) at position 399 of the resistant strains (Asp399Val, exchange 1 [E1]) and glutamine (Gln, Q) was changed to leucine (Leu, L) at position 479 (Gln479Leu, E2), as shown in Fig. 1. E3 was located in Pbp2a, where phenylalanine (Phe, F) was changed to valine (Val, V) at position 147 (Phe147Val), and the last exchange, E4, was located in Pbp2x, where alanine (Ala, A) was changed to threonine (Thr, T) at position 526 (Ala526Thr). Comparisons with Pbp from 10 other Lactobacillus species showed that both E1 and E4 are located in conserved regions (Fig. 1). E1 leads to the substitution of the fully conserved aspartic acid (charged) to valine, which is an amino acid with fundamentally different properties (hydrophobic). In the case of E4, the alanine present in the sensitive strains has properties similar to those of glycine (small), which is present in all other lactobacilli, whereas the threonine present in the resistant strains has other properties (contains a hydroxyl group). Although E2 and E3 are not located in conserved regions, these substitutions also produce changed properties. E1 to E4 can be explained by point mutations of the corresponding genes. E1 is caused by a change of a GAT codon to GTT; E2, CAA to CTA; E3, TTC to GTC; and E4, GCA to ACA. The Pbp2a- and PbpX-encoding genes did not show any differences that correlated with the resistance patterns of the strains examined.
TABLE 3.
GenBank accession numbers of Pbp compared in this study
| Strain | GenBank accession no.
|
||||
|---|---|---|---|---|---|
| Pbp1a | Pbp2b | Pbp2a | Pbp2x | PbpX | |
| ATCC 55730a,d | DQ074831 | DQ074857 | DQ074867 | DQ074915 | DQ074930 |
| ATCC 55149b,d | EU604291 | EU604292 | EU604293 | EU604294 | EU604295 |
| CF48-3Ab,d | EU604286 | EU604287 | EU604288 | EU604289 | EU604290 |
| DSM 20015b | EU604281 | EU604282 | EU604283 | EU604284 | EU604285 |
| ATCC 55148b | EU604276 | EU604277 | EU604278 | EU604279 | EU604280 |
| DSM 20016c | ABQ83182 | ABQ83468 | ABQ83526 | ABQ82854 | ABQ84156 |
FIG. 1.
Alignment of Pbp1a, Pbp2a, and Pbp2x from penicillin-sensitive and -resistant L. reuteri strains and the consensus sequences of Pbp from lactobacilli available in GenBank (10 species). Bold characters represent positions where the L. reuteri sequences are identical or have an amino acid residue with a function similar to that of the consensus sequence. Stars below the consensus sequence show positions where all Lactobacillus sequences are identical, and dots show positions where all have similar functions. The positions where resistant strains differs from sensitive strains (E1 to E4) are marked with gray and underlined characters, and the two substitutions (E1 and E4) that are located in conserved regions and lead to shifts in function are marked with a black background.
The genome sequence of L. reuteri ATCC 55730 showed four contigs that were found to harbor plasmid-related genes. Further, these contigs were built up by an elevated number of sequencing runs, indicating a higher copy number than genes located on the chromosome. The contigs could be circularized and therefore concluded to be plasmids, and the sizes were found to be 8.1 (pLR580), 12.2 (pLR581), 14.2 (pLR585), and 19.1 (pLR584) kb. The genome of ATCC 55730 was also scanned for the tet(W) and lnu(A) genes, which have previously been detected in this strain (24), and they were found as open reading frames lr1996, located on plasmid pLR581, and lr2105, located on plasmid pLR585, respectively (Table 4).
TABLE 4.
