Abstract
We have previously reported that an acid tolerance response (ATR) can be induced in Streptococcus macedonicus cells at mid-log phase after autoacidification, transient exposure to acidic pH, or acid habituation, as well as at stationary phase. Here, we compared the transcriptional profiles of these epigenetic phenotypes, by RNA arbitrarily primed PCR (RAP-PCR), and their whole-cell chemical compositions, by Fourier transform infrared spectroscopy (FT-IR). RAP-PCR fingerprints revealed significant differences among the phenotypes, indicating that gene expression during the ATR is influenced not only by the growth phase but also by the treatments employed to induce the response. The genes coding for the mannose-specific IID component, the 1,2-diacylglycerol 3-glucosyltransferase, the 3-oxoacyl-acyl carrier protein, the large subunit of carbamoyl-phosphate synthase, and a hypothetical protein were found to be induced at least under some of the acid-adapting conditions. Furthermore, principal component analysis of the second-derivative-transformed FT-IR spectra segregated S. macedonicus phenotypes individually in all spectral regions that are characteristic for major cellular constituents like the polysaccharides of the cell wall, fatty acids of the cell membrane, proteins, and other compounds that absorb in these regions. These findings provide evidence for major changes in cellular composition due to acid adaptation that were clearly different to some extent among the phenotypes. Overall, our data demonstrate the plasticity in the ATR of S. macedonicus, which reflects the inherent ability of the bacterium to adjust the response to the distinctiveness of the imposed stress condition, probably to maximize its adaptability.
The stress physiology of lactic acid bacteria (LAB) has been the target of rigorous investigation in recent years because it reveals the ability of strains to withstand and perform under the fluctuating and harsh conditions of food processes (9, 40). Among the stresses faced by LAB, acid stress is probably the most prominent, as such bacteria continuously acidify their surrounding environment by fermenting carbohydrates into acidic end products (33). Robustness under low-pH conditions is a desirable asset for industrial strains, to ensure their incessant contribution in the fermentation process that would otherwise be abrogated by this self-imposed stress (40). Aciduricity is also necessary for the survival of a strain during its transition through the digestive tract, after consumption, to exert its probiotic effects, if any (25).
Within acid stress physiology, a feature of LAB that has attracted much attention is their ability to develop an acid tolerance response (ATR) (40). The ATR reflects the inherent ability of a bacterium to adapt to a sudden drop in extracellular pH. The experimental setup most commonly used to demonstrate and quantify the ATR of a strain is the following: cells are either transiently exposed to or continuously cultivated at a suboptimal acidic pH value and then acid challenged at an extreme pH value. Even a superficial examination of the related literature reveals that the parameters of such experiments (e.g., the combination of pH values for adaptation and lethal challenge, the duration of the treatment, the medium composition, etc.) are chosen in a rather arbitrary manner. While the development of a unique framework for this type of experiment may be unfeasible, since ATR characteristics are species and, many times, subspecies dependent (9), the current status of information obscures our understanding of the phenomenon. Comparing the behaviors of different strains under acid stress conditions based on the conclusions drawn in different studies is often problematic. Most importantly, it has not yet been clearly established whether the ATR is manifested through an inimitable mechanism for a strain at a specific growth phase or, if not, to what extent the mechanism is influenced by the treatment used to induce the response. Conflicting evidence has been presented about the contribution of the proteins induced by acid stress in the overall response for Lactococcus lactis (20, 33), while the proteomes of acid-adapted Streptococcus mutans cells differed in two separate reports (21, 43). These inconsistencies have been attributed mainly to the different procedures employed to invoke the ATR in each of these studies (16, 43). Furthermore, the type of proteins induced in L. lactis transiently exposed to low pH was influenced by the pH value (16), and many of the genes that were overexpressed after a short- or a long-term acidic shock were found to be treatment specific for Streptococcus pneumoniae (27). All these examples strongly favor the notion that the mechanisms underlying ATR are affected by the distinctiveness of the environment that triggers the adaptation.
We previously assessed the acid stress physiology of Streptococcus macedonicus at the single-cell level, using flow cytometry and cell sorting (31). With this approach, we detected four differently induced acid-tolerant phenotypes that displayed dissimilarities in significant aspects of the ATR (e.g., survival rate, F-ATPase activity, cell wall biosynthesis, etc.). The aim of the current study was to investigate the effects that different treatments had on the ATR of S. macedonicus by using RNA arbitrarily primed PCR (RAP-PCR) and Fourier transform infrared spectroscopy (FT-IR). RAP-PCR has been successfully employed for detecting changes at the transcriptional level in several bacterial species exposed to stressful environments (6, 8, 35), while FT-IR has been shown to be an invaluable tool for the characterization of the global chemical composition of cells (29, 41). The main objective for implementing these approaches was to determine changes in the underlying mechanism of ATR caused by each of the treatments, as well as plausible distinctive traits of the derived phenotypes. Finally, this work reflects our interest in the acid stress physiology of food-related streptococci, since most of the knowledge about the genus stems from research performed with pathogenic strains.
MATERIALS AND METHODS
Microorganism and growth conditions.
The gram-positive bacterium S. macedonicus strain ACA-DC 198, isolated from naturally fermented Greek kasseri cheese (39), was used to study differential gene expression and global cellular biochemical changes under optimal and acid-adapting conditions. The strain was routinely cultured in MRS medium supplemented with 200 mM morpholinepropanesulfonic acid (MRS-MOPS), pH 7.0, at 37°C. The experimental setup was previously described (31). Briefly, control nonadapted cells were grown in MRS-MOPS medium until mid-log phase (approximately 109 cells/ml; optical density at 600 nm [OD600], ∼0.6). For induction of the logarithmic ATR, cells were inoculated in unbuffered MRS medium (pH 7.0) and left to progressively acidify the medium through glycolysis, until mid-log phase (approximately 109 cells/ml; OD600, ∼0.6). Alternatively, mid-log-phase cells grown in MRS-MOPS medium were recovered by centrifugation and transiently exposed in MRS medium (pH 5.5) for 1 h. For acid habituation, cells were grown in MRS medium (pH 6.0) until mid-log phase (approximately 5 × 108 cells/ml; OD600 of ∼0.5). Furthermore, for stationary-phase ATR induction, cells were obtained after final growth for 12 h in MRS-MOPS medium (approximately 1010 cells/ml; OD600, ∼1.2).
