Abstract
Viruses are known to be the most numerous biological entities in soil; however, little is known about their diversity in this environment. In order to explore the genetic diversity of soil viruses, we isolated viruses by centrifugation and sequential filtration before performing a metagenomic investigation. We adopted multiple-displacement amplification (MDA), an isothermal whole-genome amplification method with φ29 polymerase and random hexamers, to amplify viral DNA and construct clone libraries for metagenome sequencing. By the MDA method, the diversity of both single-stranded DNA (ssDNA) viruses and double-stranded DNA viruses could be investigated at the same time. On the contrary, by eliminating the denaturing step in the MDA reaction, only ssDNA viral diversity could be explored selectively. Irrespective of the denaturing step, more than 60% of the soil metagenome sequences did not show significant hits (E-value criterion, 0.001) with previously reported viral sequences. Those hits that were considered to be significant were also distantly related to known ssDNA viruses (average amino acid similarity, approximately 34%). Phylogenetic analysis showed that replication-related proteins (which were the most frequently detected proteins) related to those of ssDNA viruses obtained from the metagenomic sequences were diverse and novel. Putative circular genome components of ssDNA viruses that are unrelated to known viruses were assembled from the metagenomic sequences. In conclusion, ssDNA viral diversity in soil is more complex than previously thought. Soil is therefore a rich pool of previously unknown ssDNA viruses.
Viral abundance in the environment exceeds that of bacteria. Viruses have a significant influence on microbial communities (7, 39, 44); therefore, the ecology of environmental viral assemblages has been intensely investigated. Most of the research on viral diversity has been conducted in the aquatic environment (3, 6-8, 10, 12, 13, 39, 43, 44), and research on soil viral assemblages is limited compared to that on those found in aquatic environments (4, 18, 25, 49-51). Previous studies have revealed that viruses are the most numerous biological entities present in soil. Indeed, the viral abundance in soil and rhizosphere was previously estimated to be 1.5 × 108 g−1 (dry weight) by transmission electron microscopy (18). Virus-like particle densities were estimated to be 8.7 × 108 to 4.17 × 109 g−1 (dry weight) and 3.4 to 4.6 times more than bacteria in six Delaware soils (50); the majority of these viruses were bacteriophages, including Sipho-, Myo-, and Podoviridae (50).
Molecular analysis is essential to investigate the diversity of viral assemblages because the majority of viruses are uncultured due to a lack of suitable hosts, such as bacteria. Indeed, the cultivation of viruses that infect eukaryotes is not easy. A PCR-based approach is also not appropriate because there are no universally conserved genes or markers for viruses like the 16S rRNA gene for bacteria (17). Whole viral assemblage genome sequencing (viral metagenomics) recently overcame these limitations and became a promising method by which to investigate uncultured viral diversity (9, 10). By this approach, viruses are purified and concentrated by sequential filtrations and ultracentrifugation and whole viral genomes are extracted, amplified, and sequenced by shotgun cloning or pyrosequencing (3, 17). The advantages of sequence-independent amplification and metagenome sequencing for characterizing novel viruses are that they are simple, fast, and without bias toward any particular viral group (16). The diversity of DNA viral assemblages has been analyzed by viral metagenomics in near-shore seawater (10), marine sediment (8), human feces (9), equine feces (11), and recently soils (18).
In most previous viral metagenomic studies, linker-amplified shotgun library (LASL) techniques were used to amplify small amounts of viral DNA via PCR amplification of linker-ligated short sheared viral DNA (http://www.sci.sdsu.edu/PHAGE/LASL/index.html). Since the linkers can only be ligated to double-stranded DNA (dsDNA) and not to single-stranded DNA (ssDNA), the diversity of ssDNA viruses could not be revealed by the LASL technique (17). Recently, Angly et al. investigated marine DNA viral metagenomes from four oceanic regions by multiple-displacement amplification (MDA) and pyrosequencing (3). They could acquire metagenomic sequences from both dsDNA and ssDNA viruses by using MDA as an amplification method (3). MDA is the most widely used whole-genome amplification (WGA) method. This technique uses φ29 DNA polymerase and random hexamers (14, 28), and subnanogram quantities to 100 ng of DNA can be amplified up to ∼80 μg with relatively minimal amplification bias compared to the other WGA methods available to date (28). In contrast to PCR, MDA is an isothermal amplification method which requires the template to be denatured with heat or chemicals prior to amplification (15). If the denaturing step is excluded, dsDNA has less of a chance to bind to random hexamers, so that ssDNA is preferably amplified. Additionally, φ29 polymerase amplifies short circular DNA more efficiently than linear DNA via rolling-circle replication (15, 31). Given that most of ssDNA viruses have circular genome components, we hypothesized that ssDNA viral diversity could be selectively examined by MDA without the denaturing step. In this study, we sequenced libraries that were constructed from viral DNA amplified by MDA and found that ssDNA viral diversity could be selectively investigated by MDA without the denaturing step. This technique allowed us to demonstrate that rice paddy soil contains a diverse population of uncultured ssDNA viruses. As far as we know, this is the first report to selectively reveal the diversity of ssDNA viruses in an environment.
MATERIALS AND METHODS
Sample collection.
Soil was collected from a rice paddy field (at latitude 36°23′25‴N and longitude 127°20′35‴E) in Daejeon, Korea, in June 2007. More than 1.5 kg of soil was scraped with a clean trowel from the surface of soil (0 to 10 cm) that was submerged in water after rice planting. Subsamples from several different sites (0 to 30 cm apart) were mixed and transferred immediately to the laboratory. Sample water was removed gravimetrically before plant debris and stones were detached.
Concentration of viruses in rice paddy soil.
Viruses were extracted from soil samples with potassium citrate buffer according to previously described methods (50). Wet soil (500 g) was mixed in 1 liter of 1% potassium citrate buffer (10 g potassium citrate, 1.92 g Na2HPO4 · 12H2O, and 0.24 g KH2PO4 per liter, pH 7). Viruses were extracted from the soil particles by sonication (three times for 1 min each time at 300 W) plus 30 s of manual shaking. The suspension was centrifuged at 7,000 rpm for 10 min. The supernatant was transferred to fresh bottles and centrifuged at 7,000 rpm for 15 min before being sequentially filtered through a 0.45-μm filter and then through a 0.22-μm filter. The filtrate was concentrated with a 100-kDa polyethersulfone tangential-flow filter cartridge (Pellicon XL Filter; Millipore, Molsheim, France) equipped with the Labscale tangential-flow filter system (Millipore, Molsheim, France). The supernatant was concentrated from 850 to 60 ml. The concentrated viral suspension was filtered with a 0.22-μm syringe filter three times. The filtrate was treated with DNase I (final concentration, 20 U/ml) at 37°C for 30 min. An aliquot not treated with DNase I was used as a negative control. Viruses were stained with Sybr gold (Molecular Probes, Inc., Eugene, OR) for quantitative analysis by epifluorescence microscopy (EFM) as previously reported (37). Seven images taken by EFM were used to enumerate the viruses.
DNA extraction and WGA.