| Plasmid and gene locus | Accession no. | Function |
|---|---|---|
| pLR581 | ||
| lr1991 | EF421931 | Putative replication protein |
| lr1992 | EF421932 | Putative replication protein |
| lr1993 | DQ219980 | Transcriptional regulator |
| lr1994 | EF421933 | Phage-related protein |
| lr1995 | EF421934 | DNA integrase/recombinase |
| lr1996 | EF421935 | Tetracycline resistance protein W [tet(W)] |
| lr1997 | DQ074950 | Truncated extracellular protein |
| lr1998 | EU621694 | Arsenite efflux pump ACR3, truncated |
| lr1999 | DQ857855 | Transcriptional regulator, ArsR |
| lr2000 | EU621695 | Arsenate reductase, truncated |
| lr2001 | EU621696 | Arsenite efflux pump ACR3 |
| lr2002 | DQ857856 | Transcriptional regulator, ArsR |
| lr2003 | EU621697 | Plasmid-associated protein |
| lr2004 | DQ857857 | Replication initiator protein |
| pLR585 | ||
| lr2094 | ACC61202 | Recombinase |
| lr2095 | ACC61203 | Transcriptional regulator |
| lr2096 | DQ857862 | Polyketide antibiotic exporter |
| lr2097 | DQ857863 | Polyketide transporter, ATPase component |
| lr2098 | ACC61204 | DNA recombinase |
| lr2099 | ACC61205 | Plasmid-associated protein |
| lr2100 | ACC61206 | Plasmid-associated protein |
| lr2101 | EF421942 | Putative replication protein |
| lr2102 | EF421943 | Putative replication protein |
| lr2103 | DQ219991 | Putative transcriptional regulator |
| lr2104 | ACC61207 | Putative protease |
| lr2105 | ACC61208 | Lincomycin resistance protein [lnu(A)] |
| lr2106 | ACC61209 | Truncated mobilization protein |
| lr2107 | ACC61210 | Transposase, IS30 family |
Accession number EU583804.
Accession number EU596446.
Removal of plasmids pLR581 and pLR585 from L. reuteri ATCC 55730.
An attempt to cure L. reuteri ATCC 55730 of plasmids pLR581, harboring tet(W), and pLR585, harboring lnu(A), was made. Different methods were tested (data not shown), and a method by which the bacteria first were subjected to protoplast formation with subsequent cell wall regeneration was found to be effective in removing the plasmids from the bacteria. In a first trial, pLR581 was the target. After incubation in the protoplast buffer, 100-fold more colonies were obtained on MRS plates with sucrose than on those without sucrose, indicating efficient formation of protoplasts. Two hundred colonies from the sucrose plates were examined for growth or nongrowth on MRS and MRS-Tet agar. Seven nongrowing colonies were replated and grew on MRS-Tet, indicating that these colonies were false candidates. After 24 h of further incubation of the sucrose plates, a few more colonies appeared. Twelve such colonies were picked to MRS and MRS-Tet agar and grown overnight. One of these colonies grew well on the MRS plate but not at all on the MRS-Tet plate. This colony was replated, stored at −70°C, and designated L. reuteri ATCC 55730Tets.
L. reuteri ATCC 55730Tets was then again subjected to protoplast formation in order to obtain a strain cured of pLR585. One hundred colonies from the sucrose plates were examined for growth or nongrowth on MRS and MRS-Lin agar. One colony was found to grow well on the MRS plate but not at all on the MRS plate with lincomycin. This colony was replated, stored at −70°C, and designated L. reuteri ATCC 55730TetsLins.
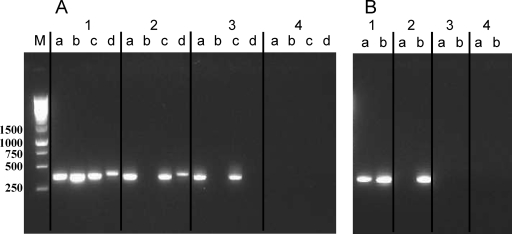
Confirmation of the loss of plasmids pLR581 and pLR585 was performed by PCR. In ATCC 55730Tets, lr2004 (gene encoding a replication protein on pLR581) and lr1996 [tet(W)] were both missing while the other ATCC 55730 plasmids and gene lr2105 [lnu(A)] could still be detected (Fig. 2A and B, lanes 2). Both lr2004 and lr2102 (gene encoding a replication protein on pLR585) were lacking in L. reuteri ATCC 55730TetsLins, whereas the genes representing the other two plasmids were still present (Fig. 2A, lane 3). Both tet(W) and lnu(A) was absent in ATCC 55730TetsLins (Fig. 2B, lane 3). This clearly showed that L. reuteri ATCC 55730 had been cured of the tet(W)- and lnu(A)-containing plasmids. The double-cured ATCC 55730TetsLins strain was later deposited at DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) as L. reuteri DSM 17938 (L. reuteri Protectis), and ATCC 55730Tets was deposited as L. reuteri DSM 17686. The genes of the removed plasmids and the putative functions of the encoded proteins are listed in Table 4.
FIG. 2.
