Abstract
To provide information on the transmission dynamics of cryptosporidial infections in domestic small ruminants and the potential role of sheep and goats as a source for human cryptosporidiosis, Cryptosporidium-positive isolates from 137 diarrheic lambs and 17 goat kids younger than 21 days of age were examined by using genotyping and subtyping techniques. Fecal specimens were collected between 2004 and 2006 from 71 sheep and 7 goat farms distributed throughout Aragón (northeastern Spain). Cryptosporidium parvum was the only species identified by restriction analyses of PCR products from small-subunit rRNA genes from all 154 microscopy-positive isolates and the sequencing of a subset of 50 isolates. Sequence analyses of the glycoprotein (GP60) gene revealed extensive genetic diversity within the C. parvum strains in a limited geographical area, in which the isolates from lambs exhibited 11 subtypes in two subtype families (IId and IIa) and those from goat kids displayed four subtypes within the family IId. Most isolates (98%) belonged to the subtype family IId, whereas only three isolates belonged to the most widely distributed family, IIa. Three of the four most prevalent subtypes (IIdA17G1a, IIdA19G1, and IIdA18G1) were previously identified in humans, and five subtypes (IIdA14G1, IIdA15G1, IIdA24G1, IIdA25G1, and IIdA26G1) were novel subtypes. All IId subtypes were identical to each other in the nonrepeat region, except for subtypes IIdA17G1b and IIdA22G1, which differed by a single nucleotide polymorphism downstream of the trinucleotide repeats. These findings suggest that lambs and goat kids are an important reservoir of the zoonotic C. parvum subtype family IId for humans.
Cryptosporidium is a protozoan parasite that causes enteric infection in many species of mammals, including humans. The genus is composed of multiple genetically distinct forms, of which C. parvum (cattle genotype, type 2) is the most common zoonotic species (41). Studies of the prevalence of Cryptosporidium in farm animals have revealed that ruminants are an important reservoir for this parasite, although most data are related to cattle. By contrast, the transmission of Cryptosporidium in domestic small ruminants has received comparatively little attention. In spite of this, the parasite is considered one of the major enteric pathogens associated with neonatal diarrhea and mortality in lambs and goat kids (11, 22). These animals represent an agricultural production sector that is even more important than cattle production in many countries. In Spain, the total sheep and goat populations in 2006 were 22.4 and 2.9 million animals, respectively, in comparison to 6.1 million head of cattle (Ministry of Agriculture, Fisheries and Food, http://www.mapa.es).
Likewise, little is known about the role of sheep and goats as zoonotic reservoirs for human cryptosporidiosis. Diarrheic lambs have been reported to excrete as many as 4.8 × 109 oocysts per gram of feces (7), which may contribute significantly to human infections by direct contact or by contamination of water catchment areas. However, the Cryptosporidium species/genotypes infecting small ruminants are not well documented. Molecular analyses have confirmed that C. parvum naturally infects lambs and goat kids (9, 15, 18, 21, 27), but some studies have revealed that sheep are more frequently infected by other apparently host-adapted Cryptosporidium spp./genotypes, mostly C. bovis and the Cryptosporidium cervine genotype, which questions the public health risk of sheep-derived isolates (13, 21, 29, 31). Additionally, a range of Cryptosporidium species (C. suis, C. andersoni, C. hominis) and genotypes (pig genotype II, marsupial genotype) have occasionally been identified in sheep (29).
In addition to the use of molecular methods for species and genotype differentiation, various molecular subtyping tools have recently been developed to characterize the transmission dynamics of Cryptosporidium infections in humans and animals. One of the tools used most commonly is based on a sequence analysis of the 60-kDa glycoprotein (GP60) gene, which enables the identification of subtype families within C. parvum and C. hominis, as well as several subtypes within each family. At least 10 subtype families have been identified among C. parvum isolates from humans and cattle (IIa to IIj), but only IId and especially the most common subtype family, IIa, appear to be zoonotic (1-4, 10, 14, 19, 23-26, 33, 34, 36, 37, 38, 42). The use of such tools for the characterization of ovine and caprine infections, however, has been scarce, and the GP60 subtyping technique has been applied only to modest numbers of specimens from sheep and goats in Italy, the United Kingdom, and Portugal (4, 10, 38). In the current study, the Cryptosporidium species and subtypes responsible for outbreaks of neonatal diarrhea in sheep and goat farms in a region located in the northeast of Spain were characterized. The results obtained were compared with those from other regions to evaluate the geographic distribution and population substructure of C. parvum.