DNA was extracted with proteinase K and phenol-chloroform/isoamyl alcohol from 10 ml of the concentrated viral suspension as described previously (41). Viral DNA was amplified with the Genomiphi kit (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. Briefly, 1 μl DNA (6.5 ng/μl) was used in each 40-μl reaction volume. One microliter of viral DNA was mixed with 19 μl of sample buffer. In order to compare the effect of DNA denaturation by heating, one sample was heated at 95°C for 3 min (designated RH) and the other sample was placed in ice for 3 min without heating (designated RX) before 18 μl reaction buffer and 2 μl φ29 DNA polymerase were added. More than 10 μg of DNA was acquired after amplification at 30°C for 1.5 h. DNA was ethanol precipitated and digested with 5 U/μl S1 nuclease in 1X buffer (Takara, Tokyo, Japan) at 30°C for 1 h. Three aliquots of DNA were amplified by MDA, digested, mixed, and used for cloning. PCR was performed with 8F and 1492R bacterial universal primers with positive and negative controls to check for bacterial DNA contamination.
Cloning and sequencing.
Cloning was performed with 6.25 μg of RH DNA and 8.3 μg of RX DNA. DNA was sheared with a HydroGene machine (speed code 3) plus sonication for 60 s. The size distribution of sheared DNAs was viewed by agarose gel electrophoresis. DNA was ethanol precipitated, phosphorylated, and blunted by blunt kinase of the BKL reagent set (Takara, Tokyo, Japan). DNAs (381 ng of RH DNA and 929 ng of RX DNA) were incubated with pUC118 vector (6 ng) and ligase mixture at 16°C for 3.5 h. Ligated DNA was extracted with phenol and transfected into DH5α competent cells by electroporation. After incubation for 1 h in SOC medium (41), an equal volume of 30% glycerol was added and cultures were stored at −80°C. The insert was amplified by colony PCR after white-blue screening. Amplicons were sequenced with a forward primer, and a total of 396 and 389 sequences were obtained for RH and RX samples, respectively.
Analysis of viral metagenome sequences.
Three different database searches were performed in order to analyze the clone sequences, i.e., TBLASTX and BLASTX analyses against the GenBank database and a TBLASTX analysis against the Phage Sequence Databank (http://scums.sdsu.edu/phage/). We downloaded 510 complete phage and prophage genome sequences from the Phage Sequence Databank and analyzed them by TBLASTX comparison with the standalone BLAST program (2). Sequences containing any hits with an E value of <0.001 in at least one search were considered to be “known” sequences. In order to classify the known sequences into biological and taxonomic groups, we compared all of the results from three searches. Sequences were considered to be viral hits if there were any virus-positive hits within three to five of the best hits from the TBLASTX and BLASTX analyses of the GenBank database. When different searches resulted in conflicting classifications, the results of the TBLAST analysis against the Phage Sequence Databank had priority. Several bacterial hits in the TBLASTX search of GenBank were considered to be viral hits according to the results of the TBLASTX search of the Phage Sequence Databank. The categories of the proteins were determined on the basis of the BLASTX results.
Contig assembly.
Sequence assembly was performed with the SeqMan program (DNAStar, Madison, WI) with a minimum stringency of 98% identity on a sequence with a minimum overlap of 20 bp. This parameter was previously determined during the construction and assembly of the in silico shotgun library to discriminate between even closely related phage genomes (10).
Analysis of replication-related genes.
Partial and full open reading frames (ORFs) containing putative replication-related genes were extracted from the metagenomic sequences that possessed significant hits with replication-related genes with the ORF finder in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Conserved domains were analyzed from these ORFs by the NCBI Conserved Domain Search (33). The ORFs containing Viral_Rep and Gemini_AL1 domains were aligned with reference sequences collected from the protein family (Pfam) database (19). Alignment and tree reconstruction were performed with the Cn3D and CDTree programs from the NCBI Conserved Domain Database (33). The block alignment algorithm of the Cn3D program was used for the alignment of the conserved domains. The parameters for reconstructing the tree with the CDTree program were as follows: alignment usage, normal alignment only; clustering method, neighbor joining; distance matrix, score of aligned residue; scoring matrix, BLOSUM62.
Real-time PCR of frequently detected sequences from unamplified and amplified DNAs.
Two primer sets were designed to target contigs C005 and C112, which represent the most frequently detected contigs of 10 and 16 sequences, respectively. The sizes of the C005 and C112 amplicons were 438 and 380 bp, respectively (Fig. 1A). PCR products from viral DNA were purified and used to make a standard curve. The PCR and real-time PCR conditions were 5 min at 94°C; 33 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and a 10-min final extension at 72°C. Real-time PCR was performed with the DyNAmo HS Sybr green quantitative PCR kit (FINNZYMES, Seoul, Korea) and the Opticon 2 thermal cycler (MJ Research). Unamplified and MDA-amplified viral DNAs were used as templates.
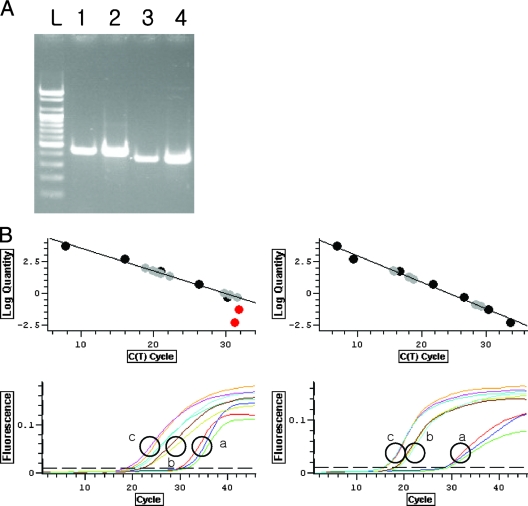
FIG. 1.
(A) Results of real-time PCR with specific primers. (B) Real-time PCR of C005 (left) and C112 (right). (A) Specific primers for C005 (lanes 1 and 2) and C112 (lanes 3 and 4) produced 438- and 380-bp amplicons, respectively. Viral DNA extracted from concentrated virus preparations (lanes 1 and 3) and DNA amplified by MDA without a denaturing step (lanes 2 and 4) were used as templates. Lane L, 100-bp ladder. (B) Circle a, DNA after MDA without the denaturing step (used for the RX library); circle b, DNA after MDA with the denaturing step (RH library); circle c, DNA before MDA. Similar concentrations of DNA (1.4, 1.3, and 1.0 ng/μl for circles a, b, and c, respectively) were used as templates. Nonlinear points indicated by red filled circles in the left-hand standard curve were not included in the standard curve. R2 values are 0.982 (left) and 0.988 (right).
Preparation of circular and linear DNAs.
Circular DNA was prepared by cloning viral DNA into the T&A Cloning Vector (Real Biotech Corp., Taipei, Taiwan). Two plasmids, S11 and S36, containing different inserts (ca. 600 bp in length) were selected and purified from transformed Escherichia coli strains. The plasmids were cut at the same site by a restriction enzyme to prepare linear DNA (Fig. 2A). The Eam1105I restriction enzyme (Takara, Tokyo, Japan) was used to cut the site opposite the insert region in the circular structure to prevent terminal underrepresentation (uneven amplification of terminal sequences during MDA) (24) by real-time PCR. The linear DNA was purified with PCR purification kits (Solgent, Seoul, Korea).
