Abstract
Symbiotic relationships between microbes and plants are common and well studied in terrestrial ecosystems, but little is known about such relationships in aquatic environments. We compared the phylogenetic diversities of leaf- and root-attached bacteria from four species of aquatic angiosperms using denaturing gradient gel electrophoresis (DGGE) and DNA sequencing of PCR-amplified 16S rRNA genes. Plants were collected from three beds in Chesapeake Bay at sites characterized as freshwater (Vallisneria americana), brackish (Potomogeton perfoliatus and Stuckenia pectinata), and marine (Zostera marina). DGGE analyses showed that bacterial communities were very similar for replicate samples of leaves from canopy-forming plants S. pectinata and P. perfoliatus and less similar for replicate samples of leaves from meadow-forming plants Z. marina and V. americana and of roots of all species. In contrast, bacterial communities differed greatly among plant species and between leaves and roots. DNA sequencing identified 154 bacterial phylotypes, most of which were restricted to single plant species. However, 12 phylotypes were found on more than one plant species, and several of these phylotypes were abundant in clone libraries and represented the darkest bands in DGGE banding patterns. Root-attached phylotypes included relatives of sulfur-oxidizing Gammaproteobacteria and sulfate-reducing Deltaproteobacteria. Leaf-attached phylotypes included relatives of polymer-degrading Bacteroidetes and phototrophic Alphaproteobacteria. Also, leaves and roots of three plant species hosted relatives of methylotrophic Betaproteobacteria belonging to the family Methylophilaceae. These results suggest that aquatic angiosperms host specialized communities of bacteria on their surfaces, including several broadly distributed and potentially mutualistic bacterial populations.
Most plants form intimate relationships with microorganisms, ranging from mutualistic to parasitic. Plant-bacterium and plant-fungus symbioses in the terrestrial environment are extremely common and have been well studied. Leguminous plants host symbiotic nitrogen-fixing bacteria in their roots, while most angiosperms (flowering plants) and members of the Pinaceae (pine family) have root-associated symbiotic mycorrhizal fungi that facilitate plant uptake of soil nutrients (2). Plant leaves also host complex assemblages of bacteria (76), some of which produce compounds that promote plant growth (17, 29, 59), including the plant growth-regulating auxin indole-3-acetic acid, a compound involved in plant development (37, 43).
In contrast, very little is known about microbes associated with aquatic plants. Surfaces of aquatic plant roots and rhizomes host active sulfate-reducing and nitrogen-fixing bacterial communities (18, 54). Similarly, leaves of aquatic plants support very active bacterial communities that are thought to be influenced by plant primary production (24, 68). Most research on aquatic plant-bacterium interactions, however, describes broader relationships with general sediment bacteria in the rhizosphere. These sediments contain active bacterial communities that consume molecules excreted by plant roots, including organic molecules (amino acids, sugars, and organic acids) and gasses (O2, N2, and CO2) (8, 28, 49, 57). In return, microbial processes, including organic matter mineralization (64), phosphorous solubilization (75), and nitrogen fixation, provide nutrients for the plants. The final process, nitrogen fixation, is the only well-studied aquatic plant-bacterium interaction in which bacteria are directly attached to the plant surface (11, 44). These diazotrophic bacteria grow on both the roots and the leaves of aquatic plants and potentially provide as much as 50% of the nitrogen demand of the plant (9). However, aside from diazotrophs, research on symbiotic aquatic plant-associated organisms lags far behind parallel research on terrestrial plants.
A few studies using molecular techniques describe the diversity of bacteria in plant-colonized sediments (5, 12, 13), but very few studies describe the diversity of organisms attached to aquatic plant surfaces. One pair of studies revealed broad microbial diversity associated with the seagrass Halophila stipulacea in the northern Gulf of Elat (71, 72), and another study identified epiphytic bacterial communities on three seagrass species from the East African coast (70). One recent study identified bacteria attached to roots of Zostera marina (33), and another identified methanotrophic bacteria associated with aquatic plants growing in methanogenic sediments (66). Most of these studies suggested that plant-attached bacterial communities are different than the bacterial communities in surrounding environments (e.g., sediments and water column), and such studies are beginning to reveal what may be a unique assemblage of aquatic plant-associated microorganisms.
The goals of this study were to describe variability in the composition of bacterial communities attached to surfaces of leaves (phylloplane) and roots (rhizoplane) of four aquatic angiosperms in the Chesapeake Bay and to identify common and potentially mutualistic bacterial populations. These plants inhabit environments ranging from freshwater (Vallisneria americana) to brackish water (Potomogeton perfoliatus and Stuckenia pectinata) to marine water (Z. marina). We found evidence of plant-specific bacterial communities on leaves and roots and observed that most bacterial phylotypes were restricted to single plant species. However, among 154 bacterial phylotypes identified by DNA sequencing, we found 12 phylotypes that were associated with more than one plant species and determined that many of these phylotypes were abundant in clone libraries and denaturing gradient gel electrophoresis (DGGE) banding patterns.
MATERIALS AND METHODS
For each plant species, 10 individual plants were collected by hand randomly from ∼100-m2 areas in different regions of Chesapeake Bay (Table 1) and dissected with presterilized equipment. Leaves and roots were triple rinsed by vigorous shaking with filter-sterilized seawater (pore size, 0.2 μm) in sterile 50-ml tubes, frozen in tubes on dry ice, and stored at −80°C for later DNA extraction. A separate set of whole-plant samples was collected and stored in seawater for plant tissue culture.
TABLE 1.
Plant species, sampling dates, and sampling locations
| Plant species | Common name | Sampling date | Latitude | Longitude | Water column salinity | Site description |
|---|---|---|---|---|---|---|
| Z. marina | Eelgrass | 18 May 2005 | 37°50′37″N | 75°59′15″W | 13 | Northeast side of Tangier Island |
| P. perfoliatus | Redhead grass | 14 June 2005 | 39°01′53″N | 76°31′31″W | 7 | North bank of Severn River |
| S. pectinata | Sago pond weed | 27 June 2005 | 39°01′53″N | 76°31′31″W | 7 | North bank of Severn River |
| V. americana | Wild celery | 26 July 2005 | 39°23′20″N | 76°02′27″W | 1 | Mouth of Sassafrass River |
Following each field collection, tissue cultures were established using standard techniques (37, 48). Briefly, meristems of the different plant species were dissected, surface sterilized, and allowed to grow under controlled axenic conditions. P. perfoliatus, S. pectinata, and V. americana became established in tissue culture, but Z. marina did not. Leaves and roots of axenically grown plants were dissected and frozen at −80°C.
DNA was extracted from plant material using an UltraClean fecal DNA kit (MoBio) by following the manufacturer's instructions, except that bead tubes containing plant material were gently shaken with a titer plate shaker (LabLine) rather than a vortex mixer so that the leaves and roots remained intact during bead beating.
Bacterial communities attached to plant leaves and roots were characterized and compared by performing PCR-DGGE analyses of the 16S rRNA gene using previously described laboratory and statistical techniques (16, 51), with the following modifications. The touchdown PCR conditions consisted of an initial 5 min of incubation at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 65 to 55°C (with the temperature reduced by 1°C per cycle for 10 cycles plus 20 cycles at 55°C), and 1 min at 72°C followed by 1 h at 72°C. Temperature reductions during PCR were done at a rate of 0.3°C per s was used. PCR products were separated into bands by electrophoresis (CBS Scientific) for 24 h at 75 V on acrylamide (8%) gels prepared with 30% acrylamide/bisacrylamide (37.5:1; Bio-Rad) and 0.5× TAE buffer (1× TAE buffer is 40 mmol liter−1 Tris [pH 8.0], 20 mmol liter−1 acetic acid, and 1 mmol liter−1 EDTA) and containing 35 to 50% denaturant (urea and formamide) gradients. Gel images were assembled from images collected for a range of exposure times, but no modifications were made within the banding pattern of each sample. DGGE bands were scored when the density was at least 5% that of the darkest band in the sample pattern.