(A) Detection of plasmids by PCR. Strains were analyzed for the presence of plasmids as follows: a, pLR580; b, pLR581; c, pLR584; d, pLR585. Lanes: 1, L. reuteri ATCC 55730; 2, L. reuteri ATCC 55730Tets; 3, L. reuteri DSM 17938; 4, L. reuteri DSM 20016 (negative control). (B) Detection of tet(W) (a) and lnu(A) (b) by PCR. Lanes 1 to 4 are as in Fig. 1A. Lane M, molecular size markers (sizes are shown in base pairs on the left).
Comparative studies on L. reuteri ATCC 55730 and DSM 17938.
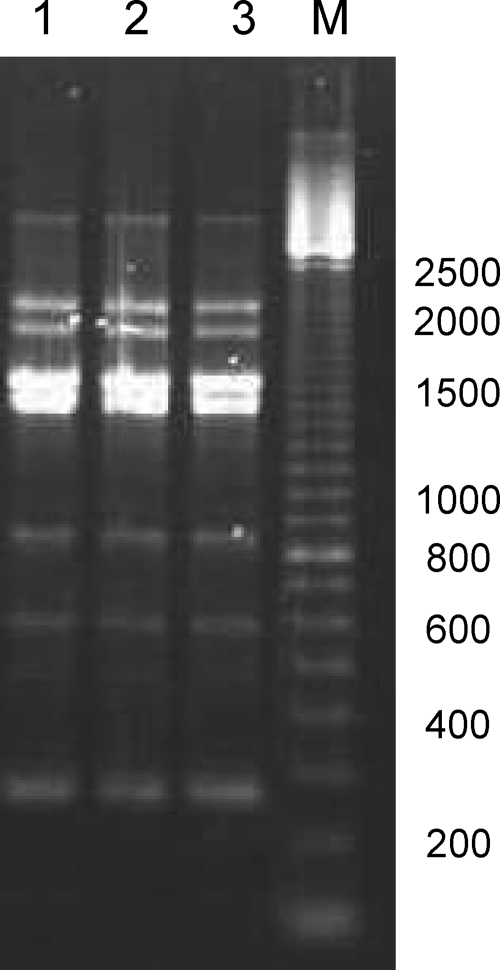
The rep-PCR patterns obtained from strains ATCC 55730 and DSM 17938 were not possible to distinguish (Fig. 3), which shows not only that DSM 17938 is a true variant of ATCC 55730 and not a contaminant but also that removal of pLR581 and pLR585 did not affect the rep-PCR fingerprint.
FIG. 3.
(GTG)5-PCR (rep-PCR)-generated genomic fingerprints of L. reuteri DSM 17938 (lane 1), L. reuteri ATCC 55730Tets (lane 2), and L. reuteri ATCC 55730 (lane 3). Lane M, molecular size markers (sizes are shown in base pairs on the right).
The MICs of tetracycline for the DSM 17938 and ATCC 55730Tets variants were found to be 12 to 16 μg ml−1 (Table 5), showing the natural intrinsic sensitivity of this species to tetracycline (18), compared to >256 μg ml−1 for ATCC 55730. This shows that the tet(W) gene is responsible for the unusually high tetracycline resistance of ATCC 55730. Strains ATCC 55730 and ATCC 55730Tets were both found to be resistant to lincomycin but not to clindamycin (a derivative of lincomycin). Removal of pLR585 resulted in a decrease in the lincomycin MIC for DSM 17938 from >16 to 0.25 μg ml−1, while the sensitivity to clindamycin was unchanged. This confirms that the lincomycin resistance of ATCC 55730 was mediated by the pLR585 plasmid-borne lnu(A) gene. All other intrinsic resistances described for ATCC 55730 (18, 24) were unchanged in DSM 17938 (data not shown).
TABLE 5.
MICs of tetracycline, lincomycin, and clindamycin for L. reuteri strains
| Strain | MIC (μg ml−1)
|
||
|---|---|---|---|
| Tetracycline | Lincomycin | Clindamycin | |
| ATCC 55730 | >256 | >16 | <0.125 |
| ATCC 55730Tets | 12-16 | >16 | <0.125 |
| DSM 17938 | 12-16 | 0.25 | <0.125 |
Fermentation patterns.
The fermentation patterns showed that ATCC 55730 and DSM 17938 fermented l-arabinose, ribose, galactose, glucose, maltose, lactose, melibiose, saccharose, raffinose, and gluconate and were negative for the rest of the substrates. The production of reuterin was of the same magnitude for both strains, and there were no detectable differences in either colony or cell morphology or in mucus binding (data not shown).