MATERIALS AND METHODS
Sample collection.
Fecal specimens from diarrheic lambs and goat kids up to 21 days old collected between 2004 and 2006 from farms from three provinces across Aragón (northeastern Spain) were used in this study. Whole fecal samples were collected directly from the rectum of the animal by using disposable gloves and plastic containers. A rectal cotton swab was taken in those animals in which freshly voided feces were not recovered. Both fresh feces and cotton swabs were examined for the presence of Cryptosporidium oocysts by microscopy of carbol fuchsin-negative-stained smears, and positive specimens were selected for molecular examination. Altogether, 154 fecal specimens were available for molecular analyses, including 137 specimens from lambs and 17 specimens from goat kids. These animals were randomly selected on 71 sheep and 7 goat farms distributed throughout 45 and 5 municipalities, respectively. One animal was sampled on 30 farms, and between two and six animals were sampled on the remaining 48 farms (mean, 1.97 ± 1.12 animals per farm). On most of the latter farms, specimens were collected during a single diarrheic outbreak, except for 11 sheep farms where two different outbreaks were investigated from 2 to 8 months apart.
DNA extraction.
Oocyst suspensions from whole fecal specimens were prepared as previously described (12). Briefly, oocysts were purified from the feces by using a saturated NaCl solution and centrifugation for 8 min at 1,100 × g (30). The supernatant containing the oocysts was washed with distilled water, and the oocysts were resuspended in 1 ml of distilled water. Oocyst suspensions were stored at 4°C prior to use. Total DNA was extracted from 200-μl oocyst suspensions by three cycles of freezing with liquid nitrogen (1 min) and thawing at 100°C (5 min), followed by incubation at 56°C for 30 min in lysis buffer containing proteinase K and purification over a spin column according to the manufacturer's instructions (QIAamp DNA minikit; Qiagen). Oocysts were removed from rectal swabs by immersing and vigorously rubbing the head of the swab against the wall of a 2-ml microcentrifuge tube containing 1 ml of distilled water. An aliquot of 200 μl of each suspension was used for DNA extraction as described above. DNA extracts were stored at −20°C.
Cryptosporidium species/genotypes differentiation.
Cryptosporidium species and genotypes were determined by nested PCR of a small-subunit (SSU) rRNA gene fragment and restriction fragment length polymorphism (RFLP) analysis with the endonucleases SspI and VspI, using the primers described previously (40). For the primary PCR step, the reaction contained 5 μl of DNA template, 1× PCR buffer, 6 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (dNTP), 0.2 μM of each primer, and 2.5 U of Taq polymerase in a total reaction volume of 50 μl. A total of 35 cycles, each consisting of 94°C for 45 s, 55°C for 45 s, and 72°C for 1 min, were performed, with an initial denaturation of 94°C for 3 min and a final extension step of 72°C for 7 min. The PCR mixture and cycling conditions in the secondary PCR were identical to those used in the primary PCR, except that a concentration of 3 mM MgCl2 and 5 μl of primary PCR product were used. PCR products and restriction fragments were subjected to electrophoresis in 1% or 2% agarose gels, respectively, and visualized by staining with ethidium bromide. A subset of 50 representative positive isolates, including 43 from lambs and 7 from goat kids collected at 36 and 4 farms, respectively, were selected to confirm the RFLP results by DNA sequence analysis. This subset included at least one of each subtype identified at the GP60 locus, as well as the isolates which did not amplify on the GP60 gene.
Subtyping of C. parvum.