DNA extracted from axenically grown plants (P. perfoliatus, S. pectinata, and V. americana) was also analyzed to identify the DGGE band representing the 16S rRNA gene from the chloroplasts of each plant. This was done so that bands representing chloroplasts could be omitted from the DGGE analysis of bacterial communities. The chloroplast band for Z. marina was identified by DNA sequencing of DGGE bands (see below).
Dominant bacterial populations from one plant of each species (root and leaf) were identified using two different techniques, DGGE band sequencing and PCR-clone library sequencing. 16S rRNA genes were PCR amplified using DGGE-PCR primers as described above and run on a denaturing gradient gel. Samples of DNA fragments from DGGE bands were obtained with sterile pipette tips, and the DNA was PCR amplified using the same protocol and run on another DGGE gel along with the original natural samples in order to identify the appropriate bands from the reamplification reactions. This procedure was repeated until each reamplification reaction produced only one strong band. Samples of the bands were removed, and the DNA was PCR amplified with M13-linked primers (primers M13f-357f [TGTAAAACGACGGCCAGTCTACGGGAGGCAGCAG] and M13r-519r [GGAAACAGCTATGACCATGACCGCGGCTGCTGGCAC]), purified, and sequenced bidirectionally with M13 primers using an ABI-3100 automated DNA sequencer.
For PCR-clone library analysis, DNA samples were PCR amplified with general bacterial primers 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGCTACCTTGTTACGACTT-3′) in 50-μl reaction mixtures under the following conditions: 4 min at 94°C, followed by 27 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min and then 5 min at 72°C. PCR products were then diluted 1:10 in fresh PCR cocktail in 10 separate tubes and subjected to three more PCR cycles. PCR products were combined and purified with a MoBio PCR purification kit, ligated into TOPO-XL plasmids (Invitrogen), and used to transform TOP-10 chemically competent cells by following manufacturer's instructions. Forty-eight clones from each library were selected at random and grown overnight at 37°C in 1 ml of SB broth (33 g tryptone, 20 g yeast extract, 2.5 g NaCl, 1 liter water; autoclaved) in 96-well deep-well plates. Cells were centrifuged for 20 min at 3,000 × g, and plasmids were extracted as follows. Pellets were resuspended in 50 μl of GTE solution (50 mM glucose, 25 mM Tris buffer [pH 8.0], 10 mM EDTA; autoclaved). Then 100 μl of an NaOH-sodium dodecyl sulfate solution (0.2 M NaOH, 1% [wt/vol] sodium dodecyl sulfate; filter sterilized) and 50 μl of KAc solution (29.5 ml glacial acetic acid, KOH pellets added to bring the pH to 4.8, H2O to bring the volume to 100 ml; prepared on ice and filter sterilized) were added, and each preparation was shaken for 2 min and centrifuged for 15 min at 1,200 × g. One-hundred-microliter portions of the supernatants were transferred to sterile 96-well shallow-well plates, 75 μl of 100% isopropanol was added to each well, and the plates were sealed, shaken, incubated for 30 min at −20°C, and centrifuged for 15 min at 1,200 × g. The supernatants were poured and blotted off, the pellets were washed once with 150 μl of 70% ethanol and once with 150 μl of 95% ethanol and were dried in a roto-evaporator and resuspended in 100 μl of Tris buffer (10 mM, pH 8.0). Plasmid inserts were sequenced bidirectionally with primers 27f and 907r (5′-CCGTCAATTCCTTTRAGTTT3′).
DNA sequences from DGGE bands and clone libraries were assembled, quality checked, aligned using the ARB sequence alignment program (www.arb-home.de), screened for chimera formation using BELLEROPHON (30), CHIMERA_CHECK (14), and visual inspection of secondary structure base pair matches, and analyzed with the Basic Local Alignment Search Tool (BLAST) (www.ncbi.nlm.nih.gov) to determine the closest relatives and to group sequences by major phylogenetic taxa. Fifteen chimeras were identified and excluded from further analysis.
Aligned clone sequences (670 bp) were exported from ARB after applying a 50% base pair frequency filter to remove nonhomologous sequences. Phylogenetic distances were calculated with DNADIST using the Jukes-Cantor model, and sequences were assigned to operational taxonomic units (OTUs) based on 97% sequence similarity using DOTUR (61). In order to place DGGE band sequences into these OTUs, DGGE band sequences and clone sequences were exported from ARB after applying 50% base pair frequency filters constructed for each major bacterial phylum or subphylum. This was done to account for variability in sequence length of the DGGE band sequences in the different phyla (121 to 158 bp was analyzed, depending on the phylum). DGGE band sequences were assigned to OTUs based on 99% sequence similarity.
Nucleotide sequence accession numbers.
The DNA sequences have been deposited in the GenBank database under accession numbers EU542029 to EU542414.
RESULTS
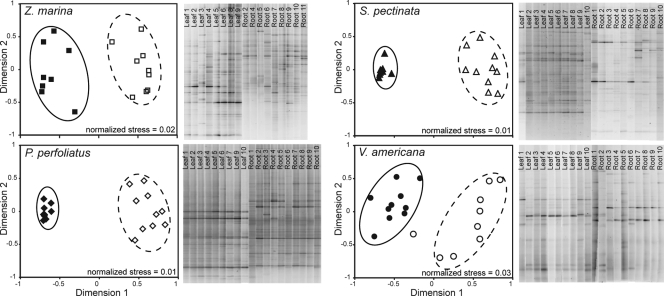
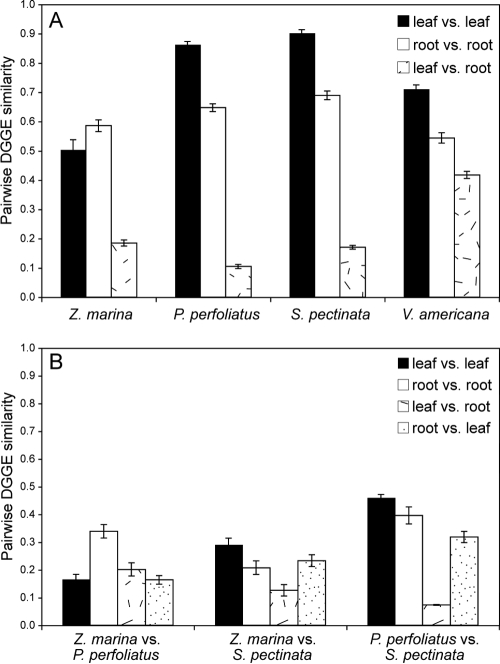
Bacterial communities attached to the surfaces of aquatic angiosperms were similar for replicates of each plant species, suggesting that there was a native, adapted, plant-associated microflora (Fig. 1). Replicate P. perfoliatus and S. pectinata plants hosted very similar, ubiquitous communities of leaf-attached bacteria (average levels of similarity, 86 and 90%, respectively) (Fig. 2A), whereas replicate Z. marina and V. americana plants hosted less similar leaf-attached communities (average levels of similarity, 50 and 71%, respectively). Root-attached communities were also less similar among replicates for each plant species (average levels of similarity ranged from 54 to 69%). In contrast, leaf- and root-attached bacterial communities were very different from one another for most plant species, and the levels of similarity ranged from 11 to 42% (Fig. 1 and 2A).