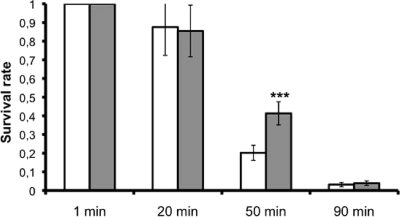
Comparison of the growth of strains DSM 17938 and ATCC 55730 showed no difference in generation time, but DSM 17938 did grow to a significantly higher density than ATCC 55730 (P < 0.01). The final ODs of ATCC 55730 and DSM 17938 were 4.78 ± 0.13 and 6.00 ± 0.26, respectively, after 510 min of growth. Acidic challenge at pH 2.0 showed no difference after 20 and 90 min, but at the interim 50-min point, DSM 17938 survived significantly better than ATCC 55730, with survival rates of 41 and 20%, respectively (Fig. 4). Both ATCC 55730 and DSM 17938 tolerated bile well when they were in stationary phase. In different experiments, 30 to 90% of the cells grew on MRS agar with 6% bile. Survival and growth of both strains were lower in the exponential phase (0.1 to 1% growing cells). However, no difference in bile tolerance could be detected between the strains (data not shown).
FIG. 4.
Acid tolerance of parent strain L. reuteri ATCC 55730 (light bars) and daughter strain L. reuteri DSM 17938 (dark bars). The columns show the proportion of bacteria surviving at pH 2.0 (mean of quadruplicates ± standard deviation). ***, P < 0.001 for 55730 versus 17938 (Student's t test).
Pathogen inhibition.
Strains ATCC 55730 and DSM 17938 were both able inhibit the growth of C. albicans, E. sakazakii, S. enterica serovar Typhimurium, and C. difficile, and there was no difference between the strains in strength of inhibition in the presence or absence of glycerol (Table 6). In the absence of glycerol, neither strain showed any inhibitory effect on C. albicans growth.
TABLE 6.
Pathogen inhibition by L. reuteri strains with and without glycerol in the substrate
| Indicator organism | Inhibition zone ± SD (mm)
|
|||
|---|---|---|---|---|
| ATCC 55730
|
DSM 17938
|
|||
| With glycerol | Without glycerol | With glycerol | Without glycerol | |
| S. enterica serovar Typhimurium | 43.7 ± 2.1 | 40.0 ± 2.0 | 42.7 ± 2.1 | 39.7 ± 2.1 |
| E. sakazakii | 42.0 ± 1.0 | 32.7 ± 1.5 | 41.0 ± 2.0 | 33.0 ± 1.0 |
| C. difficile | 41.0 ± 1.0 | 29.7 ± 0.6 | 40.7 ± 1.5 | 30.0 ± 1.0 |
| C. albicans | 41.0 ± 1.0 | NIa | 42.3 ± 1.5 | NI |
NI, no inhibition.
Clinical trial.
Compliance (participant self-reported) was 99% between days 1 and 14 and 97.5% between days 14 and 28. General health examinations revealed no alterations in weight, pulse, blood pressure, or body temperature in any group during the supplementation period, and blood safety and metabolic parameters were unchanged in any of the groups (data not shown). Blood taken directly after supplementation on day 28 was analyzed in 10 subjects, including 4 subjects from the high-dose 17938 group. All were negative for bacteremia. Thus, supplementation with L. reuteri DSM 17938 daily for 28 days at doses of up to 6.5 × 1010 CFU/day was well tolerated during the trial.
Three of the 16 individuals had detectable L. reuteri in their baseline fecal samples (Table 7). One of these isolates was 55730 like, while the others were unidentifiable L. reuteri strains. Standard ingested doses of L. reuteri ATCC 55730 and DSM 17938 led to similar levels of L. reuteri detection in the feces (Table 7). Increasing the dose of L. reuteri DSM 17938 100-fold led to 100-fold higher levels in the feces (Table 7). The levels detected were maximal at day 7 in all L. reuteri-supplemented groups and remained at this level during the period of supplementation. After a washout period of at least 2 weeks, there was no evidence of L. reuteri in the feces of any of the supplemented subjects (Table 7). Identification of fecal isolates by plasmid analysis obtained from samples taken during supplementation were found to correspond to the DSM 17938 plasmid pattern for all of the subjects ingesting this strain, with the exception of three samples where the strain was either ambiguous or resembled 55730. Isolates from the 55730 samples were all 55730 like, with the exception of two, which were ambiguous. Placebo-supplemented subjects showed no fecal colonization with L. reuteri, except for two observations of unknown L. reuteri in one individual.