Subtyping was achieved by sequence analysis of the GP60 gene, using a protocol described previously (3). A fragment of the GP60 gene (800 to 850 bp) was amplified by a nested PCR with the primers AL3531 (5′-ATA GTC TCC GCT GTA TTC-3′) and AL3535 (5′-GGA AGG AAC GAT GTA TCT-3′) in the primary PCR and with primers AL3532 (5′-TCC GCT GTA TTC TCA GCC-3′) and AL3534 (5′-GCA GAG GAA CCA GCA TC-3′) in the secondary PCR. The PCR mixture consisted of 1 μl of DNA template (for primary PCR) or 1 μl of primary PCR product (for secondary PCR), 1× PCR buffer, 3 mM MgCl2, 200 μM of each dNTP, 0.2 μM of the forward and reverse primers, and 5 U of Taq polymerase in a 50-μl reaction mixture. Each PCR was subjected to 40 cycles consisting of 95°C for 45 s, 52°C for 45 s, and 72°C for 1 min, with an initial denaturation step at 95°C for 3 min and a final extension at 72°C for 10 min. Selected SSU rRNA products, as well as all GP60 products, were purified and sequenced in both directions with the forward and reverse primers used in the secondary PCRs on a MegaBACE 500 sequencer (Amersham Biosciences) according to the manufacturer's instructions. The consensus sequences of both the sense and antisense strands were analyzed using BLASTN searches at the NCBI databases. Subtypes were named based on the number of TCA and TCG repeats in the trinucleotide repeat region and the mutations in the nonrepeat regions, as well as the number of copies of the sequence ACATCA immediately after the repeat region for subtypes belonging to the family IIa (34). For phylogenetic analysis, nucleotide sequences were aligned against each other and with reference sequences retrieved from GenBank with Clustal W and edited with BioEdit version 7.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). A neighbor-joining tree was generated using TreeCon (http://www.psb.rug.ac.be/bioinformatics/psb/Userman/treeconw.html) based on genetic distances calculated by the Kimura two-parameter model. The tree was rooted with a GP60 sequence of C. parvum subtype IIcA5G3a (accession no. AY738195). The reliability of branches was assessed by bootstrap analysis using 1,000 replicates.
Nucleotide sequence accession numbers.
Nucleotide sequences generated in this study were deposited in the GenBank database under accession numbers EU549712 to EU549719.
RESULTS
Cryptosporidium species/genotypes.
PCR products of the SSU rRNA locus were obtained for all 154 Cryptosporidium-positive isolates. Restriction analysis yielded SspI and VspI banding patterns indicative of C. parvum for all PCR products, including visible bands of approximately 108, 254, and 449 bp and 104 and 628 bp, respectively. Sequence analysis of the secondary PCR products of a subset of 50 isolates from both lambs and goat kids confirmed the identification of C. parvum. Two types of C. parvum sequences were found. Thirty-seven specimens showed 100% similarity to the reference sequence AF093490 (39) from a calf in the United States, and 13 specimen sequences were identical to the reference sequence AF308600 (17) from calves experimentally infected with bovine strain KSU-1 in the United States.
GP60 subtypes.
Nested PCR at the GP60 locus yielded products of the expected size for 151 of the 154 specimens analyzed. All these products were sequenced successfully, except for three, which had underlying signals in the electropherogram that prevented the accurate readout of sequences. Alignment of the sequences obtained with reference sequences downloaded from GenBank revealed that most isolates from lambs (n = 128) and all the isolates from goat kids (n = 17) belonged to the C. parvum subtype family IId and only three isolates from lambs were identified as members of subtype family IIa.