FIG. 1.
DGGE banding patterns and cluster analyses (multidimensional scaling) of pairwise similarity values (Dice) calculated for each plant species based on the presence or absence of each DGGE band. Filled symbols indicate leaf samples, and open symbols indicate root samples.
FIG. 2.
Average DGGE similarity values (Dice) for pairwise comparisons of DGGE banding patterns within (A) and between (B) plant species. Plants are listed in order from low salinity to high salinity from left to right. Error bars indicate standard errors.
Different plant species hosted different bacterial communities, as demonstrated by very low DGGE similarity values for comparisons of leaf- and root-attached communities of different plant species (Fig. 2B). However, these similarity values were somewhat higher for communities attached to P. perfoliatus and S. pectinata collected from the same bed.
For each plant species and surface type, most bacterial populations represented by DGGE bands could be categorized as ubiquitous (present on all replicates) or rare (present on fewer than one-half of the replicates). Table 2 shows that most populations detected with DGGE were not present on all replicate plants. In fact, for most plants, fewer than 20% of the DGGE bands were found on all replicates. However, the bands found on all replicates were generally the darkest bands in the patterns, suggesting that ubiquitous bacterial populations dominated the attached bacterial communities.
TABLE 2.
Numbers of DGGE bands
| Species | Surface | No. of plants analyzed | Total no. of DGGE bands | No. of ubiquitous DGGE bands | No. of bands found in <5 replicates |
|---|---|---|---|---|---|
| Z. marina | Leaf | 8 | 63 | 8 | 45 |
| Root | 9 | 63 | 4 | 42 | |
| P. perfoliatus | Leaf | 9 | 34 | 15 | 6 |
| Root | 10 | 58 | 11 | 33 | |
| S. pectinata | Leaf | 10 | 30 | 14 | 4 |
| Root | 10 | 40 | 6 | 23 | |
| V. americana | Leaf | 10 | 28 | 4 | 17 |
| Root | 10 | 28 | 2 | 19 |
DNA sequencing of DGGE bands revealed a broad diversity of bacteria attached to plant leaves and roots (Table 3 and Fig. 3). The communities were dominated by Proteobacteria and included members of the Bacteroidetes, Spirochaeta, and Cyanobacteria, and some populations were related to water column and sediment bacteria also found in other estuaries. Two DGGE bands from Z. marina roots (SAV02Z02 and SAV02Z05) and two DGGE bands from Z. marina leaves (SAV01Z07 and SAV01Z08) were closely related to organisms associated with Z. marina roots in a study conducted in Denmark (33), suggesting that there is a cosmopolitan Z. marina-associated microbial community. P. perfoliatus and S. pectinata collected from the same bed shared bacterial taxa belonging to the Deltaproteobacteria, the Burkholderiales and Methylophilales orders of the Betaproteobacteria, and the unicellular algae. V. americana hosted bacteria related to organisms from freshwater systems. Also, the locations of single bands containing plant plastid DNA sequences within the complex DGGE banding patterns were confirmed for three of the four species (all species but Z. marina) through DGGE analysis of DNA extracted from axenic plant tissue cultures.
TABLE 3.
Phylogenetic information for bacterial populations identified by sequencing DNA from DGGE bands, including closest database matches and closest cultivated relatives based on MEGABLAST analysisa
| Band | Plant | Source | Closest BLAST match
|
Closest cultivated relative
|
||||
|---|---|---|---|---|---|---|---|---|
| Phylum or subphylum | Accession no. | % Identity | Taxon | Accession no. | % Identity | |||
| SAV01Z01 | Z. marina | Leaf | Bacteroidetes | AY828430 | 99 | Marine bacterium SCRIPPS_413 | AF359548 | 98 |
| SAV01Z02 | Z. marina | Leaf | Bacteroidetes | AF235111 | 96 | Cytophaga sp. strain RP8 | EU375128 | 96 |
| SAV01Z03 | Z. marina | Leaf | Bacteroidetes | AY678493 | 92 | Lewinella agarilytica | AM286229 | 92 |
| SAV01Z04 | Z. marina | Leaf | Gammaproteobacteria | AF448465 | 97 | Methylotenera mobila | DQ287786 | 96 |
| SAV01Z05 | Z. marina | Leaf | Plastid | EF036309 | 100 | Chloroplast of Yucca schidigera | EU016691 | 100 |
| SAV01Z06 | Z. marina | Leaf | Alphaproteobacteria | DQ659412 | 100 | Rhodobacteraceae sp. strain BS110 | DQ659412 | 100 |
| SAV01Z07 | Z. marina | Leaf | Alphaproteobacteria | DQ436521 | 100 | Thalassobacter sp. strain GA2-M15 | EU342372 | 100 |
| SAV01Z08 | Z. marina | Leaf | Alphaproteobacteria | DQ103624 | 100 | Roseobacter sp. strain CSQ-2 | EF512125 | 99 |
| SAV02Z02 | Z. marina | Root | Epsilonproteobacteria | EF028999 | 100 | Arcobacter nitrofigilis | EU106662 | 100 |
| SAV02Z03 | Z. marina | Root | Gammaproteobacteria | DQ234105 | 96 | Dibenzofuran-degrading bacterium DBF-MAK | AB086228 | 93 |
| SAV02Z04 | Z. marina | Root | Gammaproteobacteria | AY711846 | 100 | Gammaproteobacterium SP5 | AM884361 | 95 |
| SAV02Z05 | Z. marina | Root | Gammaproteobacteria | DQ462284 | 97 | Oceanospirillaceae bacterium JAMM 1548 | AB330882 | 96 |
| SAV02Z06 | Z. marina | Root | Gammaproteobacteria | EF028933 | 100 | Gammaproteobacterium M41 | AM156910 | 96 |
| SAV03Z01 | P. perfoliatus | Leaf | Plastid, diatom | DQ906722 | 96 | Chloroplast of Phaeodactylum tricornutum | EF067920 | 96 |
| SAV03Z02 | P. perfoliatus | Leaf | Betaproteobacteria | AY678527 | 94 | Bacterium HTCC4045 | EF628482 | 93 |