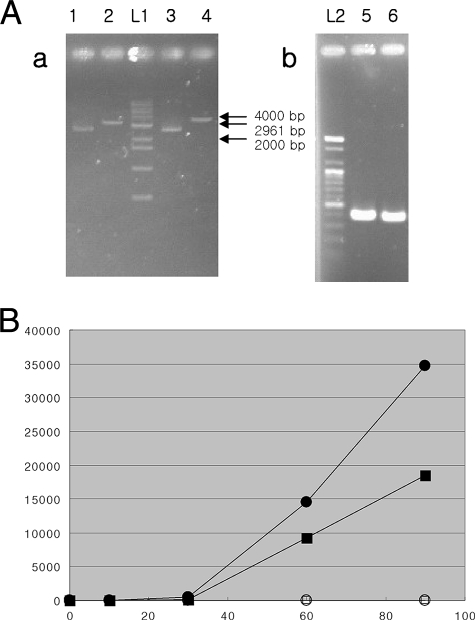
FIG. 2.
(A) Preferential amplification of circular DNA during MDA without the denaturing step. (a) Plasmids left uncut (circular DNA) and cut with the Eam1105I restriction enzyme (linear DNA). Lane 1, S11 and uncut DNA; lane 2, S11 and cut DNA; lane 3, S36 and uncut DNA; lane 4, S36 and cut DNA; lane L1, 1,000-bp ladder. (b) PCR products amplified from plasmids cut with Eam1105I. Specific primers for S11 (lane 5) and S36 (lane 6) produced 391- and 378-bp amplicons, respectively. Lane L1, 100-bp ladder. (B) Quantification of circular and linear DNAs during MDA without the denaturing step. A 0.037-ng/μl concentration of each circular or linear DNA was mixed with 10 ng/μl lambda DNA, and MDA was performed without the heating step. Concentrations of circular and linear DNAs were estimated by real-time PCR. Filled circles, circular S11; filled squares, circular S36; empty circles, linear S11; empty squares, S16. Circular DNA was amplified much more efficiently than linear DNA. x axis, time in minutes; y axis, DNA concentration in picograms per microliter.
Comparison of amplification of circular and linear DNAs during MDA.
Linear and circular DNAs were heated at 95°C for 3 min and cooled on ice immediately to denature dsDNA. Each DNA (0.037 ng/μl) was mixed with 10 ng/μl control lambda DNA and amplified by MDA without a denaturing step for 1.5 h as described above. Small aliquots were collected periodically during the MDA reaction, and the amounts of amplified circular and linear DNAs were measured by real-time PCR. Primer sets were designed for the inserted sequence of each plasmid to produce amplicons of 391 and 378 bp for S11 and S36, respectively. Standard curves were generated with purified PCR products as templates and used to quantify the amount of DNA during MDA. PCR and real-time PCR were performed under the same conditions, as described above.
Nucleotide sequence accession numbers.
Contig and metagenome sequences have been deposited at DDBJ/EMBL/GenBank under the project accession no. ABQX00000000 as a whole-genome shotgun project. The version described in this paper is the first version, ABQX01000000. The accession numbers are ABQX01000001 to ABQX01000093 for contigs and ABQX01000094 to ABQX01000878 for single reads.
RESULTS
Use of MDA for the amplification of ssDNA viruses.
We performed quantitative real-time PCR assays with the primer sets targeting two metagenomic sequences (C005 and C112) derived from ssDNA viruses (Fig. 1). Based on the real-time PCR results, the concentrations of the C005 and C112 amplicons were 0.05% ± 0.02% and 0.008% ± 0.002% of the total viral DNA concentration before MDA, respectively; however, the concentrations were increased by up to 5.2% ± 1.8% (C005) and 4.7% ± 0.23% (C112) of the total amplified DNA concentration after MDA without the denaturing step and by up to 2.8% ± 0.9% (C005) and 1.7% ± 0.3% (C112) after MDA with the denaturing step. Although these values were not considered to be strictly quantitative, they showed that MDA amplified C005 and C112 more selectively by 2 to 3 orders of magnitude. The values are in concordance with results showing that the detected frequencies of C005 and C112 in the RX library among metagenomic sequences are 2.3 and 2.7%.
The amplification efficiencies of circular and linear DNAs during MDA without the denaturing step were compared with those of cut and uncut plasmids containing different inserts. From the results, we found that circular DNA (plasmids denatured by heat; the sizes were ca. 2,700 bp) was amplified much more efficiently than linear DNA during MDA without the denaturing step (Fig. 2).
Abundance and diversity of viruses from rice paddy soil.
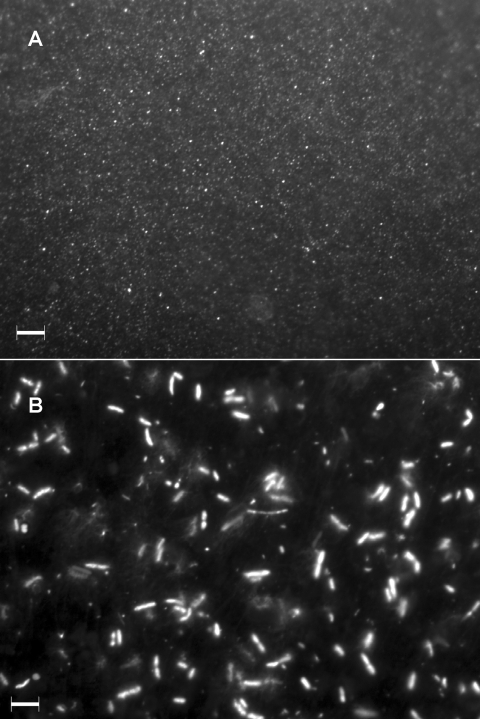
The abundance of viruses in a rice paddy soil sample was estimated by directly counting virus-like particles by EFM after sequential filtration. We obtained 2.77 × 108 ± 0.47 × 108 viruses from 1 g (wet weight) of soil. We did not observe any bacterial cells by microscopy after isolation of the viruses (Fig. 3A), and the 16S rRNA gene was not amplified by PCR with the 8F and 1492R primers from the filtered samples (data not shown).
FIG. 3.
Virus-like particles (A) and bacteria (B) from rice paddy soil stained with Sybr gold and observed by EFM. Scale bars, 5 μm. (A) No bacterial contamination is observed.