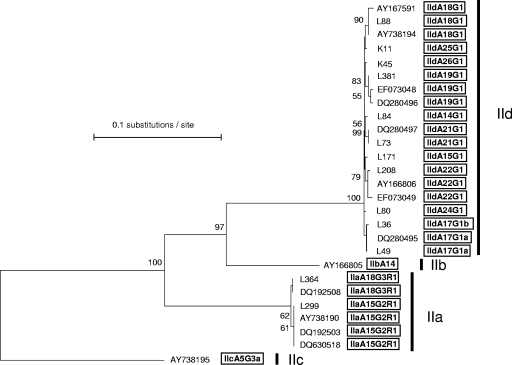
Using the recently proposed nomenclature system (34), we found 11 different subtypes in lambs and 4 subtypes in goat kids. Overall, 11 subtypes within the family IId and 2 subtypes within the family IIa were identified (Table 1). All of the IId subtypes were identical to each other in the nonrepeat region, except for subtypes IIdA17G1b and IIdA22G1, which differed from the remaining subtypes by a single nucleotide polymorphism downstream of the trinucleotide repeats. Namely, subtype IIdA17G1b exhibited a variation from A to G resulting in a change from Asn to Asp in amino acid 57, whereas subtype IIdA22G1 showed a variation from G to A resulting in a change from Gly to Asp in amino acid 78. The remaining subtypes, including those of the family IIa, were homologous with the reference sequences, except for subtypes IIdA14G1, IIdA15G1, IIdA24G1, IIdA25G1, and IIdA26G1, which differed in the number of TCA repeats and thus were novel C. parvum subtypes. The phylogenetic analyses of the GP60 gene showed a close relatedness among the isolates examined in the current study and with some reference isolates previously identified as subtype allele family IId or IIa (Fig. 1).
TABLE 1.
C. parvum GP60 subtype distribution among diarrheic lambs and goat kids younger than 21 days of age in Aragón, Spain
| Subtype | No. of GP60 subtypes in lambs from:
|
No. of GP60 subtypes in goat kids from:
|
||||
|---|---|---|---|---|---|---|
| Samples (n = 131) | Farms (n = 71) | Municipalities (n = 45) | Samples (n = 17) | Farms (n = 7) | Municipalities (n = 5) | |
| IIdA17G1a | 44 | 22 | 18 | 8 | 3 | 3 |
| IIdA17G1b | 26 | 17 | 14 | 0 | ||
| IIdA19G1 | 33 | 20 | 16 | 4 | 3 | 3 |
| IIdA18G1 | 15 | 9 | 5 | 0 | ||
| IIdA14G1 | 2 | 1 | 1 | 0 | ||
| IIdA15G1 | 3 | 1 | 1 | 0 | ||
| IIdA21G1 | 1 | 1 | 1 | 0 | ||
| IIdA22G1 | 2 | 1 | 1 | 0 | ||
| IIdA24G1 | 2 | 1 | 1 | 0 | ||
| IIdA25G1 | 0 | 2 | 1 | 1 | ||
| IIdA26G1 | 0 | 3 | 1 | 1 | ||
| IIaA15G2R1 | 2 | 2 | 2 | 0 | ||
| IIaA18G3R1 | 1 | 1 | 1 | 0 | ||
FIG. 1.
Phylogenetic relationships of the isolates examined in the current study to different C. parvum subtypes as inferred by neighbor-joining analysis of the GP60 gene, based on genetic distances calculated by the Kimura two-parameter model. Bootstrap values over 50% from 1,000 pseudoreplicates are indicated at the left of the supported node.
Frequency and distribution of GP60 subtypes.
Subtypes IIdA17G1a and IIdA19G1 were the most common and widely distributed in both lambs and goat kids, identified in 32% and 29.5% of the farms, respectively, followed by subtypes IIdA17G1b and IIdA18G1, which were seen in 24% and 12.7% of the sheep farms but not were found in goat farms. Other subtypes to one three samples from one or two farms and were identified only in lambs, except for subtypes IIdA25G1 and IIdA26G1, which were restricted to goat kids (Table 1). Most farms (42/48) where two or more animals were analyzed had only one C. parvum subtype, including 8 of the 11 sheep farms where two different diarrheic outbreaks were investigated. Three of the sheep farms where specimens were collected during a single outbreak harbored two different subtypes.