| SAV03Z03 | P. perfoliatus | Leaf | Alphaproteobacteria | AM157616 | 99 | Porphyrobacter tepidarius | AY568467 | 99 |
| SAV03Z04 | P. perfoliatus | Leaf | Betaproteobacteria | DQ064982 | 99 | Betaproteobacterium IMCC1762 | DQ664233 | 96 |
| SAV03Z05 | P. perfoliatus | Leaf | Plastid | EF036309 | 100 | Chloroplast of Yucca schidigera | EU016691 | 100 |
| SAV03Z06 | P. perfoliatus | Leaf | Alphaproteobacteria | AB220125 | 100 | Sphingomonadaceae bacterium PB208 | AB220125 | 100 |
| SAV03Z07 | P. perfoliatus | Leaf | Alphaproteobacteria | EF029025 | 100 | Rhodobacteraceae sp. strain DG1295 | DQ486507 | 100 |
| SAV03Z08 | P. perfoliatus | Leaf | Bacteroidetes | DQ229334 | 90 | Lewinella persicus | EU371935 | 84 |
| SAV04Z01 | P. perfoliatus | Root | Gammaproteobacteria | AF165907 | 97 | Endosymbiont of Lamellibrachia satsuma | AB073120 | 97 |
| SAV04Z02 | P. perfoliatus | Root | Gammaproteobacteria | AY129090 | 97 | Endosymbiont of Lamellibrachia barhami | AY129113 | 97 |
| SAV04Z03 | P. perfoliatus | Root | Bacteroidetes | AF141549 | 98 | Symbiont b-Z43 of Tuber borchii | AF233292 | 90 |
| SAV04Z04 | P. perfoliatus | Root | Betaproteobacteria | AB240458 | 97 | Azoarcus sp. strain CC-11 | AB033745 | 96 |
| SAV04Z05 | P. perfoliatus | Root | Betaproteobacteria | AB264575 | 94 | Azoarcus anaerobius | Y14701 | 93 |
| SAV04Z06 | P. perfoliatus | Root | Plastid | EF036309 | 100 | Chloroplast of Yucca schidigera | EU016691 | 100 |
| SAV04Z07 | P. perfoliatus | Root | Betaproteobacteria | DQ988325 | 100 | Hydrogenophaga sp. strain BAC306 | EU130968 | 100 |
| SAV04Z08 | P. perfoliatus | Root | Betaproteobacteria | AB179685 | 99 | Methyloversatilis universalis | DQ923115 | 93 |
| SAV05Z01 | S. pectinata | Leaf | Bacteroidetes | DQ154800 | 90 | Cytophaga sp. strain RP8 | EU375128 | 86 |
| SAV05Z02 | S. pectinata | Leaf | Bacteroidetes | AY171331 | 93 | Cytophaga sp. strain I-545 | AB073573 | 86 |
| SAV05Z03 | S. pectinata | Leaf | Cyanobacteria | DQ269094 | 97 | Aphanocapsa sp. strain 0MI27S1 | DQ264196 | 96 |
| SAV05Z05 | S. pectinata | Leaf | Cyanobacteria | DQ072927 | 98 | Phormidium sp. strain LMECYA 214 | EU078511 | 96 |
| SAV05Z06 | S. pectinata | Leaf | Betaproteobacteria | DQ064982 | 99 | Betaproteobacterium IMCC1762 | DQ664233 | 96 |
| SAV05Z07 | S. pectinata | Leaf | Bacteroidetes | AY711389 | 92 | Bacteroidetes sp. strain K3 | AM749788 | 88 |
| SAV05Z08 | S. pectinata | Leaf | Alphaproteobacteria | AJ298351 | 99 | Rhodobacteraceae sp. strain DG1295 | DQ486507 | 99 |
| SAV06Z01 | S. pectinata | Root | Spirochaeta | DQ521098 | 94 | Spirochaeta isovalerica | M88720 | 94 |
| SAV06Z02 | S. pectinata | Root | Bacteroidetes | DQ456238 | 96 | Bacteroides uniformis | EU136680 | 94 |
| SAV06Z03 | S. pectinata | Root | Gammaproteobacteria | AF165907 | 96 | Endosymbiont of Lamellibrachia satsuma | AB073120 | 96 |
| SAV06Z04 | S. pectinata | Root | Bacteroidetes | EF028993 | 94 | Marinilabilia salmonicolor | M62422 | 94 |
| SAV06Z06 | S. pectinata | Root | Deltaproteobacteria | AM418397 | 91 | Desulfovibrio pangongensis | AM418397 | 91 |
| SAV06Z07 | S. pectinata | Root | Betaproteobacteria | AB265944 | 99 | Methyloversatilis universalis | DQ923115 | 92 |
| SAV06Z08 | S. pectinata | Root | Betaproteobacteria | AB265944 | 99 | Methyloversatilis universalis | DQ923115 | 92 |
| SAV07Z01 | V. americana | Leaf | Betaproteobacteria | AJ318162 | 96 | Bacterium HTCC4045 | EF628482 | 93 |
| SAV07Z02 | V. americana | Leaf | Betaproteobacteria | DQ315717 | 98 | Leptothrix ginsengisoli | AB271046 | 89 |
| SAV07Z03 | V. americana | Leaf | Plastid, diatom | DQ906722 | 98 | Chloroplast of Phaeodactylum tricornutum | EF067920 | 97 |
| SAV07Z04 | V. americana | Leaf | Betaproteobacteria | AF448465 | 99 | Methylotenera mobila | DQ287786 | 98 |
| SAV07Z05 | V. americana | Leaf | Alphaproteobacteria | EF016501 | 100 | Novosphingobium pentaromativorans | EU167958 | 100 |
| SAV07Z06 | V. americana | Leaf | Bacteroidetes | EF686991 | 93 | Salegentibacter sp. strain BH206 | EF520007 | 91 |
| SAV07Z07 | V. americana | Leaf | Plastid | EF036277 | 96 | Chloroplast of Croomia pauciflora | DQ629458 | 96 |
| SAV07Z08 | V. americana | Leaf | Betaproteobacteria | AY144233 | 96 | Leptothrix mobilis | X97071 | 96 |
| SAV08Z01 | V. americana | Root | Bacteroidetes | AM159414 | 95 | Bacterium HTCC4101 | EF628490 | 87 |
| SAV08Z03 | V. americana | Root | Betaproteobacteria | EF107794 | 97 | Methylophilus sp. strain u33 | EU375653 | 95 |
| SAV08Z04 | V. americana | Root | Betaproteobacteria | EF107794 | 97 | Methylophilus sp. strain u33 | EU375653 | 95 |
| SAV08Z05 | V. americana | Root | Alphaproteobacteria | EF016501 | 100 | Novosphingobium pentaromativorans | EU167958 | 100 |
| SAV08Z06 | V. americana | Root | Plastid, diatom | AY711816 | 100 | Chloroplast of Asterionella glacialis | AJ319828 | 99 |
| SAV08Z07 | V. americana | Root | Plastid | EF036277 | 96 | Chloroplast of Croomia pauciflora | DQ629458 | 96 |
| SAV08Z08 | V. americana | Root | Alphaproteobacteria | EF027003 | 100 | Agrobacterium tumefaciens strain CCBAU | EU256457 | 100 |
See www.ncbi.nlm.nih.gov.
FIG. 3.
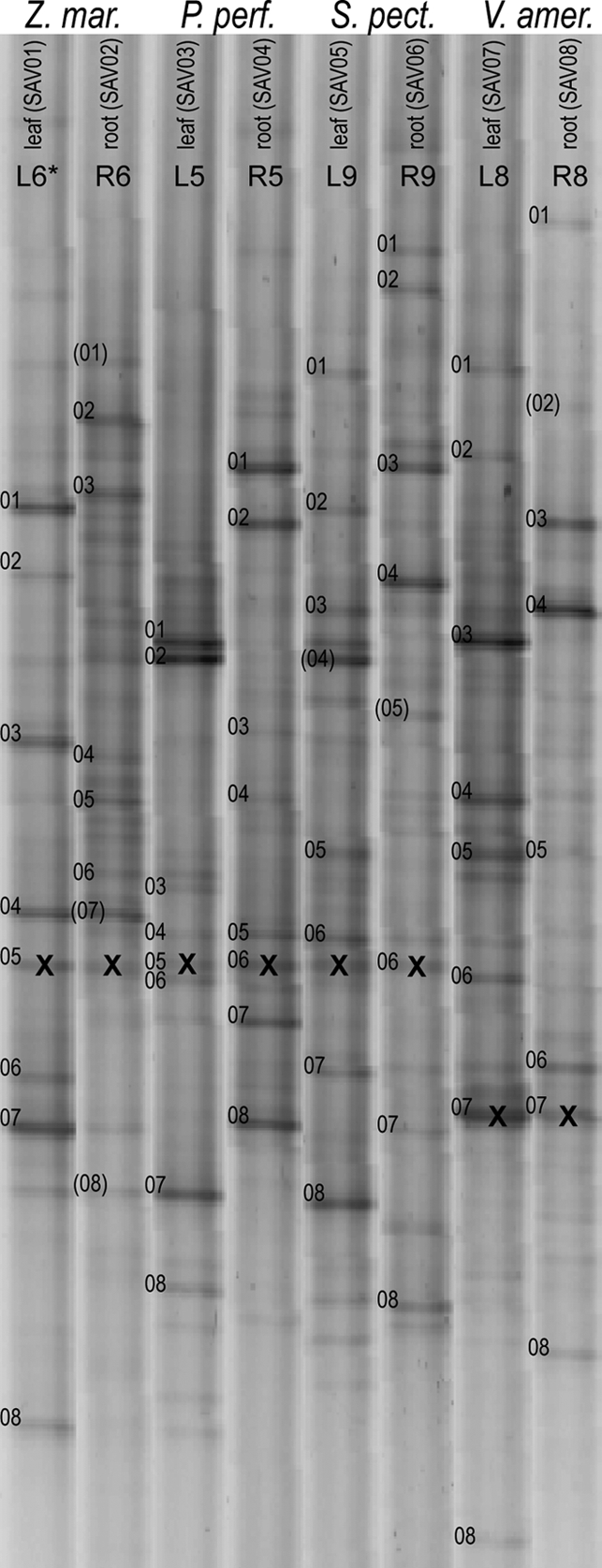
DGGE banding patterns for leaf and root samples of four plant species. The numbered DGGE bands were selected for DNA sequencing. The numbers in parentheses indicate bands that were not successfully sequenced. An X indicates a band for a sequence from a plant chloroplast 16S rRNA gene.