RNA extraction and preparation.
Total RNA was isolated with RNeasy and RNAprotect kits (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions. Residual contaminating genomic DNA was removed by on-column digestion with DNase I (Qiagen Inc.) during total RNA isolation. RNA concentration and purity were determined spectrophotometrically by measuring the A260/A280 ratio of a 1:200 dilution in RNase-free water. The integrity of total RNA was assessed electrophoretically.
RAP-PCR.
Basically, RAP-PCR was performed according to the optimized protocol for gram-positive bacterial RNA, as previously described, with some modifications (35). In brief, first-stand cDNA synthesis was performed using 1 μg of heat-denatured (10 min, 70°C) total RNA in reaction mixtures containing 1.0 μM of the arbitrarily chosen primer (5′-CGTATTCATAAGCTTCTCCCGA-3′), 0.5 mM of each deoxynucleoside triphosphate, 40 U of RNasin Plus RNase inhibitor (Promega Corp., Madison, WI), and 1× reverse transcription (RT) reaction buffer. After the mixture was equilibrated at 37°C for 5 min, 4 U of Omniscript reverse transcriptase (Qiagen Inc.) was added to a final volume of 20 μl, and the reaction mixture was incubated at 37°C for 1 h. Subsequently, the reverse transcriptase was heat inactivated at 90°C for 5 min, and the reaction mixture was placed on ice for 10 min. For second-strand synthesis, 10 μl of a 1:10 dilution of the cDNA preparation was used. PCR was performed with Taq PCR Master Mix (Qiagen Inc.) at a final volume of 100 μl, with 1.0 μΜ of the same arbitrarily chosen primer. PCR thermal cycling was performed with one low-stringency cycle of 94°C for 3 min, 37°C for 5 min, and 72°C for 5 min, followed by 39 high-stringency cycles consisting of 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min. RAP-PCR products (8 μl) were separated on a 7% nondenaturing polyacrylamide sequencing gel, and cDNA bands were visualized after SYBR Gold staining (Molecular Probes Inc., Eugene, OR) with a Fluorchem 8800 (Alpha Innotech, CA) image analysis system. Pattern similarity analysis among the RAP-PCR fingerprints of the acid-tolerant S. macedonicus phenotypes and the nonadapted control was performed with GelCompar version 4.0 software (Applied Maths, Kortrijk, Belgium). Analysis was done using Dice's similarity coefficient, and linkage levels were determined using the unweighted-pair group method using average linkages (UPGMA) clustering algorithm.
Isolation, cloning, and sequencing of RAP-PCR products.
Individual DNA bands representing differentially expressed products were excised from the gel. cDNA extraction from the polyacrylamide was performed with a Qiaex II kit (Qiagen Inc.). The eluates obtained were reamplified with a Taq PCR Master Mix and the same primer used for the RAP-PCR. The parameters followed for this reaction were 94°C for 1 min, 47°C for 1 min, and 72°C for 1 min for 35 cycles and a final extension at 72°C for 10 min. Reamplified cDNA fragments were cloned with a TOPO TA cloning kit into Mach1 Escherichia coli T1 cells (Invitrogen, Carlsbad, CA). Plasmids containing the RAP-PCR product of interest were purified with a NucleoSpin Plasmid QuickPure kit (Macherey-Nagel GmbH and Co. KG, Düren, Germany) and sequenced (Biogenomica, Chalandri, Greece). The nucleotide and the deduced amino acid sequences were used for BLASTN and BLASTX similarity searches.
Validation of differential gene expression.
Based on the nucleotide sequence of the differentially expressed RAP-PCR products, new sets of primers specific for each product were designed (Table 1). For the RT-PCR, the Titan one-tube RT-PCR system was used according to the manufacturer's instructions (Roche Diagnostics GmbH, Penzberg, Germany) with 1 μg of total RNA as the template. The absence of contaminating residual genomic DNA was verified in PCRs, using Taq PCR Master Mix and the same amount of RNA. Total RNA from control and acid-adapted cultures was extracted from two independent experiments and served as the template in two separate reactions for each gene. Samples were resolved on 12% nondenaturing polyacrylamide gels in duplicate, and bands were visualized with ethidium bromide. Bands were quantified using Gel-Pro Analyzer software (version 4.0). Analysis of variance (ANOVA) and Tukey's test were applied to determine the significance (P < 0.05) of the differences between the relative intensities of bands, using Statgraphics Centurion software (version XV).
TABLE 1.
Primers used in the RT-PCR experiments
| Primer | Sequence (5′ to 3′) | Target gene product |
|---|---|---|
| PTS_F | AACCTTACTTGCTTCTCAC | Mannose-specific IID component |
| PTS_R | TTGAAGAAAGCTGTGTAAG | |
| RFAG_F | CTATCTCCAAGATTTGAAAG | 1,2-Diacylglycerol 3-glucosyltransferase |
| RFAG_R | GGTCAATCCTTGTGTCTCAC | |
| FAB_F | CTGTTGAAGCTGTTGCTAC | 3-Oxoacyl-acyl carrier protein |
| FAB_R | GCTCTGCTTCTTGACCTTG | |
| CARB_F | CTGGTCTTAATATGGCAATG | Large subunit of carbamoyl-phosphate synthase |
| CARB_R | GTATTGACGATTTCAGATTC | |
| ARM_F | TGGTAGAGTACTAAACTGC | Hypothetical protein involved in acid resistance of S. macedonicus |
| ARM_R | TTACGTCTGTAGAATCGAG | |
| 16SMac_F | GCAGTGGCTTAACCATTGTTCG | 16S RNA subunit |
| 16SMac_R | GCCTAACACCTAGCACTCATCG | |
| 23SMac_F | CCTGGGAAGGTAAGCCAAAGAG | 23S RNA subunit |
| 23SMac_R | CACGGTACTGGTTCACTATCGG |
FT-IR sample preparation and measurements.