DISCUSSION
The present study was not designed to determine the occurrence of cryptosporidial infections in small ruminants. Nevertheless, the large number of sheep and goat farms where positive specimens were found suggests that Cryptosporidium is a common and widespread pathogen for newborn lambs and goat kids in this geographical area, in agreement with a previous study conducted on sheep farms which showed prevalence rates as high as 82% in diarrheic lambs under 21 days of age (8). Isolates identified in this previous study were suspected to be C. parvum, based on the size of the oocysts, but no molecular data supported this hypothesis. Results of the current molecular analyses reveal that C. parvum is indeed the major species responsible for diarrheic outbreaks on small-ruminant farms in this area of Spain, since it was the only Cryptosporidium species identified by both restriction analyses of all the positive isolates and sequencing of a subset of these isolates.
These findings are consistent with most restricted molecular studies conducted in the United Kingdom, where C. parvum was seen in specimens from both diarrheic and healthy preweaned lambs and goat kids (9, 10, 18, 21, 27). Likewise, C. parvum was identified in limited numbers of isolates from both young and adult sheep and goats from Australia, Cyprus, Spain, the Czech Republic, and Zambia (15, 16, 20, 28). However, our results appear to contrast with those of some reports that question the role of sheep as a zoonotic reservoir for human cryptosporidiosis. These studies revealed that other Cryptosporidium spp. are much more prevalent than C. parvum, although all of the studies were conducted with either asymptomatic neonate or adult sheep, suggesting that the presence of clinical symptoms or the age of the animals may correlate with the Cryptosporidium spp. harbored by small ruminants. In western Australia, Ryan et al. (29) identified eight distinct species/genotypes in isolates from postweaned lambs or adult sheep, and, with the exception of one C. hominis specimen, the majority were isolates not commonly found in humans. Similar results were reported more recently for asymptomatic lambs and ewes from two farms in the United States and the United Kingdom, where most specimens were found to be infected with C. bovis and the Cryptosporidium cervine genotype (13, 31).
The variability of C. parvum subtypes in sheep or goats is not well documented, since no studies with a large sample size have previously been conducted. GP60 typing results have been published to date for only five isolates from lambs in the United Kingdom, two specimens from sheep in Portugal, and one isolate from a goat in Italy (4, 10, 38). The current study showed extensive genetic diversity within C. parvum isolates in a limited geographical area, in which the isolates from lambs exhibited 11 subtypes in two alleles (IId and IIa), and those from goat kids displayed 4 subtypes within the allele IId. The higher parasite diversity seen in specimens from lambs, than in those from goat kids, was probably due to the more extensive sampling of sheep farms. It is worth noting that all subtypes were recovered from diarrheic animals, and thus, all of them appear to be pathogenic to lambs and goat kids. Additionally, most farms where two or more animals were sampled had only one C. parvum subtype circulating in lambs and goat kids, including almost all the farms where two different diarrheic outbreaks were investigated, revealing the endemicity of cryptosporidial infections in small-ruminant farms.
These last observations highlight the usefulness of the GP60 subtyping technique for tracking the infection sources of outbreaks from sheep or goat farms and reveal differences from the transmission dynamics of cryptosporidiosis in cattle. In the United States and Canada, concurrent infections with mixed C. parvum subtypes have been reported to be common on cattle farms as a result of the introduction of new subtypes to the native cattle population through the purchase of calves from local or national markets (25, 37, 42). This practice also results in the greater subtype diversity seen on cattle farms in Turkey (35). By contrast, the majority of lambs or goat kids on Spanish farms are home bred and born indoors at restricted periods of time, resulting in large groups of neonates that are highly susceptible to infection, in close proximity, and frequently overcrowded, which provides ideal conditions for the rapid spread of a single C. parvum subtype from the first lambs to be infected.