TABLE 7.
Fecal L. reuteri levels in subjects given either L. reuteri ATCC 55730 or DSM 17938 for 28 days
| Group (no. of subjects) | Amt of L. reuteria
|
|||||
|---|---|---|---|---|---|---|
| Day 0 | Day 7 | Day 14 | Day 28 | Day 42 | Day 56 | |
| 17938 high (5) | <2b | 7.14 (4) | 6.23 (5) | 7.26 (4) | <2 | <2 |
| 17938 standard (4) | <2b | 4.92 (3) | 4.15 (4) | 5.19 (2) | <2 | <2b |
| 55730 (3) | <2 | 4.91 (3) | 4.26 (3) | 4.40 (1) | <2 | <2 |
| Placebo (4) | <2b | <2 | <2b | <2 | <2 | <2 |
Log10 CFU g−1 (wet weight) feces. Values are the mean of the samples in which L. reuteri was detected (the number of samples is in parentheses). Variation between samples was large but similar for all data points.
L. reuteri was detected in one of the subjects at 3.7 to 6.9 log10 CFU g−1.
DISCUSSION
β-Lactam resistance is conferred mainly by two mechanisms: drug inactivation by β-lactamases and target site (i.e., Pbp) alterations. For both types, transferable resistance determinants have been identified, e.g., ampC and mecA, respectively (21, 30). When the nature of the β-lactam resistance of L. reuteri ATCC 55730 was investigated, point mutations in Pbp1a, Pbp2a, and Pbp2x were identified. Alterations in both Pbp1a and Pbp2x are often reported to cause resistance to β-lactams in streptococci, and high-level resistance is often caused by alterations in more than one of the proteins (14, 22). The Pbp are “housekeeping” proteins found in all bacteria. In L. reuteri ATCC 55730, the corresponding genes are all located on the chromosome and are not coupled to any mechanisms for transfer to other bacteria. In addition, there is no other gene present in the genome sequence with similarities to known penicillin resistance genes. We therefore suggest that the β-lactam resistance of L. reuteri ATCC 55730 is caused by a number of point mutations in the genes encoding Pbp1a, Pbp2a, and/or Pbp2x and that this resistance can be regarded as nontransferable.
The ribosome protection-type resistance gene tet(W) (5, 35) is one of the most widespread tetracycline resistance genes in environmental samples (4, 44). Lincosamide nucleotidyltransferases encoded by lnu (formerly lin) genes inactivate lincosamides by adenylation (9, 11), and the gene has been shown to occur on plasmids (29). Lincosamide antibiotics include lincomycin and clindamycin, a semisynthetic derivative of lincomycin (10). Coupled resistances to lincomycin and clindamycin seem to be associated with the lnu(F) gene but not with the lnu(A) gene, which only confers lincomycin resistance (2). Our finding that L. reuteri ATCC 55730 is not resistant to clindamycin is in accordance with others (18, 26) but not with the result of Kastner et al. (24). The reason for the difference is not known, but the method validation of Egervärn et al. (19) in accordance with the recent ACE-ART project in Europe indicates the reliability of this study. The resistance to lincomycin in L. reuteri ATCC 55730 is caused by lnu(A), located on pLR585, but both L. reuteri ATCC 55730 and L. reuteri DSM 17938 are sensitive to clindamycin.
Our data clearly indicate that L. reuteri ATCC 55730 harbors four plasmids and that two of them harbored the tet(W) and lnu(A) genes, respectively. The total number of plasmids is not in agreement with the report of Klein et al. (26), who concluded that the strain harbors six plasmids with sizes in the range of 5 to 24 MDa, which corresponds to approximately 7.7 to 37 kb. The difference is probably due to the methods used. Klein et al. detected the plasmids by agarose gel electrophoresis, which can give an overestimation of the number of plasmids since plasmids can be present in different forms and can thus give rise to several bands. Different curing techniques have been used on Lactobacillus species, including chemical curing with agents like novobiocin, sodium dodecyl sulfate, acridine dyes, and ethidium bromide or using high temperature or protoplast formation (12, 28, 32, 33, 36, 42, 43). We have tested all of these methods, and protoplast formation was the only one that succeeded in curing ATCC 55730 of the plasmids. Also, protoplast formation was considered to be an appropriate method since it does not involve genetically modifying the organism and does not induce mutations in the bacterial chromosome that might be a consequence of the use of other curing agents.