Samples of ∼109 cells were removed from control and acid-adapted cultures, washed twice with sterile saline solution (0.9% [wt/vol] ΝaCl), and resuspended in 50 μl of double-distilled water. Ten microliters of the resulting suspension was spotted at the center of an IR-transparent ZnSe optical disc. The samples were further vacuum dried in a desiccator in the presence of anhydrous silica. Bacterial IR spectra were obtained using a Nicolet 6700 spectrometer (Nicolet Instrument Corp., Madison, WI), and measurements were recorded in the range of 4,000 to 500 cm−1, with an interval of 4 cm−1. The final spectrum of each sample was achieved by averaging 80 scans.
FT-IR data analysis.
Before data were analyzed, the spectra were preprocessed, since FT-IR spectral features are often overlapped (41). Spectral processing was performed using Omnic software (Thermo Electron Inc., San Jose, CA). The signal was smoothed and baseline corrected using algorithms built into the software. Furthermore, the second derivative of each spectrum was calculated with the Savitzky-Golay derivative filter and default parameters (12). To compare global biochemical changes among control and differently induced acid-adapted S. macedonicus cells, we examined the spectral regions that offered the maximum information and discriminatory power for characteristic cellular macromolecules (29), independently. In detail, we compared the second derivatives of the FT-IR spectra of the different phenotypes in the region that is typical for polysaccharides of the cell wall (1,200 to 900 cm−1), the mixed region governed by proteins, fatty acids, and other phosphate-carrying compounds (1,500 to 1,200 cm−1), the region influenced by the amide groups from proteins and peptides (1,800 to 1,500 cm−1), and, finally, the region dominated by the fatty acids of the bacterial cell membrane (3,000 to 2,800 cm−1). Data from these regions were exported to Statgraphics Centurion software (version XV) and subjected to the multivariate statistical technique of principal component analysis (PCA). PCA has been widely used in the interpretation of IR spectra, showing whether there are natural clusters in the data and describing similarities or differences from multivariate data sets (18, 24).
Nucleotide sequence accession numbers.
Sequences were deposited in the EMBL nucleotide sequence database under accession numbers AM939571 to AM939575 (Table 2).
TABLE 2.
RAP-PCR products identified to encode proteins
| RAP-PCR producta | Predicted protein (EC no./organism) | No. of identical residues/ total residues (% identity) | E value | EMBL accession no. |
|---|---|---|---|---|
| B1 | Mannose-specific IID component (2.7.1.69/S. mutans UA159) | 82/89 (92) | 6e−43 | AM939571 |
| B2 | 1,2-Diacylglycerol 3-glucosyltransferase (2.4.1.157/S. pyogenes MGAS10750) | 62/81 (76) | 3e−30 | AM939572 |
| B3 | 3-Oxoacyl-acyl carrier protein (2.3.1.41/S. mutans UA159) | 62/73 (84) | 6e−22 | AM939573 |
| B4 | Large subunit of carbamoyl-phosphate synthase (6.3.5.5/S. mutans UA159) | 79/84 (94) | 2e−36 | AM939574 |
| B5 | Hypothetical protein (L. lactis subsp. cremoris SK11) | 20/42 (47) | 0.004 | AM939575 |
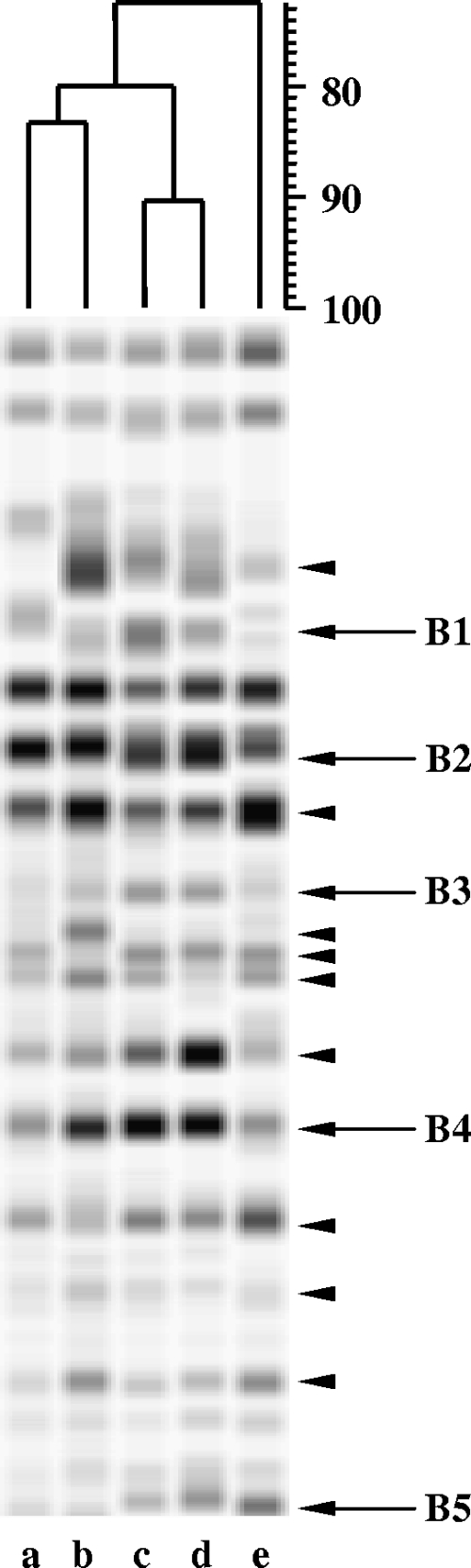
RAP-PCR products (B1 to B5) correspond to the annotation of arrows described in the legend to Fig. 1.
RESULTS
RAP-PCR analysis of S. macedonicus acid-tolerant phenotypes.
To identify changes at the transcriptional level of S. macedonicus cells during acid adaptation, we analyzed four differently induced acid-tolerant phenotypes by using RAP-PCR. Three of the phenotypes were derived from mid-log-phase cells (i.e., after autoacidifying unbuffered medium, being transiently exposed in acidic medium, or becoming acid habituated by continuous growth under acidic conditions), while one phenotype was derived from cells grown until stationary phase (31). Mid-log-phase cells cultivated in buffered medium served as a control. Total RNA extracted from control and acid-adapted cultures was inspected electrophoretically to ensure integrity of rRNA bands and that no overt degradation had occurred during the isolation steps (data not shown). First- and second-strand syntheses were performed according to a RAP-PCR protocol optimized for gram-positive bacteria (35). The arbitrarily chosen primer used in the reaction mixtures had been employed in previously reported RAP-PCR with Streptococcus pyogenes (8). Amplified cDNA samples were resolved by polyacrylamide gel electrophoresis, resulting in typical RAP-PCR fingerprints (Fig. 1).