Another significant observation was the difference between the distribution of C. parvum subtypes reported for small ruminants and the distribution data reported for cattle. Most of the parasites seen in the current study (98%) belonged to the subtype family IId, found so far only in modest numbers of specimens from humans, cattle, sheep, and water samples in Kuwait, Portugal, Serbia, and Hungary (3-5, 19, 26, 34). By contrast, only three isolates from lambs belonged to the most widely distributed subtype family IIa. Namely, two isolates were subtyped as IIaA15G2R1, reported worldwide to be one of the predominant subtypes in calves and the major subtype responsible for zoonotic cryptosporidiosis (1, 3, 4, 6, 25, 32, 34, 37, 42). This subtype was also previously seen in one sheep in Portugal and three lambs linked to a human infection in the United Kingdom (4, 10). One isolate belonged to the subtype IIaA18G3R1, considered the most prevalent subtype in calves in Northern Ireland and also seen in waterborne outbreaks of human cryptosporidiosis in the same country (14, 36). A molecular study of Cryptosporidium isolates from diarrheic calves in the north of Spain has also revealed the clear dominance of subtype IIaA15G2R1, which was seen in more than 75% of the animals, as well as the presence of subtype IIaA18G3R1 on two farms. It is noteworthy that one of these two farms was located in the same municipality where the lamb infected by subtype IIaA18G3R1 was detected, suggesting the possibility of cross-transmission between sheep and cattle farms or transmission through water (J. Quílez, E. Torres, R. M. Chalmers, G. Robinson, E. del Cacho, and C. Sánchez-Acedo, unpublished data).
The three most prevalent subtypes in the current study, IIdA17G1, IIdA19G1, and IIdA18G1, were previously identified in humans and animals and thus should be considered potential zoonotic subtypes, although mutations of single or various nucleotides downstream of the trinucleotide repeat region of the GP60 gene were seen, compared to some of the reference isolates. Thus, two different sequences were seen within the subtype IIdA17G1, identified in over half of the lambs and goat kids. Two-thirds of these isolates, named subtype IIdA17G1a, demonstrated 100% sequence identity to the Portuguese human isolate CT1, also reported in four cattle and one water sample in the same country (4, 5). By contrast, the subtype designated IIdA17G1b differed from the Portuguese isolates by a single base transition and is reported here for the first time. The sequences of isolates subtyped IIdA19G1 were identical to the sequence of isolate CT15, identified in three human specimens from Portugal, but showed three base transitions in comparison to the sequence of isolate HU-72, recently reported in calves in Hungary (4, 26). Similarly, isolates subtyped IIdA18G1 had 100% identity to the isolate 8897 detected in children in Kuwait but differed from the isolate 42 found in two calves with diarrhea in Serbia and the isolate GI1 from a goat in Italy by one and two nucleotide polymorphisms, respectively (19, 34, 38).
The much less prevalent subtypes within the family IId have not been previously identified anywhere and thus should be considered novel subtypes (IIdA14G1, IIdA15G1, IIdA24G1, IIdA25G1, and IIdA26G1), except for subtype IIdA21G1, which was seen in a sample from a lamb and was identical to the isolate OI4 from one sheep in Portugal and two isolates subtyped IIdA22G1, which differed from the isolate 4759 from a Portuguese human immunodeficiency virus-seropositive patient and the isolate HU-27 from a calf in Hungary by one and three base transitions, respectively (3, 4, 26).
In conclusion, results of the study have revealed the uniqueness of cryptosporidial infections on small-ruminant farms, where the C. parvum subtype family IId is strongly adapted to both lambs and goat kids, which thus seem to be one of the most important reservoirs for this zoonotic group of C. parvum isolates. Nevertheless, the low occurrence of the most common zoonotic allele, IIa, suggests that young sheep and goats do not represent a potential source for human cryptosporidiosis that is as important as other farm animals such as cattle. This is the first published description of Cryptosporidium subtypes detected in animals in Spain, as well as the first large surveillance study about the molecular characterization of Cryptosporidium in small ruminants.
Acknowledgments
This work was supported by funds from Spanish (AGL2004-03233) and regional (DGA-A46) research programs. E. Torres is a fellowship holder of the Diputación General de Aragón.
We thank Guy Robinson, Kristin Elwin, and J. Pedro Bueso for helpful assistance.