Besides the tet(W) gene, eliminated plasmid pLR581 harbors 13 other genes. Most of them fall into two categories: replication protein-encoding genes and arsenic resistance genes (several of the latter are truncated). The pLR585 plasmid also harbors 13 genes besides lnu(A), 2 of which encode polyketide antibiotic exporters (lr2096 and lr2097). Importantly, none of the genes on the plasmids have any connection or function of importance for the known probiotic characteristics of this strain.
L. reuteri DSM 17938 containing only two plasmids (pLR580 and pLR584) has the same rep-PCR profile, fermentation pattern, reuterin production, morphology, growth rate, adhesion to mucus, pathogen inhibition profile, and bile tolerance as L. reuteri ATCC 55730. The abilities of the daughter strain to grow to a higher density and to survive better under acidic conditions are probably related to a decreased burden with respect to DNA replication and thereby a higher competitive ability compared to the parent strain. L. reuteri ATCC 55730 has earlier been shown to tolerate passage through the human gastrointestinal tract, as shown by fecal shedding, and attaches and grows in the stomach, duodenum, and ileum of subjects ingesting the strain (20, 39). Laboratory analysis of both parent strain L. reuteri ATCC 55730 and daughter strain L. reuteri DSM 17938 showed at least similar acid and bile tolerance and mucus binding, predicting that DSM 17938 should not have a changed ability to colonize the entire human gastrointestinal tract compared to ATCC 55730. Fecal analysis after ingestion confirmed this and shows that L. reuteri DSM 17938 survives throughout the human gastrointestinal tract in the same way as L. reuteri ATCC 55730 and further that colonization is only temporary, as is common in probiotic strains. L. reuteri DSM 17938 could be unequivocally identified by the plasmid pattern, which showed a direct correlation between the ingested and shed strains. The blood safety parameters studied demonstrate that L. reuteri DSM 17938 has a safety profile similar to that of the L. reuteri ATCC 55730 strain. Furthermore, an analysis of the L. reuteri DSM 17938 genome annotation did not reveal any further gene or gene cluster known to be involved in virulence or antibiotic resistance (not shown).
In conclusion, we have demonstrated that L. reuteri ATCC 55730 could be cured of two independent plasmids carrying unwanted antibiotic resistance traits and that the resultant L. reuteri DSM 17938 strain did not lose any of its probiotic characteristics. This may be a valuable alternative for other true probiotic strains that are found to harbor plasmid-borne resistances.
Acknowledgments
We thank Noris Carbajal and Uma Nathan for performing the pathogen inhibition assays, Dan Nilsson for fecal analyses, and Tor Melin and Karin Diderot for coordination and assistance in the performance of the clinical trial.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Abrahamsson, T. R., T. Jakobsson, M. F. Böttcher, M. Fredrikson, M. C. Jenmalm, B. Björkstén, and G. Oldaeus. 2007. Probiotics in prevention of IgE-associated eczema: a double blind randomised placebo-controlled trial. J. Allergy Clin. Immunol. 119:1174-1180. [DOI] [PubMed] [Google Scholar]
- 2.Achard, A., C. Villers, V. Pichereau, and R. Leclercq. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 49:2716-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurrence of tet(O) in ruminal bacteria. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 6.Barlow, S., A. Chesson, J. D. Collins, E. Dybing, A. Flynn, C. Fruijtier-Pölloth, A. Hardy, A. Knaap, H. Kuiper, P. Le Neindre, J. Schans, J. Schlatter, V. Silano, S. Skerfving, and P. Vannier. 2007. Introduction of a qualified presumption of safety (QPS) approach for assessment of selected microorganisms referred to EFSA. Opinion of the Scientific Committee. Eur. Food Saf. Authority J. 587:1-16. [Google Scholar]
- 7.Båth, K., S. Roos, T. Wall, and H. Jonsson. 2005. The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253:75-82. [DOI] [PubMed] [Google Scholar]