FIG. 1.
RAP-PCR patterns of total RNA from nonadapted and acid-adapted S. macedonicus cells. The profiles were obtained from nonadapted control cells (lane a), autoacidified cells (lane b), transiently adapted cells (lane c), acid-habituated cells (lane d), and stationary-phase cells (lane e). Bands exhibiting higher abundance than the control bands were excised from the gel, reamplified, cloned, and sequenced. cDNA fragments that were found to correspond to protein-encoding genes (B1 to B5) and internal regions of either 16S or 23S rRNAs are indicated by arrows and arrowheads, respectively. The dendrogram (% similarity) was inferred by the Dice similarity coefficient and the UPGMA clustering algorithm to identify pattern similarities among the RAP-PCR fingerprints.
The relative amount of a specific product amplified by RAP-PCR closely parallels the relative amount of the specific corresponding RNA molecule in a given sample of total RNA (42). In this way, differences in gene expression can be inferred after a comparison of the RAP-PCR fingerprints is performed (35). As expected, the majority of the RAP-PCR products were common throughout the cDNA profiles of all samples analyzed (Fig. 1). Nevertheless, a considerable number of cDNA bands clearly differed in intensity, not only between the RAP-PCR profiles of the control and the acid-tolerant phenotypes but also among the profiles of the acid-tolerant phenotypes themselves. Additionally, a very small number of bands appeared specifically under one or two of the conditions tested and were basically absent from the others. Clustering the RAP-PCR fingerprints clearly segregated the RNA patterns of stationary-phase cells from those of mid-log-phase cells (approximately 72.5% similarity) (Fig. 1). The latter were further separated into two subclusters (approximately 80% similarity), one containing the nonadapted control and the autoacidified cells (approximately 83.3% similarity) and the other containing cells adapted after transient exposure to low pH or after acid habituation (approximately 90.3% similarity). This distinction among the acid-tolerant phenotypes according to their RAP-PCR profiles clearly indicates that the treatments employed to trigger acid adaptation influenced S. macedonicus transcriptional response.
Identification, characterization, and validation of differentially expressed RAP-PCR products.
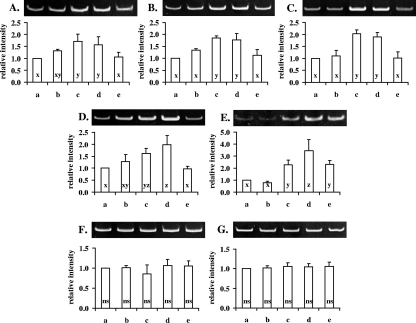
To identify inducible genes of S. macedonicus in response to acid adaptation, distinct bands that exhibited higher abundance or were present solely in the RAP-PCR fingerprints of at least one of the acid-tolerant phenotypes in comparison to that of the control were recovered and reamplified by standard PCR, with the same arbitrary primer as that used in the RAP-PCR. Reamplified bands were subsequently cloned and sequenced. Five out of 14 cDNA fragments corresponded to protein-encoding genes, while the rest were identical with parts of the 16S or 23S rRNA molecules of the species. The proteins deduced from the sequence of the five fragments exhibited significant levels of identity with counterparts of LAB species closely related to S. macedonicus (Table 2). Thus, the genes encoding the mannose-specific IID component, the 1,2-diacylglycerol 3-glucosyltransferase, the 3-oxoacyl-acyl carrier protein, the large subunit of carbamoyl-phosphate synthase, and a hypothetical protein were found to be overexpressed under acid-adapting conditions. Differential expression of all protein-encoding genes was validated by RT-PCR, revealing expression patterns in accordance with those observed in the RAP-PCR fingerprints (Fig. 2). This demonstrates that, indeed, transcriptional changes due to acid adaptation are affected by the treatment employed to trigger the response. As expected, the expression levels of 16S and 23S RNA, as determined by RT-PCR, were found to be equal among all samples tested (Fig. 2), since the upregulation of these transcripts indicated by the RAP-PCR analysis is a typical artifact of the technique due to the high abundance of both molecules in bacterial total RNA extracts (28).
FIG. 2.
Confirmation of differential gene expression by RT-PCR analysis. RT-PCR was performed with RNA extracted from nonadapted or acid-adapted cells, using primers specific for each of the transcripts coding for the mannose-specific IID component (A), the 1,2-diacylglycerol 3-glucosyltransferase (B), the 3-oxoacyl-acyl carrier protein (C), the large subunit of carbamoyl-phosphate synthase (D), a hypothetical protein (E), and 16S (F) and 23S RNA (G). The intensity of each band relative to that of the control (first lane of each gel) was calculated after densitometric analysis. Lanes of each gel are identified in accordance with the columns of the corresponding charts: nonadapted control cells (a), autoacidified cells (b), transiently adapted cells (c), acid-habituated cells (d), and stationary-phase cells (e). The relative intensities of bands of a specific transcript that do not differ significantly (Tukey's test, P < 0.05) are noted with the same letter (x, y, or z). For 16S (F) or 23S RNA (G) transcripts, ANOVA (at P < 0.05) revealed no significant difference (ns) among the relative band intensities.
FT-IR analysis of S. macedonicus acid-tolerant phenotypes.