In order to construct viral clone libraries, viral DNA was extracted from about 4.25 × 1010 viral particles. Two shotgun libraries, RH (heat treatment) and RX (no heat treatment), were constructed from DNA samples that were amplified by different MDA methods with or without a heat-denaturing step (95°C for 3 min), respectively. Cloned sequences (396 RH and 389 RX) with an average length of 641 bp were retrieved from the viral clone libraries. The resulting sequences from each library were analyzed with TBLASTX and BLASTX against the GenBank database and the Phage Sequence Databank. Classification of the results that were obtained from the database searches are summarized in Table 1. The numbers of sequences that were found to be significant hits (E value, <0.001) in the RH and RX libraries were 141 (36%) and 127 (33%), respectively. More than 60% of the sequences had no hits, which implies that the soil viral genomic diversity has not been sufficiently characterized thus far. Of the known sequences in the RH library, 109 (77%) were related to viruses and 19% were related to bacteria. The results of the TBLASTX searches of GenBank and the Phage Sequence Databank were slightly different. About 40% of the sequences that resulted in bacterial hits from GenBank were identified as hits from virus or prophage in the Phage Sequence Databank; in this study, we regarded these as virus or prophage hits. The discrepancy between database searches appears to result from the fact that genomic sequence data from soil viruses has not sufficiently accumulated in the databases, whereas the databases contain multiple bacterial sequences that often have prophage sequences in their genomes (20). The average identity value of the sequences from the TBLASTX searches was relatively low (34% ± 14%), which could also be evidence of an insufficient volume of viral genomic data.
TABLE 1.
Classification of sequences from RH and RX libraries based on hits from database searches
| Type of hits | No. (%) of reads
|
|
|---|---|---|
| RH library | RX library | |
| Known | 141 (36) | 127 (33) |
| Unknown | 255 (64) | 262 (67) |
| Total | 396 (100) | 389 (100) |
| Biological groups of known hits | ||
| Virus related | 109 (77) | 119 (94) |
| Archaea | 1 (1) | |
| Bacteria | 27 (19) | 5 (4) |
| Eucarya | 1 (1) | 2 (2) |
| Plasmid | 4 (3) | |
| Types of virus-related hits | ||
| dsDNA phage | 33 (30) | 7 (6) |
| dsDNA eu-virusa | 2 (2) | |
| ssDNA phage | 19 (17) | 16 (13) |
| ssDNA eu-virusa | 40 (37) | 90 (75) |
| ssRNA (metagenome) | 1 (1) | 1 (1) |
| Prophage | 15 (14) | 2 (2) |
| Provirus | 1 (1) | 1 (91) |
| Classification of dsDNA viruses | ||
| Caudoviralesb | 1 (3) | |
| Myoviridaeb | 8 (24) | 2 (22) |
| Podoviridaeb | 8 (24) | 4 (44) |
| Siphoviridaeb | 15 (45) | |
| Tectiviridaeb | 1 (3) | |
| Unclassifiedb | 1 (11) | |
| Poxviridae | 2 (22) | |
| Classification of ssDNA viruses | ||
| Inoviridaeb | 1 (1) | |
| Microviridaeb | 19 (32) | 15 (14) |
| Circoviridae | 17 (29) | 47 (44) |
| Geminiviridae | 6 (10) | 17 (16) |
| Nanoviridae | 17 (29) | 26 (25) |
In this case, eu-virus means nonprokaryotic virus.
These viral classifications belong to bacteriophages (prokaryotic viruses).
In the RH library, 30% of the sequences were dsDNA phage and 14% were prophage. A large portion of the viral hits (37%) were related to ssDNA eukaryotic viruses (eu-viruses) and ssDNA phages (17%). The ratio of ssDNA viruses (eu-viruses plus phages) to dsDNA viruses was about 1.8:1. Hits that were identified as dsDNA phages were classified into Siphoviridae (45%), Myoviridae (24%), Podoviridae (24%), and Tectiviridae (3%) on the basis of their taxonomic annotations in the GenBank database. The families Siphoviridae, Myoviridae, and Podoviridae were previously reported to be major entities of soil viral assemblages (50). ssDNA viruses were classified into Microviridae (32%), Circoviridae (29%), Nanoviridae (29%), and Geminiviridae (10%). Microviridae is a family that belongs to bacteriophage groups, whereas the remaining groups are eu-viruses that infect animal and plants. All of the above-mentioned ssDNA viruses had short circular ssDNA genomes.
In the RX library, 94% of the known sequences were virus-related hits and 88% of these were from ssDNA viruses (75% eu-viruses and 13% ssDNA phage). Circoviridae (44%), Nanoviridae (25%), Microviridae (14%), and Geminiviridae (16%) were the majority of ssDNA viral hits in the RX library; these results are similar to those of the RH library.
Significantly hit viral proteins from the metagenomic library were classified on the basis of a BLASTX search against the GenBank database (Table 2). We divided the proteins into two categories, (i) a dsDNA group (dsDNA phages and prophages from the RH and RX libraries) and (ii) a ssDNA group (ssDNA viral proteins from the RH and RX libraries). We found that the kinds of ssDNA viral proteins were not diverse compared to those of dsDNA viral proteins. The majority (76%) of the ssDNA viral proteins were similar to replication-related proteins, and 15% were similar to structural proteins. This is because major components of ssDNA viral genomes are replication-related and structural proteins and ssDNA viral genomes are very short and contain only a few ORFs in their genomes (38).
TABLE 2.
Protein types with significant viral hits from two metagenomic libraries
| Protein type | No. of sequences from:
|
|
|---|---|---|
| RH DNA library | RX DNA library | |
| dsDNA phage + prophage from RH and RX libraries | ||
| Hypothetical | 14 | 1 |
| Helicase | 3 | |
| Capsid | 2 | |
| DNA polymerase | 2 | |
| Methyltransferase | 2 | |
| Replication associated | 2 | |
| Terminase | 2 | 1 |
| ATPase | 1 | |
| Cell wall hydrolyzer | 1 | |
| Collagen precursor | 1 | |
| Deaminase | 1 | |
| Hydrolase | 1 | |
| Ligase | 1 | |
| Packing | 1 | |
| Portal | 1 | |
| Primase | 1 | |
| Recombination | 1 | 1 |
| Rep | 1 | |
| Replication initiation | 1 | |
| Surface layer protein | 1 | |
| Tail fiber | 1 | |
| Transcription regulator | 1 | |
| Total | 41 | 4 |
| ssDNA viruses from RH and RX libraries | ||
| Replication related | 37 | 85 |
| Replication associated | 19 | 39 |
| Rep | 10 | 21 |
| Replicase | 2 | 11 |
| Rep and coat | 3 | 8 |
| Replication initiation | 3 | 6 |
| Structural | 16 | 8 |
| Capsid | 14 | 4 |
| Structural | 2 | 4 |
| Helicase | 2 | 7 |
| Hypothetical | 1 | 1 |
| Nonstructural | 1 | 1 |
| Polymerase | 1 | |
| Total | 58 | 102 |
Sequences from the two libraries were assembled separately, and their contigs were analyzed to estimate the diversity of the viral assemblages. We assembled 396 (RH), 389 (RX), and 785 sequences (total) into 34, 53, and 93 contigs and 292, 182, and 425 singletons, respectively. The proportions of sequences to contigs were 26, 53, and 46% (Table 3).
TABLE 3.