Footnotes
Published ahead of print on 11 July 2008.
REFERENCES
- 1.Abe, N., M. Matsubayashi, I. Kimata, and M. Iseki. 2006. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol. Res. 99:303-305. [DOI] [PubMed] [Google Scholar]
- 2.Akiyoshi, D. E., J. K. Tumwine, S. Bakeera-Kitata, and S. Tzipori. 2006. Subtype analysis of Cryptosporidium isolates from children in Uganda. J. Parasitol. 92:1097-1100. [DOI] [PubMed] [Google Scholar]
- 3.Alves, M., L. Xiao, I. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves, M., L. Xiao, F. Antunes, and O. Matos. 2006. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol. Res. 99:287-292. [DOI] [PubMed] [Google Scholar]
- 5.Alves, M., A. M. Ribeiro, C. Neto, E. Ferreira, M. J. Benoliel, F. Antunes, and O. Matos. 2007. Distribution of Cryptosporidium species and subtypes in water samples in Portugal: a preliminary study. J. Eukaryot. Microbiol. 53:S24-S25. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn, B. G., J. M. Mazurek, M. Hlavsa, J. Park, M. Tillapaw, M. Parrish, E. Salehi, W. Franks, E. Koch, F. Smith, L. Xiao, M. Arrowood, V. Hill, A. da Silva, S. Johnston, and J. L. Jones. 2006. Outbreak of cryptosporidiosis associated with consumption of ozonated apple cider. Emerg. Infect. Dis. 12:684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukhari, Z., and H. V. Smith. 1997. Cryptosporidium parvum: oocyst excretion and viability patterns in experimentally infected lambs. Epidemiol. Infect. 119:105-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causapé, A. C., J. Quílez, C. Sánchez-Acedo, E. del Cacho, and F. López-Bernad. 2002. Prevalence and analysis of potential risk factors for Cryptosporidium parvum infection in lambs in Zaragoza (northeastern Spain). Vet. Parasitol. 104:287-298. [DOI] [PubMed] [Google Scholar]
- 9.Chalmers, R. M., K. Elwin, W. J. Reilly, H. Irvine, A. L. Thomas, and P. R. Hunter. 2002. Cryptosporidium in farmed animals: the detection of a novel isolate in sheep. Int. J. Parasitol. 32:21-26. [DOI] [PubMed] [Google Scholar]
- 10.Chalmers, R. M., C. Ferguson, S. Caccio, R. B. Gasser, Y.G Abs El-Osta, L. Heijnen, L. Xiao, K. Elwin, S. Hadfield, M. Sinclair, and M. Stevens. 2005. Direct comparison for selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397-410. [DOI] [PubMed] [Google Scholar]
- 11.de Graaf, D. C., E. Vanopdenbosch, L. M. Ortega-Mora, H. Abbassi, and J. E. Peeters. 1999. A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol. 29:1269-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elwin, K., R. M. Chalmers, R. Roberts, E. C. Guy, and D. P. Casemore. 2001. Modification of a rapid method for the identification of gene-specific polymorphisms in Cryptosporidium parvum and its application to clinical and epidemiological investigations. Appl. Environ. Microbiol. 67:5581-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elwin, K., and R. M. Chalmers. 2008. Contemporary identification of previously reported novel Cryptosporidium isolates reveals Cryptosporidium bovis and the cervine genotype in sheep (Ovis aries). Parasitol. Res. 102:1103-1105. [Google Scholar]
- 14.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. G. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goma, F. Y., T. Geurden, J. Siwila, I. G. K. Phiri, S. Gabriel, E. Claerebout, and J. Vercruysse. 2007. The prevalence and molecular characterisation of Cryptosporidium spp. in small ruminants in Zambia. Small Rumin. Res. 72:77-80. [Google Scholar]