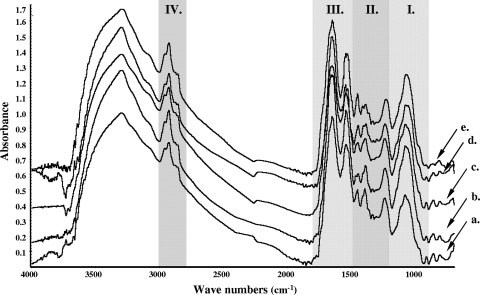
Figure 3 shows the FT-IR spectra (4,000 to 600 cm−1) of the four differently induced acid-tolerant phenotypes and the nonadapted control. The spectrum of S. macedonicus cells was comparable to the spectra reported for other bacteria (11, 24, 29, 41). Strong absorptions were detected in all four spectral regions that characterize the major cellular constituents. In detail, in the first region (Fig. 3, region I, 1,200 to 900 cm−1), the broad band at ∼1,100 to 950 cm−1 arises from the stretching vibrations of the C-O of polysaccharides present within the cell wall. The second region (Fig. 3, region II, 1,500 to 1,200 cm−1), also called the mixed region, is informative mostly for the cell's proteins, fatty acids, and other phosphate-carrying compounds. Three major bands were observed in this region. Absorption at ∼1,453 cm−1 can be assigned to bending of CH2 in lipids or proteins and lipids, while absorption at ∼1,400 cm−1 is due to C(CH3)2 vibrations in proteins or carbohydrates. The peak at ∼1,240 cm−1 is attributed to the asymmetric stretching mode of phosphate groups in phospholipids or nucleic acids. In the third region (Fig. 3, region III, 1,800 to 1,500 cm−1) the vibrations of amide I and II bonds result in two intense peaks at ∼1,650 and ∼1,550 cm−1, respectively, reflecting the proteinaceous content of the cell. Finally, in the fourth region (Fig. 3, region IV, 3,000 to 2,800 cm−1), overlapped bands at around 2,900 cm−1 are assigned to the C-H stretching modes of the cell membrane fatty acids. FT-IR spectra of S. macedonicus exhibited differences among all phenotypes tested (including the nonadapted control) within the four spectral regions.
FIG. 3.
FT-IR spectra of S. macedonicus nonadapted and acid-adapted cells. The spectra are presented in the following order: nonadapted control cells (a), autoacidified cells (b), transiently adapted cells (c), acid-habituated cells (d), and stationary-phase cells (e). Gray-shaded areas mark the characteristic spectral regions in which major constituents of bacterial cells absorb 1,200 to 900 cm−1 (region I); 1,500 to 1,200 cm−1 (region II); 1,800 to 1500 cm−1 (region III); and 3,000 to 2,800 cm−1 (region IV). Each spectrum presented is the average of spectra recorded from 12 independent experiments.
PCA of FT-IR spectra deriving from nonadapted and acid-adapted S. macedonicus cells.
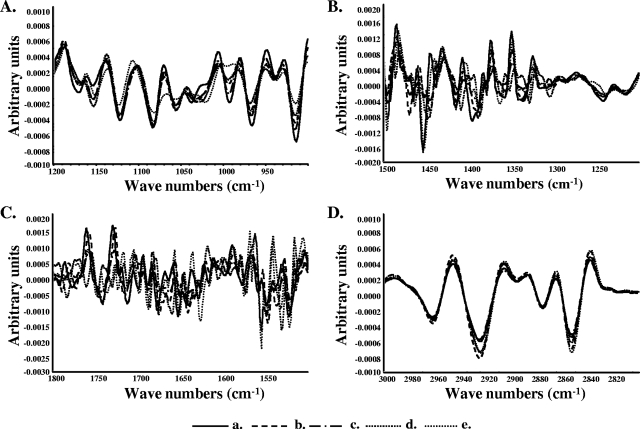
FT-IR spectra of intact bacteria are usually composed of broad and superimposed absorbance bands deriving from the multitude of cellular components, and, thus, most of the information is more or less hidden (41). Figure 4 shows overlaid second-derivative-transformed spectra of S. macedonicus acid-tolerant phenotypes and the nonadapted control for each spectral region. Second-derivative transformation separates absorption bands, removes baseline shifts, and increases apparent spectral resolution (24). Indeed, this type of signal processing made differences between the spectral features of acid-adapted and nonadapted S. macedonicus cells much more prominent.
FIG. 4.
Overlaid second-derivative-transformed spectra in the four characteristic spectral regions (panels A to D, corresponding to regions I to IV) reveal differences in whole-cell chemical composition among the S. macedonicus acid-tolerant phenotypes and that of the nonadapted control. Each transformed spectrum presented is the average of spectra recorded from 12 independent cultures of nonadapted (a), autoacidified (b), transiently adapted (c), acid-habituated (d), and stationary-phase (e) cells.
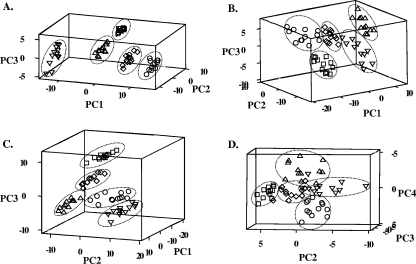
To appreciate the significance of this multivariate data set and to determine whether the method used to induce the ATR influences the overall response, we proceeded with PCA of the second-derivative-transformed spectra. PCA converts the original set of variables to a new set of uncorrelated variables called principal components (PCs) (18). In the resulting plots of the PCs, the distance between points is the measurement for the similarity of the samples. Clear segregations with distinct sample clusters were observed among the acid-tolerant phenotypes and the nonadapted control (Fig. 5). Discrete grouping of samples originating from different treatments in three out of four spectral regions (i.e., I, II, and III) was readily evident within the first three PCs, accounting for 75 to 85% of the variability in the original data. For the fourth spectral region (IV), unambiguous clustering was achieved only after plotting PC2 to PC4 that described 19% of the cumulative variation of the samples. This result indicated subtler differences among the groups of samples in this particular region of the spectrum than among groups in other regions tested. Taken as a whole, PCA revealed unique features of the FT-IR spectra among the acid-tolerant phenotypes and the nonadapted control. Our findings show evidence for not only the presence of significant alterations in major chemical constituents of S. macedonicus cells due to acid adaptation but also a clear dependence of such alterations on the particular method employed to induce the response.
FIG. 5.
PCA of second-derivative-transformed spectra in the four characteristic spectral regions (panels A to D, corresponding to regions I to IV) of S. macedonicus acid-tolerant phenotypes and the nonadapted control. For each of the phenotypes, 12 independent samples were analyzed. In all cases, nonadapted (○), autoacidified (□), transiently adapted (⋄), acid-habituated (▵) and stationary-phase (▿) cells formed distinct groups.