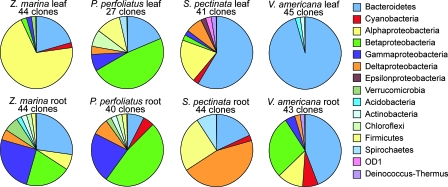
DNA sequencing of PCR-based clone libraries provided another comparison of bacterial communities. Analysis of the DNA sequences demonstrated that there was great variability in diversity between roots and leaves and among plant species (Fig. 4). Z. marina leaves were dominated by typical marine Alphaproteobacteria that were rare or absent elsewhere, while Z. marina roots hosted a diverse microbial assemblage. P. perfoliatus leaves and roots were dominated by several types of Betaproteobacteria, many of which were also found on roots of other plant species in this study. S. pectinata leaves were dominated by several groups of Bacteroidetes, and the roots were dominated by organisms related to sulfate-reducing Deltaproteobacteria. These Deltaproteobacteria were also found on the roots of P. perfoliatus and V. americana. The communities on P. perfoliatus and S. pectinata shared only a few OTUs even though they were collected from the same bed in the Severn River. V. americana leaves were dominated by Bacteroidetes, but these organisms were not closely related to those that dominated S. pectinata leaves. However, many of the Betaproteobacteria on V. americana leaves were similar to those attached to P. perfoliatus leaves. V. americana roots were completely dominated by one large group of Bacteroidetes that were not related to organisms from other plants.
FIG. 4.
Pie diagrams showing the major phylogenetic taxa of bacteria represented in PCR-based clone libraries of 16S rRNA genes from leaf and root samples.
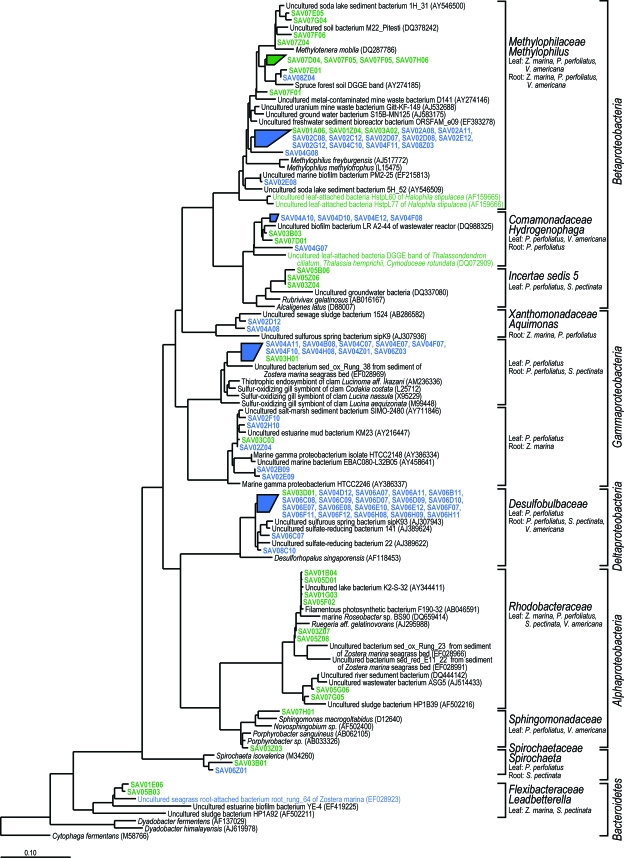
Bacterial 16S rRNA gene sequences collected from all plants clustered into 152 OTUs, 23 of which included both clone library sequences and DGGE band sequences. Most of these OTUs were associated with single plant species, but 12 OTUs included sequences from multiple plant species, comprising a total of 78 clone sequences (24%) and 13 DGGE bands (26%) ( Fig. 5).
FIG. 5.
Bacterial rRNA gene sequences found on more than one plant species arranged in a neighbor-joining tree calculated with the Jukes-Cantor model (Escherichia coli positions 104 to 894; 666 bp), aligned using ARB, and subjected to a 50% base pair frequency filter in which gaps were treated as valid base pairs. Short sequences were added to the neighbor-joining tree with ARB_Parsimony. Phylum, subphylum, family and genus assignments were made with Naive Bayesian rRNA Classifier v. 2.0 of the Ribosomal Database Project using an 80% confidence threshold (70a). Green indicates leaf-attached bacteria, and red indicates root-attached bacteria. Sequences used in this study are indicated by bold type (see Table 3).
DISCUSSION
It is becoming evident that aquatic angiosperms host unique and potentially mutualistic assemblages of microorganisms on their surfaces. Recent research on several aquatic angiosperms demonstrated that root-attached bacteria are different than rhizosphere sediment bacteria (33, 41, 56), and leaf-attached bacteria are unlike typical bacterioplankton (70, 72). A number of different microbial metabolic processes are elevated on plant surfaces, including sulfide oxidation (42), methane oxidation (66), iron reduction (34), sulfate reduction, and nitrogen fixation (54), all of which are influenced by plant activity and some of which are arguably beneficial to the plant. Moreover, aquatic angiosperms produce antimicrobial agents, including zosteric acid, that limit bacterial and fungal colonization of plant surfaces and allow only certain microbes to become established (7, 25, 32, 53). Our study shows that four different species of aquatic angiosperms in the Chesapeake Bay each host species-specific microbial communities attached to root and leaf surfaces and that they share several of the same types of bacteria despite growing under a broad range of environmental conditions.
Leaf-attached bacterial communities and root-attached bacterial communities were very different from one another for most plant species (Fig. 2), which is not surprising given the obvious differences in environmental conditions faced by the microbes (3). Leaf communities included Cyanobacteria and relatives of phototrophic Alphaproteobacteria in the Rhodobacteraceae, as well as close relatives of aerobic organisms, including the alphaproteobacterium Loktanella vestfoldensis and the Bacteroidetes species Fluviicola taffensis and Leadbetterella byssophilla. Root communities included relatives of obligately anaerobic organisms in the Deltaproteobacteria, Spirochaeta, and Clostridiales. However, root- and leaf-attached communities on V. americana plants were more similar based on DGGE banding patterns (average level of similarity, 42%), and although most clones clustered into leaf- and root-specific clusters, two large clusters of root-attached clones included clones from V. americana leaves.