Contig formation of metagenomic sequences from two libraries
| No. of reads assembled in a contig | No. of contigs in library
|
||
|---|---|---|---|
| RH | RX | RX + RH | |
| 1a | 292 | 182 | 425 |
| 2 | 21 | 29 | 49 |
| 3 | 7 | 9 | 17 |
| 4 | 1 | 3 | 7 |
| 5 | 1 | 3 | 6 |
| 6 | 0 | 2 | 2 |
| 7 | 2 | 1 | 2 |
| 8 | 1 | 0 | 1 |
| 9 | 0 | 0 | 0 |
| 10 | 1 | 0 | 2 |
| 11 | 0 | 2 | 0 |
| 12 | 0 | 1 | 1 |
| 13 | 0 | 1 | 1 |
| 14 | 0 | 1 | 2 |
| 15 | 0 | 1 | 2 |
| 16 | 0 | 0 | 1 |
| Total no. of contigs | 34 | 53 | 93 |
A contig that contains one read is unassembled.
The average genome size of ssDNA viruses detected in the RX library was calculated on the basis of their best hit from the GenBank database and their genome size. The average genome size of ssDNA viruses detected in the RX library was calculated to be 2,110 bp (a total of 428 ssDNA viral genomes in the GenBank database were averaged to 3,390 bp).
Phylogenetic analysis of replication-related genes from uncultured viruses.
In total, 122 viral DNA sequences showed significant hits with replication-related genes. After excluding short sequences and merging redundant sequences, we obtained 85 partial or full ORFs coding putative replication-related genes from the contig and singletons. Most of the sequences had distant relationships with known viral genes; therefore, the identities from the BLASTX search were lower than 35% in most cases. An NCBI Conserved Domain Database search was performed to investigate the phylogenetic relationship of these putative replication-related protein sequences. We found that 58 peptides contained at least one conserved domain; these included the putative viral replication protein domain (Pfam accession no. PF02407, Pfam ID Viral_Rep, 39 peptides), the viral replication domain C terminus (PF08419, Viral_Rep_C, 14 peptides), the Geminivirus Rep protein catalytic domain (PF00799, Gemini_AL1, 6 peptides), the Geminivirus Rep protein central domain (PF08283, Gemini_AL1_M, 3 peptides), the RNA helicase domain (PF00910, RNA_helicase, 4 peptides), and two COG domains; 28 ORFs showed no conserved domain in the NCBI Conserved Domain Database search.
Forty Viral_Rep and six Gemini_AL1 domain-containing peptides were aligned with the representative sequences for each conserved domain, and an additional four replication protein-like sequences from protozoa and plasmids containing conserved regions relating to Viral_Rep domains (21) were added to the alignment. Only properly aligned regions of the conserved domains were used for the phylogenetic trees. After excluding partially aligned sequences, 28 and 4 peptides were used for reconstructing phylogenetic trees for the RH and RX groups, respectively (Fig. 4; see Fig. S2 and S3 in the supplemental material).
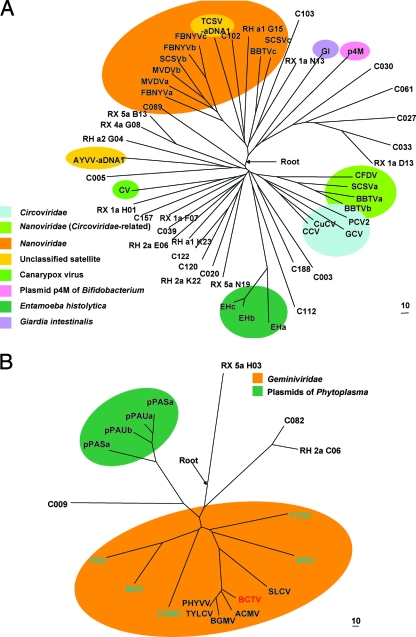
FIG. 4.
Phylogenetic trees of the amino acid sequences from the Rep_Viral (A) and Gemini_AL3 (B) domains obtained in this study and related members with those domains. The alignment lengths for tree construction were 75 and 108 amino acids for trees A and B, respectively. The scale bars indicate the distance score calculated with the scoring matrix BLOSUM62. The prefix C in the sequence name means that the sequence is from a contig, and the prefixes RH or RX indicates the library where the sequence was retrieved. (A) Reference sequences: CFDV, Coconut foliar decay virus (accession no. Q66005); SCSVa, -b, and -c, Subterranean clover stunt virus (accession no. Q87009, Q87013, and Q9ICP7); BBTVa, -b, and -c, Banana bunchy top virus (accession no. Q8QTK9, Q83026, and Q65378); PCV2, Porcine circovirus 2 (accession no. Q8BB16); GCV, Goose circovirus (accession no. Q8AYY2); CCV, Canary circovirus (accession no. Q912W1); CuCV, Columbid circovirus (accession no. Q91GA3); EHa, -b, and -c, Entamoeba histolytica HM-1 (accession no. XP_648754, XP_648748, XP_650115); CV, Canarypox virus (accession no. NP_955176); AYVV-aDNA1, Ageratum yellow vein virus-associated DNA 1 (accession no. Q8QME8); MVDVa and -b, Milk vetch dwarf virus; FBNYVa, -b, -c, Fava bean necrotic yellows virus (accession no. O91250, O91250, and Q66862); TCSV-aDNA1, Tobacco curly shoot virus-associated DNA 1 (accession no. Q7T7M5); GI, Giardia intestinalis (accession no. Q9NJY0); p4M, plasmid of Bifidobacterium pseudocatenulatum (accession no. NP_613078). (B) Reference sequences: CSMV, Chloris striate mosaic virus (accession no. P18921); PSV, Panicum streak virus (accession no. Q00338); MSV, Maize streak virus (accession no. P03568); WDV, Wheat dwarf virus (accession no. P06847); TYDV, Tobacco yellow dwarf virus (accession no. P31617); TYLCV, Tomato yellow leaf curl virus (accession no. P36279); PHYVV, Pepper huasteco yellow vein virus (accession no. Q06923); BGMV, Bean golden mosaic virus (accession no. P05175), ACMV, African cassava mosaic virus (accession no. P14982); SLCV, Squash leaf curl virus (accession no. P2904); BCTV, Beet curly top virus (accession no. P14991); pPASa and -b and pPAUa and -b, plasmids of Candidatus P. asterisasteris (accession no. Q2NIE5, Q2NIE4, Q0QLC1, and Q0QLC5). For the accession numbers of sequences from this study in the figures and alignments of the translated sequences, see the supplemental material.
The peptides from the Viral_Rep group were widely distributed throughout the phylogenetic trees (Fig. 4A). Some members were related to the established families Nanoviridae and Circoviridae. The tree showed that some reference sequences from nanoviruses were more closely related to Circoviridae than Nanoviridae as previously reported (22); however, the majority of the Viral_Rep group did not fall within the Nanoviridae and Circoviridae families. Five peptides showed a distant but monophyletic relationship to the putative replication-associated protein REP2 from Giardia intestinales, a parasitic protozoan that causes diarrhea in humans and other mammals (1) and to plasmid p4M from a Bifidobacterium strain (22). One peptide was related to the proviral protein of Entamoeba histolytica HM-1:IMSS, an intestinal protozoan parasite and the causative agent of amoebiasis (42). Eleven peptides made a distinct clade that included a Rep-like protein from the canarypox virus (47), a dsDNA virus that causes canarypox in birds. Four peptides made a clade with ageratum yellow vein virus-associated DNA 1, an autonomously replicating DNA associated with the Begomovirus Ageratum yellow vein virus (42).