DISCUSSION
We previously reported that an ATR is induced in S. macedonicus cells at mid-log phase after autoacidification, transient exposure to nonlethal low pH, or acid habituation at a suboptimal acidic pH, as well as after the entrance of cells into stationary phase (31). Here, we focused on the direct comparison of these acid-tolerant phenotypes, looking for probable differences at the transcriptional level and the whole-cell chemical composition. Our goal was to elucidate the effect of the growth phase on ATR and, most importantly, to determine whether the response is influenced by the different treatments employed to induce it.
Although the application of RAP-PCR to prokaryotes has been well documented, studies of gram-positive microorganisms are limited (6). RAP-PCR was originally adapted from the differential-display PCR method, and it is a powerful technique for the analysis of transcriptional changes, especially those of organisms for which very little or no genetic information is available (6, 22). Even though more robust or modern techniques have been developed for gene expression profiling, RAP-PCR has in many cases significant advantages (5, 10, 36), and it is still being used as a valid method for many studies performed with different organisms (4, 17, 23, 34, 37). In our case, we employed this technique to determine transcriptional changes of S. macedonicus due to acid adaptation. Differences in the resulting fingerprints were detected among all phenotypes tested. Clustering of the cDNA patterns revealed a clear segregation of stationary-phase cells from all of the mid-log-phase acid-adapted cells, providing evidence that the mechanisms underlying ATR induction are apparently growth phase dependent. Interestingly, the ATR at a particular growth phase was also found to be influenced by the treatment used to invoke it, since transiently adapted and acid-habituated mid-log-phase cells clustered together with a high degree of similarity, though separately from autoacidified cells. This classification of the acid-tolerant phenotypes based on their RAP-PCR fingerprints strongly parallels the survival rates the cells exhibited after undergoing an acidic lethal challenge (31). Stationary-phase cells showed the highest tolerance, followed by the transiently adapted and acid-habituated cells, which were indistinguishable in terms of their survival, and the autoacidified cells with the faintest resistance (31). The divergence of the stationary-phase-induced ATR from that induced at mid-log phase after different manipulations was more or less expected. It is well established that bacteria cells entering the multistress environment of stationary phase cease growth and develop a general stress-resistant state that also confers acid tolerance (40). In LAB, this distinct physiological condition is usually independent of external pH (1, 20, 40), but the auto-acidifying capacity of such microorganisms probably contributes to its induction by accelerating energy depletion (13, 38). On the contrary, acid tolerance is induced at mid-log phase in response to the sole stimulus of low extracellular pH to counteract its negative effects and sustain growth (9, 40). Similar to our findings, both common and unique acid-induced genes, after a short-term or a long-term acidic shock, have been reported for S. pneumoniae (27). Furthermore, the fact that the ATR of autoacidified cells was found to be even less similar to that of the other two mid-log-phase acid-tolerant phenotypes is indicative of the different effect the gradual pH reduction through lactate accumulation has on S. macedonicus cells.
The RAP-PCR approach also allowed us to identify a set of acid stress-responsive genes of S. macedonicus for the first time. The deduced amino acid sequence of the first cDNA band showed the highest similarity (92%) with the IID component of the mannose-specific phosphoenolpyruvate-phosphotransferase system (PEP-PTS) of S. mutans UA159. In most streptococci, there is no PEP-PTS specific for glucose, so the mannose-specific PEP-PTS is responsible for the transport of this sugar (7). The upregulation of this transcript verifies our previous observations in which glucose uptake through PEP-PTS activity was found to be increased in all mid-log-phase acid-tolerant phenotypes in comparison to that of the nonadapted control (31). S. macedonicus during acid adaptation also exhibited higher F-ATPase activity (31). Under these conditions, glucose utilization may be necessary to compensate for the depletion of ATP pools caused by enhanced proton extrusion from the cytoplasm through the F-ATPase. The induction of both enzymatic activities during acid adaptation demonstrates that S. macedonicus responds differently to acidic conditions than the majority of oral streptococci, which typically upregulate F-ATPase and downregulate the glucose-specific PEP-PTS (32).
The second cDNA band showed the highest similarity at the protein level (76%) to 1,2-diacylglycerol 3-glucosyltransferase of S. pyogenes MGAS10750. This enzyme is involved in the biosynthesis of the glycolipid moiety that anchors lipoteichoic acid onto the bacterial cell membrane (30). In S. mutans, inactivation of the dltC gene encoding a protein that catalyzes the incorporation of d-alanine residues into the cell wall-associated lipoteichoic acids resulted in acid sensitivity, reduced ability for ATR induction, and increased proton permeability (3). When these findings are combined, they could indicate a significant role for the teichoic acids in the ATR, since the polyanionic nature of this matrix is not only responsible for the homeostasis of ions but also assists in their trafficking (30), which is clearly disrupted in S. macedonicus cells when they are exposed to acid stress (31).
The deduced amino acid sequence of the third cDNA band showed the highest similarity (84%) with the 3-oxoacyl-acyl carrier protein synthase of S. mutans UA159. This protein is encoded by the fabF gene which belongs to the fab gene cluster that is responsible for fatty acid biosynthesis. In S. mutans, the membrane fatty acid composition is altered during acid adaptation toward an increased proportion of monounsaturated fatty acids, probably to decrease cell membrane permeability to protons (15). The fabM gene product was found to be the key player for that conversion in this microorganism, since a fabM deletion mutant did not synthesize monounsaturated fatty acids and was rendered extremely sensitive to low pH (14). In addition, S. mutans cells treated with the fatty acid biosynthesis inhibitor cerulenin were also unable to alter their membrane fatty acid profiles and survive severe acidification (15). Despite the fact that cerulenin specifically inhibits the FabF activity in an irreversible manner, the contribution of this protein to the observed compositional changes of the cell membrane has not yet been investigated. The upregulation of the fabF transcript under acid-adapting conditions along with our previous observation that cerulenin inhibited ATR induction in S. macedonicus (31), as it did in S. mutans, indicate an important role for FabF in this response. This finding is further supported by the general mechanism of fatty acid biosynthesis proposed for S. pneumoniae, in which competition between FabK and FabF acting downstream of FabM regulates the unsaturated/saturated fatty acids synthesis ratio (26).