Attached bacterial communities were always more similar within a plant species than across plant species, suggesting that each plant species hosts a unique microbial community. Uku et al. (70) compared leaf-attached bacterial communities from several aquatic plants in two separate beds and found that microbial diversity was linked to plant species rather than study site. Similar results were obtained for rhizosphere soil microbial communities of some terrestrial plants (15, 40). However, in other studies the terrestrial plant-associated microbial diversity was influenced more by soil type (63), plot typology, and local vegetation profile (55) than by individual plant species. Sediment characteristics and other environmental conditions (e.g., salinity) were clearly different for our three sampling locations and may have been responsible for the variability in attached bacterial communities for some plant species. However, P. perfoliatus and S. pectinata were collected from the same bed, and their associated bacterial communities were more similar within species than between species (Fig. 2). This might have resulted from a community shift during the 13 days between the sampling dates for the two plant species, but it may also be explained by the presence of plant-species-specific microbial communities.
Bed morphology appeared to influence the heterogeneity of leaf-attached bacterial communities. DGGE similarity values for replicates of leaf-attached communities of P. perfoliatus and S. pectinata were much higher than those for Z. marina and V. americana (Fig. 2). P. perfoliatus and S. pectinata are canopy-forming species, meaning that they grow all the way to the water surface. These dense beds greatly slow water flow over and through the bed and create a very-low-energy environment in which leaf-attached communities may develop. In contrast, Z. marina and V. americana are meadow-forming species that reach the water surface only when they are reproductive, and, although they reduce the hydrodynamic energy of their environment, their leaves are always exposed to overlying currents and waves. Additionally, our sampling sites for these meadow-forming species were more exposed to wind and waves than the sampling sites for the canopy-forming species. Such conditions allow more water and suspended material to move over and through the beds and may result in greater heterogeneity in leaf-attached bacterial communities.
Despite differences in environmental conditions, bed morphology, overall bacterial community composition, and sample collection date, we identified 12 bacterial OTUs that appeared on more than one plant species. We hypothesized that these ubiquitous populations comprise unique, adapted, and potentially mutualistic communities of plant-attached bacteria. Rare populations may have been part of these adapted communities but may also have been sediment or planktonic bacterial populations that randomly associated with the dominant plant-attached bacterial communities. Bacterial groups associated with more than one plant species were members of the Proteobacteria, Spirochaetes, and Bacteroidetes (Fig. 5). For many of these groups, attachment to plant surfaces seems to be appropriate based on the physiology and metabolism of cultivated relatives. Bacteroidetes related to uncultivated bacteria from Z. marina roots (33) were found attached to leaves of Z. marina and S. pectinata. This bacterial phylum includes many organisms capable of producing a wide array of enzymes for polymer degradation and are commonly found attached to surfaces (58).
Deltaproteobacteria related to the Desulfurobulbaceae were found on roots of P. perfoliatus, S. pectinata, and V. americana. These organisms are obligately anaerobic sulfate reducers and are found in salt marsh rhizospheres (6, 35) and other marine sediments. Sulfate-reducing bacteria are known colonizers of aquatic plant roots (4, 19, 41) and are thought to be responsible for most nitrogen fixation in seagrass and salt marsh sediments (10, 73).
Gammaproteobacteria related to sulfur-oxidizing isolates and to uncultivated bacteria from seagrass sediments (33) were identified on roots of P. perfoliatus and S. pectinata. Sulfur-oxidizing bacteria have been isolated from seagrass roots (41) and may contribute to chemical and plant-mediated detoxification of sulfides produced by sulfate-reducing bacteria (38).
Populations of Roseobacter sp. were identified on the leaves of Z. marina, P. perfoliatus, and S. pectinata. Roseobacter sp. is thought to use a photoheterotrophic metabolism in which heterotrophy is supplemented with a light-harvesting system to produce ATP (62, 67), an ability appropriate for life attached to a plant leaf. This genus of Alphaproteobacteria includes cultivated organisms capable of metabolizing organic sulfur compounds, such as the common algal osmolyte dimethylsulfoniopropionate (22). Organic osmolytes in seagrasses include organic acids, soluble carbohydrates, and amino acids but apparently do not include sulfur-containing compounds (26, 50, 69, 77).
Populations belonging to the order Methylophilales were found on the leaves or roots of Z. marina, P. perfoliatus, and V. americana. Cultivated organisms belonging to this order of Betaproteobacteria metabolize single-carbon organic molecules, such as methanol and methylamine, and some are capable of denitrification (21). Distantly related methylotrophic bacteria belonging to the Alphaproteobacteria, such as Methylobacterium extorquens, are well-known plant-growth-promoting microorganisms for terrestrial plants (1, 27, 46, 59) and are thought to consume methanol released by plants as a by-product of metabolism (45, 52). These bacteria express regulator proteins involved in epiphytic growth (23) and are also capable of producing plant hormones, such as cytokinins, including trans-zeatin (31, 39). It is not known whether methylotrophic Betaproteobacteria play a similar role when they are attached to aquatic angiosperms.
A global effort is under way to restore coastal and estuarine ecosystems, and a major part of this effort is the reestablishment of aquatic angiosperm beds. Aquatic angiosperms have several physiological traits, such as oxygenation of the rhizosphere (60) and production of antimicrobial agents (7), that influence the composition of attached microbial communities and may encourage the growth of mutualistic microbial populations. Such mutualistic relationships are common in terrestrial plants and have been exploited for the development of plant-growth-promoting microorganisms and biocontrol agents to improve crop growth (36, 47, 74) and to aid terrestrial plant restoration (20, 65). A similar approach could be used for aquatic angiosperm restoration efforts. Application of these naturally occurring microbes during propagation and transplantation of aquatic angiosperms may improve the success of aquatic angiosperm restoration.
Acknowledgments
We thank M. Suzuki for determining DNA sequences of DGGE band fragments, B. Severn and A. Bajak for lab and field assistance, E. Kiss for lab assistance, and H. Adams, J. Apple, K. Mielcarek, C. Pratt, and C. Wicks for assistance with field sampling.
This research was supported by grant NA04NMF4570415 from the National Oceanic and Atmospheric Administration Chesapeake Bay Office.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Abanda-Nkpwatt, D., M. Musch, J. Tschiersch, M. Boettner, and W. Schwab. 2006. Molecular interaction between Methylobacterium extorquens and seedlings: growth promotion, methanol consumption, and localization of the methanol emission site. J. Exp. Bot. 57:4025-4032. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulos, C. J., and C. W. Mims. 1979. Introductory mycology, 3rd ed. Wiley & Sons, New York, NY.