The Gemini_AL1 group appeared to be included in the established family Geminiviridae; however, the sequences were notably divergent from known members of this family (Fig. 4B). One sequence exhibited close relationships with plasmids from Phytoplasma sp. bacteria, phytopathogens like Geminivirus (5, 30).
Viral_Rep and Gemini_AL1 families belong to the Rep-like domain clan (CL0169, Rep), which contains eight protein family members that are related to replication proteins for viruses and plasmids. A clan contains two or more protein family members derived from a single evolutionary origin. We aligned these sequences and used them for rooting the tree; we proposed and indicated the rooting position in the trees (Fig. 4A and B).
Reconstruction of complete circular ssDNA viral genomes.
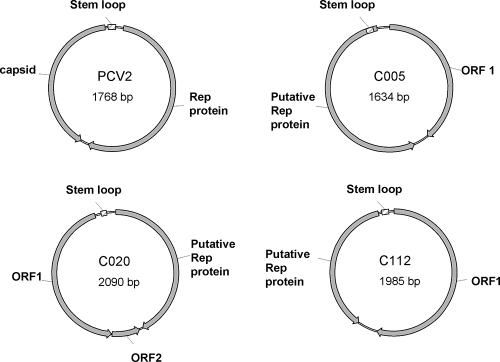
Among the contigs that were assembled from the total sequences of the two libraries, we found that 19 of them contained repeated sequences or circular sequences which had the same sequences in front (the start) and at the rear (the end). The sizes of these repeated or circular sequences varied from 290 to 2,495 bp (Table 4). Of these repeated or circular sequences, four contig sequences (C020, C112, C005, and C132, with sizes of 2,090, 1,984, 1,634, and 1,108 bp, respectively) showed significant hits with ssDNA eu-viruses in the TBLASTX and BLASTX searches of the GenBank database; the other contigs were not related to any known sequences. In order to confirm that the circular DNA contigs really had circular structures, we designed inverse PCR primers in opposite directions against two contigs (C005 and C112) that are thought to contain the putative circular genomes. This resulted in the amplification of PCR amplicons of the expected size from the MDA-amplified viral sample. Sequencing of the amplicons (data not shown) showed that they had the same sequences as the circular genomes. In the case of C005, the chimera sequence that existed in the contig sequence and was excluded during construction of the circular genome was not found in the sequence of the PCR amplicon. With the exception of replication gene-related ORFs, all of the other ORFs in the putative genomic components showed no significant hits to known proteins. The viral replication protein of C020 and C112 had two conserved domains from the putative viral replication protein (PF02407) and the viral replication domain C terminus (PF08419). The viral replication protein of C005 had one conserved domain of the viral replication domain C terminus (PF08419). No conserved domains were identified for C132. Analyses of these putative viral genomic components were performed (Fig. 5). C132 was excluded from the analyses because this contig was obtained from only two reads (Table 4), and potential sequencing errors and/or chimeric sequences can result in highly biased analyses. The genome organizations of the three putative genome components were similar to that of PCV2, a circovirus (Fig. 5A), as well as to those of other circoviruses, nanoviruses, and geminiviruses (35, 46). All of the components had a putative stem-loop structure in their intergenic regions. The loop region of this structure contains a conserved nonanucleotide motif that is found in plant geminiviruses and plant nanoviruses and that corresponds to the site of viral DNA replication (34). The stem-loop sequences were aligned with that of PCV2 (see Fig. S1 in the supplemental material). Except for the putative Rep protein, each putative genome component had only one or two additional ORFs (overlapped ORFs were not considered). If the circular component constitutes a viral genome, these ORFs could be the putative capsid protein but none of these ORFs gave significant hits with known proteins. Because various kinds of capsid proteins from circoviruses, nanoviruses, and geminiviruses have a high frequency of arginine/lysine residues in their amino-terminal regions (35), we investigated the arginine/lysine frequency in the amino-terminal regions of the ORFs of the putative genome components. We found that one of the ORFs of each component had an arginine/lysine-rich amino-terminal region (see Fig. S1 in the supplemental material), although this was only weakly so in the case of C112.
TABLE 4.
Contigs forming circular DNA or containing repeated sequences
| Contig | Length (bp) | Size of circular or repeated sequence (bp) | Hits of circular or repeated sequences | No. of assembled sequences | No. of sequences from:
|
|
|---|---|---|---|---|---|---|
| RH library | RX library | |||||
| C119 | 2,652 | 2,495 | Unknown | 10 | 3 | 7 |
| C020 | 2,477 | 2,090 | ssDNA eu-virus | 15 | 8 | 7 |
| C112 | 2,219 | 1,984 | ssDNA eu-virus | 16 | 5 | 11 |
| C005 | 2,896a | 1,634 | ssDNA eu-virus | 10 | 1 | 9 |
| C076 | 2,971a | 1,230 | Unknown | 14 | 1 | 13 |
| C138 | 1,231 | 1,207 | Unknown | 6 | 0 | 6 |
| C071 | 1,554 | 1,186 | Unknown | 8 | 7 | 1 |
| C132 | 1,172 | 1,108 | ssDNA eu-virus | 2 | 1 | 1 |
| C047 | 1,489 | 1,090 | Unknown | 5 | 1 | 4 |
| C145 | 1,541 | 969 | Unknown | 4 | 1 | 3 |
| C075 | 1,838 | 890 | Unknown | 5 | 0 | 5 |
| C078 | 936 | 860 | Unknown | 2 | 1 | 1 |
| C077 | 1,312 | 850 | Unknown | 2 | 0 | 2 |
| C027 | 1,957a | 820 | Unknown | 5 | 1 | 4 |
| C001 | 1,421a | 690 | Unknown | 13 | 0 | 13 |
| C175 | 786 | 540 | Unknown | 2 | 1 | 1 |
| C154 | 1,053 | 440 | Unknown | 2 | 2 | 0 |
| C002 | 1,653 | 320 | Unknown | 14 | 0 | 14 |
| C116 | 1,268 | 290 | Unknown | 15 | 1 | 14 |
These contigs were suspected to contain chimera sequences outside the circular DNA fragment; chimeras was recognized at a point at which interruption of circular sequences occurs.
FIG. 5.
Genome organizations of the three putative circular genomic components reconstructed from soil viral metagenomic sequences. The components show a genomic organization similar to that of Porcine circovirus2 (PCV2), a representative circovirus.
DISCUSSION
Use of MDA for the amplification of ssDNA viruses.