- 8.Borriello, S. P., W. P. Hammes, W. Holzapfel, P. Marteau, J. Schrezenmeir, M. Vaara, and V. Valtonen. 2003. Safety of probiotics that contain lactobacilli or bifidobacteria. Clin. Infect. Dis. 36:775-780. [DOI] [PubMed] [Google Scholar]
- 9.Bozdogan, B., L. Berrezouga, M. Kuo, D. Yurek, K. Farley, B. Stockman, and R. Leclercq. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brisson-Noël, A., and P. Courvalin. 1986. Nucleotide sequence of gene linA encoding resistance to lincosamide in Staphylococcus haemolyticus. Gene 43:247-253. [DOI] [PubMed] [Google Scholar]
- 11.Brisson-Noël, A., P. Delrieu, D. Samain, and P. Courvalin. 1988. Inactivation of lincosamide O-nucleotidyltransferases and comparison of the corresponding resistance gene. J. Biol. Chem. 263:15880-15887. [PubMed] [Google Scholar]
- 12.Chin, S. C., N. Abdullah, T. W. Siang, and H. Y. Wan. 2005. Plasmid profiling and curing of Lactobacillus strains isolated from the gastrointestinal tract of chicken. J. Microbiol. 43:251-256. [PubMed] [Google Scholar]
- 13.Chung, T. C., L. Axelsson, S. E. Lindgren, and W. J. Dobrogosz. 1989. In vitro studies on reuterin synthesis by Lactobacillus reuteri. Microb. Ecol. Health Dis. 2:137-144. [Google Scholar]
- 14.Coffey, T. J., C. G. Dowson, M. Daniels, and B. G. Spratt. 1995. Genetics and molecular biology of beta-lactam-resistant pneumococci. Microb. Drug. Resist. 1:29-34. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 16.Courvalin, P. 2006. Antibiotic resistance: the pros and cons of probiotics. Dig. Liver Dis. Suppl. 2:S261-S265. [DOI] [PubMed] [Google Scholar]
- 17.Curragh, H. J., and M. A. Collins. 1992. High levels of spontaneous drug resistance in Lactobacillus. J. Appl. Bacteriol. 73:31-36. [Google Scholar]
- 18.Egervärn, M., M. Danielsen, S. Roos, H. Lindmark, and S. Lindgren. 2007. Antibiotic susceptibility profiles of Lactobacillus reuteri and Lactobacillus fermentum. J. Food Protect. 70:412-418. [DOI] [PubMed] [Google Scholar]
- 19.Egervärn, M., H. Lindmark, S. Roos, G. Huys, and S. Lindgren. 2007. Effects of inoculum size and incubation time on broth microdilution susceptibility testing of lactic acid bacteria. Antimicrob. Agents Chemother. 51:394-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glintborg, V., S. Dawids, J. Preuss Hasselby, H. Winther Nielsen, and A. Mertz Nielsen. 2006. Long-term administration of Lactobacillus reuteri (ATCC55730) has no influence on gastric mucosal inflammation and colonization of Helicobacter pylori in humans. A pilot study. Int. J. Probiotics Prebiotics 1:225-232. [Google Scholar]
- 21.Hiramatsu, K., Y. Katayama, H. Yuzawa, and T. Ito. 2002. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 292:67-74. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu, K., M. Ohama, Y. Mijajima, K. Kishi, S. Mizunoe, I. Tokimatsu, H. Nagai, J. Kadota, T. Saikawa, and M. Nasu. 2004. Antimicrobial susceptibilities and analysis of genes related to penicillin or macrolide resistance in Streptococcus pneumoniae. Int. J. Antimicrob. Agents 24:125-129. [DOI] [PubMed] [Google Scholar]
- 23.Indrio, F., G. Riezzo, F. Raimondi, M. Bisceglia, L. Cavallo, and R. Francavilla. 2008. The effects of probiotics on feeding tolerance, bowel habits, and gastrointestinal motility in preterm newborns. J. Pediatr. 152:801-806. [DOI] [PubMed] [Google Scholar]
- 24.Kastner, S., V. Perreten, H. Bleuler, G. Hugenschmidt, C. Lacroix, and L. Miele. 2006. Antibiotic susceptibility patterns and resistance genes of starter cultures and probiotic bacteria used in food. Syst. Appl. Microbiol. 29:145-155. [DOI] [PubMed] [Google Scholar]
- 25.Klare, I., C. Konstabel, G. Werner, G. Huys, V. Vankerckhoven, G. Kahlmeter, B. Hilderbrandt, S. Müller-Bertling, W. Witte, and H. Goossens. 2007. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 59:900-912. [DOI] [PubMed] [Google Scholar]
- 26.Klein, G., C. Hallmann, I. A. Casas, J. Abad, J. Louwers, and G. Reuter. 2000. Exclusion of vanA, vanB and vanC type glycopeptide resistance in strains of Lactobacillus reuteri and Lactobacillus rhamnosus used as probiotics by polymerase chain reaction and hybridization methods. J. Appl. Microbiol. 89:815-824. [DOI] [PubMed] [Google Scholar]