- 3.Andrews, J. H., and R. F. Harris. 2000. The ecology and biogeography of microorganisms of plant surfaces. Annu. Rev. Phytopathol. 38:145-180. [DOI] [PubMed] [Google Scholar]
- 4.Bagwell, C. E., J. R. La Rocque, G. W. Smith, S. W. Polson, M. J. Friez, J. W. Longshore, and C. R. Lovell. 2002. Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39:113-119. [DOI] [PubMed] [Google Scholar]
- 5.Bagwell, C. E., Y. M. Piceno, A. Ashburne-Lucas, and C. R. Lovell. 1998. Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl. Environ. Microbiol. 64:4276-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahr, M., B. C. Crump, V. Klepac-Ceraj, A. Teske, M. L. Sogin, and J. E. Hobbie. 2005. Molecular characterization of sulfate-reducing bacteria in a New England salt marsh. Environ. Microbiol. 7:1175-1185. [DOI] [PubMed] [Google Scholar]
- 7.Bushmann, P. J., and M. S. Ailstock. 2006. Antibacterial compounds in estuarine submersed aquatic plants. J. Exp. Mar. Biol. Ecol. 331:41-50. [Google Scholar]
- 8.Caffrey, J. M., and W. M. Kemp. 1990. Nitrogen cycling in sediments with estuarine populations of Potamogeton perfoliatus and Zostera marina. Mar. Ecol. Prog. Ser. 66:147-160. [Google Scholar]
- 9.Capone, D. G. 1983. N2 fixation in seagrass communities. Mar. Technol. Soc. J. 17:32-37. [Google Scholar]
- 10.Capone, D. G. 1982. Nitrogen fixation (acetylene reduction) by rhizosphere sediments of the eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 10:67-75. [Google Scholar]
- 11.Capone, D. G., P. A. Penhale, R. S. Oremland, and B. F. Taylor. 1979. Relationship between productivity and N2 (C2H2) fixation in a Thalassia testudinum community. Limnol. Oceanogr. 24:117-125. [Google Scholar]
- 12.Cifuentes, A., J. Antón, S. Benlloch, A. Donnelly, R. A. Herbert, and F. Rodríguez-Valera. 2000. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl. Environ. Microbiol. 66:1715-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cifuentes, A., J. Anton, R. de Wit, and F. Rodriguez-Valera. 2003. Diversity of Bacteria and Archaea in sulphate-reducing enrichment cultures inoculated from serial dilution of Zostera noltii rhizosphere samples. Environ. Microbiol. 5:754-764. [DOI] [PubMed] [Google Scholar]
- 14.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa, R., M. Gotz, N. Mrotzek, J. Lottmann, G. Berg, and K. Smalla. 2006. Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56:236-249. [DOI] [PubMed] [Google Scholar]
- 16.Crump, B. C., and J. E. Hobbie. 2005. Synchrony and seasonality of bacterioplankton communities in two temperate rivers. Limnol. Oceanogr. 50:1718-1729. [Google Scholar]
- 17.Dey, R., K. K. Pal, D. M. Bhatt, and S. M. Chauhan. 2004. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 159:371-394. [DOI] [PubMed] [Google Scholar]
- 18.Donnelly, A. P., and R. A. Herbert. 1999. Bacterial interactions in the rhizosphere of seagrass communities in shallow coastal lagoons. J. Appl. Microbiol. 85:151S-160S. [DOI] [PubMed] [Google Scholar]
- 19.Finster, K., T. R. Thomsen, and N. B. Ramsing. 2001. Desulfomusa hansenii gen. nov., sp nov., a novel marine propionate-degrading, sulfate-reducing bacterium isolated from Zostera marina roots. Int. J. Syst. Evol. Microbiol. 51:2055-2061. [DOI] [PubMed] [Google Scholar]
- 20.Gemma, J. N., and R. E. Koske. 1997. Arbuscular mycorrhizae in sand dune plants of the north Atlantic Coast of the US: field and greenhouse inoculation and presence of mycorrhizae in planting stock. J. Environ. Manag. 50:251-264. [Google Scholar]
- 21.Ginige, M. P., P. Hugenholtz, H. Daims, M. Wagner, J. Keller, and L. L. Blackall. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourion, B., M. Rossignol, and J. A. Vorholt. 2006. A proteomic study of Methylobacterium extorquens reveals a response regulator essential for epiphytic growth. Proc. Natl. Acad. Sci. USA 103:13186-13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haglund, A. L., E. Tornblom, B. Bostrom, and L. Tranvik. 2002. Large differences in the fraction of active bacteria in plankton, sediments, and biofilm. Microb. Ecol. 43:232-241. [DOI] [PubMed] [Google Scholar]
- 25.Harrison, P. G. 1982. Control of microbial growth and of amphipod grazing by water-soluble compounds from leaves of Zostera marina. Mar. Biol. 67:225-230. [Google Scholar]
- 26.Hasegawa, P. M., R. A. Bressan, J. K. Zhu, and H. J. Bohnert. 2000. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:463-499. [DOI] [PubMed] [Google Scholar]
- 27.Holland, M. A., and J. C. Polacco. 1994. Ppfms and other covert contaminants—is there more to plant physiology than just plant? Annu. Rev. Plant Physiol. Plant Mol. Biol. 45:197-209. [Google Scholar]
- 28.Holmer, M., and L. Laursen. 2002. Effect of shading of Zostera marina (eelgrass) on sulfur cycling in sediments with contrasting organic matter and sulfide pools. J. Exp. Mar. Biol. Ecol. 270:25-37. [Google Scholar]
- 29.Hornschuh, M., R. Grotha, and U. Kutschera. 2006. Moss-associated methylobacteria as phytosymbionts: an experimental study. Naturwissenschaften 93:480-486. [DOI] [PubMed] [Google Scholar]
- 30.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 31.Ivanova, E. G., N. V. Doronina, A. O. Shepelyakovskaya, A. G. Laman, F. A. Brovko, and Y. A. Trotsenko. 2000. Facultative and obligate aerobic methylobacteria synthesize cytokinins. Microbiology 69:646-651. [PubMed] [Google Scholar]
- 32.Jensen, P. R., K. M. Jenkins, D. Porter, and W. Fenical. 1998. Evidence that a new antibiotic flavone glycoside chemically defends the sea grass Thalassia testudinum against zoosporic fungi. Appl. Environ. Microbiol. 64:1490-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen, S. I., M. Kuhl, and A. Prieme. 2007. Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 62:108-117. [DOI] [PubMed] [Google Scholar]
- 34.King, G. M., and M. A. Garey. 1999. Ferric iron reduction by bacteria associated with the roots of freshwater and marine macrophytes. Appl. Environ. Microbiol. 65:4393-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klepac-Ceraj, V., M. Bahr, B. C. Crump, A. P. Teske, J. E. Hobbie, and M. F. Polz. 2004. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6:686-698. [DOI] [PubMed] [Google Scholar]
- 36.Kloepper, J. W., J. Leong, M. Teintze, and M. N. Schroth. 1980. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 286:885-886. [Google Scholar]
- 37.Koch, E. W., and M. J. Durako. 1991. In vitro studies of the submerged angiosperm Ruppia maritima—auxin and cytokinin effects on plant growth and development. Mar. Biol. 110:1-6. [Google Scholar]
- 38.Koch, M. S., S. A. Schopmeyer, M. Holmer, C. J. Madden, and C. Kyhn-Hansen. 2007. Thalassia testudinum response to the interactive stressors hypersalinity, sulfide and hypoxia. Aquat. Bot. 87:104-110. [Google Scholar]
- 39.Koenig, R. L., R. O. Morris, and J. C. Polacco. 2002. tRNA is the source of low-level trans-zeatin production in Methylobacterium spp. J. Bacteriol. 184:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowalchuk, G. A., D. S. Buma, W. de Boer, P. G. L. Klinkhamer, and J. A. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 81:509-520. [DOI] [PubMed] [Google Scholar]
- 41.Kusel, K., T. Trinkwalter, H. L. Drake, and R. Devereux. 2006. Comparative evaluation of anaerobic bacterial communities associated with roots of submerged macrophytes growing in marine or brackish water sediments. J. Exp. Mar. Biol. Ecol. 337:49-58. [Google Scholar]