We adapted MDA to amplify viral DNA and found that this technique could be used to selectively amplify ssDNA viruses. The RX library was constructed by MDA without the denaturing step; 90% of these sequences were related to ssDNA viruses. In contrast, the RH library was constructed by MDA with the denaturing step; only 50% of these sequences were related to ssDNA viruses, thus implying that ssDNAs were amplified more efficiently by MDA without the denaturing step than with the denaturing step. The denaturing step converts dsDNA to ssDNA. Random hexamers anneal to ssDNA, and φ29 DNA polymerase synthesizes dsDNA. Once the priming events have occurred, the displacement activity of the φ29 polymerase continuously supplies new priming sites for unbound random hexamers (31). If the denaturation step is excluded, it is difficult for dsDNA to bind to the random hexamers because MDA is performed under isothermal conditions at 30°C. In contrast, ssDNA rapidly anneals to random hexamers at 30°C. This explains why almost all of the known sequences in the RX library were from ssDNA viruses. Another reason for the preferential amplification of ssDNA viruses might be their circular viral genomes. The ssDNA viruses detected in this study belong to the families Circoviridae, Geminiviridae, Nanoviridae, and Microviridae, all of whose members have small circular genomes ranging from about 1,000 to 9,000 bp. The φ29 polymerase used in the MDA reaction amplifies DNA via a rolling-circle amplification mechanism in which single-stranded templates can be continuously produced along with the circular DNA (31). The results showing that circular DNA was amplified much more efficiently than linear DNA (Fig. 2) also supports the notion that MDA without the denaturing step amplifies ssDNA viruses selectively. Thus, ssDNA viruses containing short circular genomes could be amplified selectively from mixed viral DNA. The fact that the proportions of the two metagenomic sequences (C005 and C112) after MDA were high (Fig. 1) could explain why the sequences of contigs C005 and C112 were detected very frequently (9 and 11 times among 389 sequences) in the RX library (Table 4). The fact that these sequences were related to those of ssDNA viruses also provides further evidence that MDA without the denaturing step preferentially amplifies ssDNA genomes. In addition, it was reported that circular DNA virus genomes could be amplified by a sequence-independent strategy by MDA (or multiply primed rolling-circle amplification). The human papillomavirus circular genome has been amplified from DNA extracted from cell lines and bovine tissues; the concentration of the papillomavirus DNA was increased by 2.4 × 104-fold (40). This method was also used to amplify the circular genomes of various viruses, such as polyomaviruses (27), anellovirus (36), circoviruses (26), and geminiviruses (23). Considering that papillomavirus and polyomavirus were dsDNA viruses and these dsDNA viruses could be amplified efficiently by MDA, the circular structure was a main reason for the preferential amplification of circular ssDNA viruses. In addition, the result that 50% of the significant hits related to ssDNA viruses was increased to 90% without the denaturing step implied that eliminating the denaturing step was necessary to investigate ssDNA viruses more selectively. A precise and detailed evaluation of the factors that affect the efficiency of the MDA technique such as the DNA length, linearity, and strand type; the reaction time; and the denaturing step should be performed in future studies.
Diversity of viruses in rice paddy soil.
When MDA was performed with the denaturing step (RH library), we acquired a number of metagenomic sequences that were related to both dsDNA and ssDNA viruses. All of the dsDNA viruses were prokaryotic viruses (phages); the majority of them (>92%) were tailed phages, and only one was a polyhedral phage (belonging to the family Tectiviridae). A large proportion of the significant hits in the RH library were related to ssDNA viruses. This abundance of ssDNA viral hits is not in concord with a previous observation suggesting that dsDNA viruses are the major entity in a soil viral assemblage (50); preferential amplification of circular DNA by MDA was suggested to be the main reason for the results. The majority of ssDNA viruses detected in this study were related to animal and plant viruses (68 to 85%) and not to phages. The proportions of animal and plant viruses were almost the same. It is suspected that the major source of plant viruses is rice or other plants that grow in the field and the source of the animal viruses seems to be wild bird feces or composted manure used as organic fertilizer. Animal and plant viruses detected in this study belong to the families Circoviridae, Nanoviridae, and Geminiviridae, in which a lot of pathogenic viruses exist (32, 45, 48); this indicates that soil could be a reservoir for viruses that are pathogenic to animals and plants. Another main group of ssDNA viruses obtained in this study was Microviridae, which is a bacteriophage family. Chp1-like ssDNA microphages belonging to the family Microviridae were also found to be abundant in the Sargasso Sea (3).
Phylogenetic analysis of replication-related genes of uncultured viruses.
Phylogenetic analyses of replication-related genes showed that the majority of the sequences acquired in this study represented previously unknown viruses. The viral family Geminviridae, containing a large number of plant-pathogenic viruses, is represented by four genera: Begomovirus, Curtovirus, Mastrevirus, and Topocuvirus. The sequences acquired in this study relating to the family Geminiviridae did not fall into any of these genera, indicating that they are new members of Geminiviridae, the largest family of ssDNA viruses. The majority of the sequences obtained in this study were distantly related to Nanoviridae and Circoviridae but did not fall into to the established families. Some of these sequences were closely related to nonviral genomic material such as satellites, bacterial plasmids, genomes of protozoa, and dsDNA viruses rather than ssDNA viruses (Fig. 2). Nevertheless, some of these sequences might have originated from ssDNA viruses and they may represent new ssDNA virus families. The fact that more than 60% of the hits in this study were nonsignificant implies that ssDNA viral diversity in a soil environment may be more complex than previously thought.
Construction of complete circular ssDNA viral genomes.
The presence of a conserved stem-loop structure in the intergenic region and two ORFs encoding a putative capsid protein and a putative Rep protein suggests that the three circular sequences are the entire genomes or genome components of unknown ssDNA viruses (some ssDNA viruses, such as Nanovirus and Geminivirus, have multiple genomic components). From this point of the view, the other circular sequences (derived from contigs) that gave no hit with any known protein in the sequence databases might also turn out to be completely novel viruses. Breitbart et al. pointed out the possibility of sequencing the entire genome of uncultured viruses by using metagenomic approaches (10). Several complete genomes of RNA viruses have been reconstructed from a coastal RNA viral metagenomic study (13). In this report, several putative circular ssDNA viral genomes were reconstructed with a relatively small amount of sequencing (<800 sequence reads). These results indicate that metagenomic research is a useful method to uncover unknown genomic entities among environmental viruses.
MDA without the denaturing step cannot displace previous methods such as the LASL method. Rather, the work described herein points to an approach that permits the investigation of ssDNA viral diversity, something that cannot be accomplished by the LASL method. MDA has several shortcomings such as chimera formation (29, 52), biased amplification (24), and contamination of external DNA (52). Nevertheless, preferential amplification of circular ssDNA viral DNA by MDA without the denaturing step could provide a new tool to explore currently unexplored viral diversity.
Although viruses are major biological entities in soil, viral diversity in this environment was largely unexplored. By using the novel culture-independent metagenomic approach, we have investigated the diversity of DNA viral assemblages. MDA is a useful approach for the investigation of both ssDNA and dsDNA viral diversity; this technique was also used to selectively investigate the diversity of ssDNA viruses. Our research also showed that unknown short circular ssDNA viral genomes or genome components can be detected without viral cultivation by sequencing the metagenome and amplifying the DNA by MDA. Further studies are needed to reveal the viral diversity of different soil samples and to quantitatively analyze soil viruses. These efforts would contribute to our understanding of the role of viral assemblages in biological soil communities.
Supplementary Material
Acknowledgments
This work was supported by the Environmental Biotechnology National Core Research Center (KOSEF: R15-2003-012-02002-0) and the 21C Frontier Microbial Genomics and Application Center Program. K.-H.K. was supported by a Korea Research Foundation grant (MOEHRD, Basic Research Promotion Fund, KRF-2006-351-D00011).