The fourth cDNA band showed the highest similarity at the protein level (94%) to the large subunit of the carbamoyl-phosphate synthase (CarB) of S. mutans UA159. Carbamoyl-phosphate is an intermediate in pyrimidine and arginine biosynthesis. In S. pneumoniae, carB along with other genes involved in the first steps of the de novo biosynthesis of pyrimidine nucleotides were found to be upregulated under acid-adapting conditions (27). The enhanced nucleotide synthesis may be necessary to support the repair of damaged DNA that has been demonstrated to take place at low pH (19). Nevertheless, carB induction could also be implicated in the production of arginine, which, after its degradation through the arginine deiminase pathway, releases NH3 in an attempt to alkalize extracellular pH (32).
Finally, the fifth cDNA band was determined to encode a hypothetical protein of L. lactis subsp. cremoris SK11. The function of this protein is unknown, so further investigation is needed.
It should be stated that the number of transcripts coding for proteins that were found to be differentially expressed is well within the average number of genes identified in other studies employing the same technique (6, 22, 35). With the RAP-PCR approach, only a number of genes responsive during acid adaptation of S. macedonicus could be identified, and it is not excluded that the expression of other genes was also influenced. The profile of the RNA fingerprints is affected by many parameters including the amount of total RNA and the conditions chosen for the overall RAP-PCR, as well as the sequence of the arbitrarily chosen primer (35). This is one of the limitations of the RAP-PCR approach, since it provides only a “snapshot” of a portion of the transcriptome. In addition, RT-PCR revealed differences in the expression patterns of the genes mentioned above among the acid-tolerant phenotypes that were in accordance with those observed in the cDNA fingerprints and, thus, validated the RAP-PCR method. Furthermore, changes in the transcriptome of S. macedonicus are very important for the ATR, since we previously demonstrated that actinomycin D and chloramphenicol (inhibitors of transcription and de novo protein synthesis, respectively) critically reduced the ability of the bacterium to induce the response (31).
In a significant number of transcriptomics and proteomics studies concerning the ATR induction in a variety of bacteria, significant changes in the chemical composition of cells have been predicted (9). This assumption is based on the fact that the expression patterns of genes and proteins involved in the central or intermediary metabolism and in the biosynthesis and/or degradation of different macromolecules and smaller molecules are altered during acid adaptation. Only some of these predicted changes have actually been verified directly (15). FT-IR spectroscopy is a physicochemical method that can determine the global chemical features of cells (29). PCA of second-derivative-transformed spectra revealed a clear segregation among the acid-tolerant phenotypes and the nonadapted control of S. macedonicus in all four spectral regions that characterize cellular composition. It is, thus, demonstrated that a wide variety of substances, like polysaccharides of the cell wall, fatty acids of the cell membrane, proteins, nucleic acids, and all other compounds of the cell that may absorb in these spectral regions, undergo significant alterations during acid adaptation. Most importantly, the observed changes were not only growth-phase dependent, but they were also influenced by the specific treatment employed for the ATR induction. Previously, we reported that acid tolerance in S. macedonicus is acquired at the single-cell level due to its intrinsic intrapopulation diversity (31). This could be one strategy employed by the bacterium to increase the likelihood of survival (2). Furthermore, the fact that none of the major mechanisms underlying acid adaptation (i.e., transcriptional changes, de novo protein synthesis, and changes of the cell membrane as well as the cell wall composition) is absolutely necessary for the induction of the response, as we have already demonstrated for S. macedonicus in the past (31), may constitute a second strategy for coping with low pH. The partial redundancy of these mechanisms ensures that acid tolerance is not totally abrogated in case anyone of them is not properly launched. Despite the fact that common aspects were detected among the acid-tolerant phenotypes, indicating that the ATR is likely to evolve around a common theme (at least for the mid-log-phase cells), the results presented in this study, in our opinion, reflect the plasticity in ATR of S. macedonicus in both the mechanisms involved and the end changes they direct. We believe that the observed plasticity may be employed by S. macedonicus as a third strategy that aids survival, since a probable inability of the bacterium to adjust the overall response to the distinctive traits of a specific acidic stress condition would hinder its effort for maximum adaptability.
Here, we elucidated that the ATR in S. macedonicus is influenced by the growth phase of the cells and the treatments employed to induce the response. Differences at the transcriptional level and in the whole-cell chemical composition were detected by RAP-PCR and FT-IR, respectively, among the acid-tolerant phenotypes tested. RAP-PCR also allowed the identification of acid stress-responsive genes that were informative about the ways S. macedonicus attempts to counteract low pH environments. We will continue with the generation of food grade acid-resistant mutants that may have important biotechnological applications.
Acknowledgments
The present work has been cofinanced by the European Social Fund and the National resources EPEAEK and YPEPTH.
Footnotes
Published ahead of print on 8 August 2008.