- 42.Lee, R. W. 1999. Oxidation of sulfide by Spartina alterniflora roots. Limnol. Oceanogr. 44:1155-1159. [Google Scholar]
- 43.Lindow, S. E., and M. T. Brandl. 2003. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipschultz, F., J. J. Cunningham, and J. C. Stevenson. 1979. Nitrogen-fixation associated with 4 species of submerged angiosperms in the central Chesapeake Bay. Estuar. Coast. Mar. Sci. 9:813-818. [Google Scholar]
- 45.MacDonald, R. C., and R. Fall. 1993. Detection of substantial emissions of methanol from plants to the atmosphere. Atmos. Environ. Part A Gen. Top. 27:1709-1713. [Google Scholar]
- 46.Madhaiyan, M., S. Poonguzhali, H. S. Lee, K. Hari, S. P. Sundaram, and T. M. Sa. 2005. Pink-pigmented facultative methylotrophic bacteria accelerate germination, growth and yield of sugarcane clone Co86032 (Saccharum officinarum L.). Biol. Fertil. Soils 41:350-358. [Google Scholar]
- 47.Mayak, S., T. Tirosh, and B. R. Glick. 2004. Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 166:525-530. [Google Scholar]
- 48.Moffler, M. D., and M. J. Durako. 1984. Axenic culture of Thalassia testudinum Banks ex. Konig (Hydrocharitaceae). Am. J. Bot. 71:1455-1460. [Google Scholar]
- 49.Moriarty, D. J. W., R. L. Iverson, and P. C. Pollard. 1986. Exudation of organic carbon by the seagrass Halodule wrightii Aschers and its effect on bacterial growth in the sediment. J. Exp. Mar. Biol. Ecol. 96:115-126. [Google Scholar]
- 50.Murphy, L. R., S. T. Kinsey, and M. J. Durako. 2003. Physiological effects of short-term salinity changes on Ruppia maritima. Aquat. Bot. 75:293-309. [Google Scholar]
- 51.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nemecek-Marshall, M., R. C. MacDonald, F. J. Franzen, C. L. Wojciechowski, and R. Fall. 1995. Methanol emission from leaves—enzymatic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development. Plant Physiol. 108:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newby, B. M. Z., T. Cutright, C. A. Barrios, and Q. W. Xu. 2006. Zosteric acid—an effective antifoulant for reducing fresh water bacterial attachment on coatings. JCT Res. 3:69-76. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen, L. B., K. Finster, D. T. Welsh, A. Donelly, R. A. Herbert, R. de Wit, and B. A. Lomstein. 2001. Sulphate reduction and nitrogen fixation rates associated with roots, rhizomes and sediments from Zostera noltii and Spartina maritima meadows. Environ. Microbiol. 3:63-71. [DOI] [PubMed] [Google Scholar]
- 55.Nunan, N., T. J. Daniell, B. K. Singh, A. Papert, J. W. McNicol, and J. I. Prosser. 2005. Links between plant and rhizoplane bacterial communities in grassland soils, characterized using molecular techniques. Appl. Environ. Microbiol. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pereg, L. L., Y. Lipkin, and N. Sar. 1994. Different niches of the Halophila stipulacea seagrass bed harbor distinct populations of nitrogen-fixing bacteria. Mar. Biol. 119:327-333. [Google Scholar]
- 57.Pollard, P. C., and D. J. W. Moriarty. 1991. Organic-carbon decomposition, primary and bacterial productivity, and sulfate reduction, in tropical seagrass beds of the Gulf of Carpentaria, Australia. Mar. Ecol. Prog. Ser. 69:149-159. [Google Scholar]
- 58.Reichenbach, H. 1989. Genus 1. Cytophaga Winogradsky 1929, 577, (AL) emend, p. 2015-2050. In J. T. Staley, M. P. Bryant, N. Pfenning, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 59.Ryu, J., M. Madhaiyan, S. Poonguzhali, W. Yim, P. Indiragandhi, K. Kim, R. Anandham, J. Yun, K. H. Kim, and T. Sa. 2006. Plant growth substances produced by Methylobacterium spp. and their effect on tomato (Lycopersicon esculentum L.) and red pepper (Capsicum annuum L.) growth. J. Microbiol. Biotechnol. 16:1622-1628. [Google Scholar]
- 60.Sandjensen, K., C. Prahl, and H. Stokholm. 1982. Oxygen release from roots of submerged aquatic macrophytes. Oikos 38:349-354. [Google Scholar]
- 61.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shiba, T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 63.Singh, B. K., S. Munro, J. M. Potts, and P. Millard. 2007. Influence of grass species and soil type on rhizosphere microbial community structure in grassland soils. Appl. Soil Ecol. 36:147-155. [Google Scholar]
- 64.Smith, G. W., S. S. Hayasaka, and G. W. Thayer. 1984. Ammonification of amino acids by the rhizoplane microflora of Zostera marina L. and Halodule wrightii Aschers. Bot. Mar. 27:23-27. [Google Scholar]
- 65.Smith, M. R., I. Charvat, and R. L. Jacobson. 1998. Arbuscular mycorrhizae promote establishment of prairie species in a tallgrass prairie restoration. Can. J. Bot. Rev. Can. Bot. 76:1947-1954. [Google Scholar]
- 66.Sorrell, B. K., M. T. Downes, and C. L. Stanger. 2002. Methanotrophic bacteria and their activity on submerged aquatic macrophytes. Aquat. Bot. 72:107-119. [Google Scholar]
- 67.Swingley, W. D., S. Sadekar, S. D. Mastrian, H. J. Matthies, J. Hao, H. Ramos, C. R. Acharya, A. L. Conrad, H. L. Taylor, L. C. Dejesa, M. K. Shah, M. E. O'Huallachain, M. T. Lince, R. E. Blankenship, J. T. Beatty, and J. W. Touchman. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J. Bacteriol. 189:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tornblom, E., and M. Sondergaard. 1999. Seasonal dynamics of bacterial biomass and production on eelgrass Zostera marina leaves. Mar. Ecol. Prog. Ser. 179:231-240. [Google Scholar]
- 69.Touchette, B. W. 2007. Seagrass-salinity interactions: physiological mechanisms used by submersed marine angiosperms for a life at sea. J. Exp. Mar. Biol. Ecol. 350:194-215. [Google Scholar]
- 70.Uku, J., M. Bjork, B. Bergman, and B. Diez. 2007. Characterization and comparison of prokaryotic epiphytes associated with three East African seagrasses. J. Phycol. 43:768-779. [Google Scholar]
- 70a.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weidner, S., W. Arnold, and A. Puhler. 1996. Diversity of uncultured microorganisms associated with the seagrass Halophila stipulacea estimated by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microbiol. 62:766-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weidner, S., W. Arnold, E. Stackebrandt, and A. Puhler. 2000. Phylogenetic analysis of bacterial communities associated with leaves of the seagrass Halophila stipulacea by a culture-independent small-subunit rRNA gene approach. Microb. Ecol. 39:22-31. [DOI] [PubMed] [Google Scholar]
- 73.Welsh, D. T. 2000. Nitrogen fixation in seagrass meadows: regulation, plant-bacteria interactions and significance to primary productivity. Ecol. Lett. 3:58-71. [Google Scholar]
- 74.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]
- 75.Wigand, C., and J. C. Stevenson. 1997. Facilitation of phosphate assimilation by aquatic mycorrhizae of Vallisneria americana Michx. Hydrobiologia 342:35-41. [Google Scholar]
- 76.Yang, C. H., D. E. Crowley, J. Borneman, and N. T. Keen. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc. Natl. Acad. Sci. USA 98:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye, C. J., and K. F. Zhao. 2003. Osmotically active compounds and their localization in the marine halophyte eelgrass. Biol. Plant. 46:137-140. [Google Scholar]