- 27.Lionetti, E., V. L. Miniello, S. P. Castellaneta, A. DeCanio, A. M. Magista, A. DeCanio, M. Giovanni, F. Ierardi, L. Cavallo, and R. Francavilla. 2006. Lactobacillus reuteri therapy to reduce side-effects during anti-Helicobacter pylori treatment in children: a randomised placebo controlled trial. Aliment. Pharmacol. Ther. 24:1461-1468. [DOI] [PubMed] [Google Scholar]
- 28.Liu, M. L., J. K. Kondo, M. B. Barnes, and D. T. Bartholomew. 1988. Plasmid-linked maltose utilization in Lactobacillus ssp. Biochimie 70:351-355. [DOI] [PubMed] [Google Scholar]
- 29.Perreten, V., N. Giampa, U. Schuler-Schmid, and M. Teuber. 1998. Antibiotic resistance genes in coagulase-negative staphylococci isolated from food. Syst. Appl. Microbiol. 21:113-120. [DOI] [PubMed] [Google Scholar]
- 30.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos, S., F. Karner, L. Axelsson, and H. Jonsson. 2000. Lactobacillus mucosae sp. nov., a new species with in vitro mucus-binding activity isolated from pig intestine. Int. J. Syst. Evol. Microbiol. 50:251-258. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Barba, J. L., J. C. Piard, and R. Jimenez-Diaz. 1991. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J. Appl. Bacteriol. 71:417-421. [DOI] [PubMed] [Google Scholar]
- 33.Sarra, P. G., M. Vescovo, and M. Fulgoni. 1986. Study on crop adhesion genetic determinant in Lactobacillus reuteri. Microbiologica 9:279-285. [PubMed] [Google Scholar]
- 34.Savino, F., E. Pelle, E. Palumeri, R. Oggero, and R. Miniero. 2007. Lactobacillus reuteri (American Type Culture Collection strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics 119:124-130. [DOI] [PubMed] [Google Scholar]
- 35.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, O., and S. Ohmomo. 2001. Efficient protoplast regeneration for some homofermentative lactobacilli and pediococci. Arch. Microbiol. 177:36-40. [DOI] [PubMed] [Google Scholar]
- 37.Teuber, M. 1999. Spread of antibiotic resistance with food-borne pathogens. Cell. Mol. Life Sci. 56:755-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 39.Valeur, N., P. Engel, N. Carbajal, E. Connolly, and K. Ladefoged. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 70:1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vankerckhoven, V., G. Huys, M. Vancanneyt, C. Vael, I. Klare, M.-B. Romond, J. M. Entenza, P. Moreillon, R. D. Wind, J. Knol, E. Wiertz, B. Pot, E. E. Vaughan, G. Kahlmeter, and H. Goossens. 2008. Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci. Technol. 19:102-114. [Google Scholar]
- 41.Versalovic, J., M. Schneider, F. de Bruijn, and J. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 42.Vescovo, M., L. Morelli, and V. Bottazzi. 1982. Drug resistance plasmids in Lactobacillus acidophilus and Lactobacillus reuteri. Appl. Environ. Microbiol. 43:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vescovo, M., L. Morelli, P. S. Cocconcelli, and V. Bottazzi. 1984. Protoplast formation, regeneration and plasmid curing in Lactobacillus reuteri. FEMS Microbiol. Lett. 23:333-334. [Google Scholar]
- 44.Villedieu, A., M. L. az-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wall, T., K. Båth, R. A. Britton, H. Jonsson, J. Versalovic, and S. Roos. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 73:3924-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weizman, Z., G. Asli, and A. Alsheikh. 2005. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics 115:5-9. [DOI] [PubMed] [Google Scholar]
- 47.Wolf, B. W., K. Garleb, D. Ataya, and I. A. Casas. 1995. Safety and tolerance of Lactobacillus reuteri in healthy adults male subjects. Microb. Ecol. Health Dis. 8:41-50. [Google Scholar]
- 48.Wolf, B. W., K. Wheeler, D. Ataya, and K. Garleb. 1998. Safety and tolerance of Lactobacillus reuteri supplementation to a population infected with the human immunodeficiency virus. Food Chem. Toxicol. 36:1085-1094. [DOI] [PubMed] [Google Scholar]