REFERENCES
- 1.Alemayehu, D., E. O'Sullivan, and S. Condon. 2000. Changes in acid tolerance of Lactococcus lactis during growth at constant pH. Int. J. Food Microbiol. 55:215-221. [DOI] [PubMed] [Google Scholar]
- 2.Booth, I. R. 2002. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 78:19-30. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, I. S., J. L. Groh, M. M. Ramsey, J. D. Ballard, and L. R. Krumholz. 2004. Differential expression of Desulfovibrio vulgaris genes in response to Cu(II) and Hg(II) toxicity. Appl. Environ. Microbiol. 70:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, K. C., B. Komm, N. B. Arnold, and M. Korc. 2007. The application of differential display as a gene profiling tool. Methods Mol. Biol. 383:31-40. [DOI] [PubMed] [Google Scholar]
- 6.Chia, J.-S., Y.-Y. Lee, P.-T. Huang, and J.-Y. Chen. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochu, A., C. Vadeboncoeur, S. Moineau, and M. Frenette. 2003. Genetic and biochemical characterization of the phosphoenolpyruvate:glucose/mannose phosphotransferase system of Streptococcus thermophilus. Appl. Environ. Microbiol. 69:5423-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collin, M., and A. Olsen. 2001. Identification of conditionally expressed genes in Streptococcus pyogenes using RNA fingerprinting. FEMS Microbiol. Lett. 196:123-127. [DOI] [PubMed] [Google Scholar]
- 9.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domachowske, B. J. 2004. Microarrays and gene expression profiling in microbiology and infectious diseases: a clinician's perspective. Clin. Microbiol. Newsl. 26:155-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ede, S. M., L. M. Hafner, and P. M. Fredericks. 2004. Structural changes in the cells of some bacteria during population growth: a Fourier transform infrared-attenuated total reflectance study. Appl. Spectrosc. 58:317-322. [DOI] [PubMed] [Google Scholar]
- 12.Essendoubi, M., D. Toubas, M. Bouzaggou, J. M. Pinon, M. Manfait, and G. D. Sockalingum. 2005. Rapid identification of Candida species by FT-IR microspectroscopy. Biochim. Biophys. Acta 1724:239-247. [DOI] [PubMed] [Google Scholar]
- 13.Even, S., N. D. Lindley, P. Loubiere, and M. Cocaign-Bousquet. 2002. Dynamic response of catabolic pathways to autoacidification in Lactococcus lactis: transcript profiling and stability in relation to metabolic and energetic constraints. Mol. Microbiol. 45:1143-1152. [DOI] [PubMed] [Google Scholar]
- 14.Fozo, E. M., and R. G. Quivey, Jr. 2004. The fabM gene product of Streptococcus mutans is responsible for the synthesis of monounsaturated fatty acids and is necessary for survival at low pH. J. Bacteriol. 186:4152-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fozo, E. M., and R. G. Quivey, Jr. 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frees, D., F. K. Vogensen, and H. Ingmer. 2003. Identification of proteins induced at low pH in Lactococcus lactis. Int. J. Food Microbiol. 87:293-300. [DOI] [PubMed] [Google Scholar]
- 17.Frias-Lopez, J., G. T. Bonheyo, and B. W. Fouke. 2004. Identification of differential gene expression in bacteria associated with coral black band disease by using RNA-arbitrarily primed PCR. Appl. Environ. Microbiol. 70:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodacre, R., E. M. Timmins, R. Burton, N. Kaderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 19.Hanna, M. N., R. J. Ferguson, Y.-H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartke, A., S. Bouche, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. The lactic acid stress response of Lactococcus lactis subsp. lactis. Curr. Microbiol. 33:194-199. [DOI] [PubMed] [Google Scholar]
- 21.Jayaraman, G. C., J. E. Penders, and R. A. Burne. 1997. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25:329-341. [DOI] [PubMed] [Google Scholar]
- 22.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:1152-1161. [DOI] [PubMed] [Google Scholar]
- 23.Le Breton, Y., C. Muller, Y. Auffray, and A. Rince. 2007. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl. Environ. Microbiol. 73:3738-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, M., M. Al-Holy, H. Al-Qadiri, D. H. Kang, A. G. Cavinato, Y. Huang, and B. A. Rasco. 2004. Discrimination of intact and injured Listeria monocytogenes by Fourier transform infrared spectroscopy and principal component analysis. J. Agric. Food Chem. 52:5769-5772. [DOI] [PubMed] [Google Scholar]
- 25.Ljungh, A., and T. Wadstrom. 2006. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 7:73-89. [PubMed] [Google Scholar]
- 26.Lu, Y. J., and C. O. Rock. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59:551-566. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Galiano, A. J., K. Overweg, M. J. Ferrandiz, M. Reuter, J. M. Wells, and A. G. de la Campa. 2005. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151:3935-3946. [DOI] [PubMed] [Google Scholar]
- 28.Nagel, A. C., J. T. Fleming, G. S. Sayler, and K. L. Beattie. 2001. Screening for ribosomal-based false positives following prokaryotic mRNA differential display. BioTechniques 30:988-990, 992, 994-996. [DOI] [PubMed] [Google Scholar]
- 29.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 30.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papadimitriou, K., H. Pratsinis, G. Nebe-von-Caron, D. Kletsas, and E. Tsakalidou. 2007. Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 73:465-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quivey, R. G., W. L. Kuhnert, and K. Hahn. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301-314. [DOI] [PubMed] [Google Scholar]
- 33.Rallu, F., A. Gruss, and E. Maguin. 1996. Lactococcus lactis and stress. Antonie van Leeuwenhoek 70:243-251. [DOI] [PubMed] [Google Scholar]
- 34.Sharma, S., Z. Szele, R. Schilling, J. C. Munch, and M. Schloter. 2006. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shepard, B. D., and M. S. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanton, L. W. 2001. Methods to profile gene expression. Trends Cardiovasc. Med. 11:49-54. [DOI] [PubMed] [Google Scholar]
- 37.Stelzer, S., S. Egan, M. R. Larsen, D. H. Bartlett, and S. Kjelleberg. 2006. Unravelling the role of the ToxR-like transcriptional regulator WmpR in the marine antifouling bacterium Pseudoalteromonas tunicata. Microbiology 152:1385-1394. [DOI] [PubMed] [Google Scholar]
- 38.Svensater, G., O. Bjornsson, and I. R. Hamilton. 2001. Effect of carbon starvation and proteolytic activity on stationary-phase acid tolerance of Streptococcus mutans. Microbiology 147:2971-2979. [DOI] [PubMed] [Google Scholar]
- 39.Tsakalidou, E., E. Zoidou, B. Pot, L. Wassill, W. Ludwig, L. A. Devriese, G. Kalantzopoulos, K. H. Schleifer, and K. Kersters. 1998. Identification of streptococci from Greek Kasseri cheese and description of Streptococcus macedonicus sp. nov. Int. J. Syst. Bacteriol. 48:519-527. [DOI] [PubMed] [Google Scholar]
- 40.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 41.van der Mei, H. C., D. Naumann, and H. J. Busscher. 1993. Grouping of oral streptococcal species using Fourier-transform infrared spectroscopy in comparison with classical microbiological identification. Arch. Oral Biol. 38:1013-1019. [DOI] [PubMed] [Google Scholar]
- 42.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]