Footnotes
Published ahead of print on 15 August 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adam, R. D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14:447-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., M. J. Day, and J. C. Fry. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, X., J. Zhang, A. Ewing, S. A. Miller, A. Jancso Radek, D. V. Shevchenko, K. Tsukerman, T. Walunas, A. Lapidus, J. W. Campbell, and S. A. Hogenhout. 2006. Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bench, S. R., T. E. Hanson, K. E. Williamson, D. Ghosh, M. Radosovich, K. Wang, and K. E. Wommack. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629-7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitbart, M., I. Hewson, B. Felts, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cann, A. J., S. E. Fandrich, and S. Heaphy. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151-156. [DOI] [PubMed] [Google Scholar]
- 12.Casas, V., and F. Rohwer. 2007. Phage metagenomics. Methods Enzymol. 421:259-268. [DOI] [PubMed] [Google Scholar]
- 13.Culley, A. I., A. S. Lang, and C. A. Suttle. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795-1798. [DOI] [PubMed] [Google Scholar]
- 14.Dean, F. B., S. Hosono, L. Fang, X. Wu, A. F. Faruqi, P. Bray-Ward, Z. Sun, Q. Zong, Y. Du, J. Du, M. Driscoll, W. Song, S. F. Kingsmore, M. Egholm, and R. S. Lasken. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 99:5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, F. B., J. R. Nelson, T. L. Giesler, and R. S. Lasken. 2001. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11:1095-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart, E. L. 2007. Viral metagenomics. Rev. Med. Virol. 17:115-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 18.Fierer, N., M. Breitbart, J. Nulton, P. Salamon, C. Lozupone, R. Jones, M. Robeson, R. A. Edwards, B. Felts, S. Rayhawk, R. Knight, F. Rohwer, and R. B. Jackson. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouts, D. E. 2006. Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34:5839-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs, M. J., V. V. Smeianov, J. L. Steele, P. Upcroft, and B. A. Efimov. 2006. Two families of rep-like genes that probably originated by interspecies recombination are represented in viral, plasmid, bacterial, and parasitic protozoan genomes. Mol. Biol. Evol. 23:1097-1100. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs, M. J., and G. F. Weiller. 1999. Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus. Proc. Natl. Acad. Sci. USA 96:8022-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haible, D., S. Kober, and H. Jeske. 2006. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 135:9-16. [DOI] [PubMed] [Google Scholar]
- 24.Hosono, S., A. F. Faruqi, F. B. Dean, Y. Du, Z. Sun, X. Wu, J. Du, S. F. Kingsmore, M. Egholm, and R. S. Lasken. 2003. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 13:954-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia, Z., R. Ishihara, Y. Nakajima, S. Asakawa, and M. Kimura. 2007. Molecular characterization of T4-type bacteriophages in a rice field. Environ. Microbiol. 9:1091-1096. [DOI] [PubMed] [Google Scholar]
- 26.Johne, R., D. Fernandez-de-Luco, U. Hofle, and H. Muller. 2006. Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification. J. Gen. Virol. 87:1189-1195. [DOI] [PubMed] [Google Scholar]
- 27.Johne, R., W. Wittig, D. Fernandez-de-Luco, U. Hofle, and H. Muller. 2006. Characterization of two novel polyomaviruses of birds by using multiply primed rolling-circle amplification of their genomes. J. Virol. 80:3523-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lasken, R. S., and M. Egholm. 2003. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 21:531-535. [DOI] [PubMed] [Google Scholar]
- 29.Lasken, R. S., and T. B. Stockwell. 2007. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liefting, L. W., M. T. Andersen, T. J. Lough, and R. E. Beever. 2006. Comparative analysis of the plasmids from two isolates of “Candidatus Phytoplasma australiense.” Plasmid 56:138-144. [DOI] [PubMed] [Google Scholar]
- 31.Lizardi, P. M., X. Huang, Z. Zhu, P. Bray-Ward, D. C. Thomas, and D. C. Ward. 1998. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 19:225-232. [DOI] [PubMed] [Google Scholar]
- 32.Mansoor, S., S. H. Khan, A. Bashir, M. Saeed, Y. Zafar, K. A. Malik, R. Briddon, J. Stanley, and P. G. Markham. 1999. Identification of a novel circular single-stranded DNA associated with cotton leaf curl disease in Pakistan. Virology 259:190-199. [DOI] [PubMed] [Google Scholar]
- 33.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meehan, B. M., J. L. Creelan, M. S. McNulty, and D. Todd. 1997. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J. Gen. Virol. 78(Pt. 1):221-227. [DOI] [PubMed] [Google Scholar]
- 35.Niagro, F. D., A. N. Forsthoefel, R. P. Lawther, L. Kamalanathan, B. W. Ritchie, K. S. Latimer, and P. D. Lukert. 1998. Beak and feather disease virus and porcine circovirus genomes: intermediates between the geminiviruses and plant circoviruses. Arch. Virol. 143:1723-1744. [DOI] [PubMed] [Google Scholar]
- 36.Niel, C., L. Diniz-Mendes, and S. Devalle. 2005. Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J. Gen. Virol. 86:1343-1347. [DOI] [PubMed] [Google Scholar]
- 37.Noble, R., and J. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 38.Olvera, A., M. Cortey, and J. Segales. 2007. Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology 357:175-185. [DOI] [PubMed] [Google Scholar]
- 39.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 40.Rector, A., R. Tachezy, and M. Van Ranst. 2004. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 78:4993-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Saunders, K., I. D. Bedford, and J. Stanley. 2002. Adaptation from whitefly to leafhopper transmission of an autonomously replicating nanovirus-like DNA component associated with ageratum yellow vein disease. J. Gen. Virol. 83:907-913. [DOI] [PubMed] [Google Scholar]
- 43.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 44.Suttle, C. A., A. M. Chan, and M. T. Cottrell. 1990. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347:467-469. [Google Scholar]
- 45.Todd, D. 2000. Circoviruses: immunosuppressive threats to avian species: a review. Avian Pathol. 29:373-394. [DOI] [PubMed] [Google Scholar]
- 46.Todd, D., J. H. Weston, D. Soike, and J. A. Smyth. 2001. Genome sequence determinations and analyses of novel circoviruses from goose and pigeon. Virology 286:354-362. [DOI] [PubMed] [Google Scholar]
- 47.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2004. The genome of canarypox virus. J. Virol. 78:353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varma, A., and V. G. Malathi. 2003. Emerging geminivirus problems: a serious threat to crop production. Ann. Appl. Biol. 142:145-164. [Google Scholar]
- 49.Williamson, K. E., M. Radosevich, D. W. Smith, and K. E. Wommack. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 9:2563-2574. [DOI] [PubMed] [Google Scholar]
- 50.Williamson, K. E., M. Radosevich, and K. E. Wommack. 2005. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71:3119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson, K. E., K. E. Wommack, and M. Radosevich. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 69:6628-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, K., A. C. Martiny, N. B. Reppas, K. W. Barry, J. Malek, S. W. Chisholm, and G. M. Church. 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24:680-686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.