- 16.Hajdušek, O., O. Ditrich, and J. Šlapeta. 2004. Molecular identification of Cryptosporidium spp. in animal and human hosts from the Czech Republic. Vet. Parasitol. 122:183-192. [DOI] [PubMed] [Google Scholar]
- 17.Le Blancq, S. M., N. V. Khramtsov, F. Zamani, S. J. Upton, and T. W. Wu. 1997. Ribosomal RNA gene organization in Cryptosporidium parvum. Mol. Biochem. Parasitol. 90:463-478. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlin, J., C. Amar, S. Pedraza-Díaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misic, Z., and N. Abe. 2007. Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 134:351-358. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, U. M., K. D. Sargent, P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, A. Elliot, and R. C. A. Thompson. 1998. Molecular characterization of Cryptosporidium from various hosts. Parasitology 117:31-37. [DOI] [PubMed] [Google Scholar]
- 21.Mueller-Doblies, D., M. Giles, K. Elwin, R. P. Smith, F. A. Clifton-Hadley, and R. M. Chalmers. 2008. Distribution of Cryptosporidium species and the first description of natural Cryptosporidium bovis infection in sheep in the UK. Vet. Parasitol. 154:214-219. [DOI] [PubMed] [Google Scholar]
- 22.Olson, M. E., B. J. Ralston, R. O'Handley, N. J. Guselle, and A. J. Appelbee. 2003. What is the clinical and zoonotic significance of cryptosporidiosis in domestic animals and wildlife. p. 51-68. In R. C. A. Thompson, A. Armson, and U. M. Ryan (ed.), Cryptosporidium: from molecules to disease. Elsevier B.V., Amsterdam, The Netherlands.
- 23.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 48(Suppl.):28S-31S. [DOI] [PubMed] [Google Scholar]
- 24.Peng, M. M., S. R. Meshnick, N. A. Cunliffe, B. D. Thindwa, C. A. Hart, R. L. Broadhead, and L. Xiao. 2003. Molecular epidemiology of cryptosporidiosis in children in Malawi. J. Eukaryot. Microbiol. 50(Suppl.):557-559. [DOI] [PubMed] [Google Scholar]
- 25.Peng, M. M., M. L. Wilson, R. E. Holland, S. R. Meshnick, A. A. Lal, and L. Xiao. 2003. Genetic diversity of Cryptosporidium spp. in cattle in Michigan: implications for understanding the transmission dynamics. Parasitol. Res. 90:175-180. [DOI] [PubMed] [Google Scholar]
- 26.Plutzer, J., and P. Karanis. 2007. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet. Parasitol. 146:357-362. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard, G. C., J. A. Marshall, M. Giles, R. M. Chalmers, and R. N. Marshall. 2007. Cryptosporidium parvum infection in orphan lambs on a farm open to the public. Vet. Rec. 161:11-14. [DOI] [PubMed] [Google Scholar]
- 28.Ryan, U., C. Read, P. Hawkins, M. Warnecke, P. Swanson, M. Griffith, D. Deere, M. Cunningham, and P. Cox. 2005. Genotypes of Cryptosporidium from Sydney water catchment areas. J. Appl. Microbiol. 98:1221-1229. [DOI] [PubMed] [Google Scholar]
- 29.Ryan, U. M., C. Bath, I. Robertson, C. Read, A. Elliot, L. Mcinnes, R. Traub, and B. Besier. 2005. Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl. Environ. Microbiol. 71:4992-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryley, J. F., R. Meade, J. Hazelhurst, and T. E. Robinson. 1976. Methods in coccidiosis research: separation of oocysts from faeces. Parasitology 73:311-326. [DOI] [PubMed] [Google Scholar]
- 31.Santín, M., J. M. Trout, and R. Fayer. 2007. Prevalence and molecular characterization of Cryptosporidium and Giardia species and genotypes in sheep in Maryland. Vet. Parasitol. 146:17-24. [DOI] [PubMed] [Google Scholar]
- 32.Stantic-Pavlinic, M., L. Xiao, S. Glaberman, A. A. Lal, T. Orazen, A. Rataj-Verglez, J. Logar, and I. Berce. 2003. Cryptosporidiosis associated with animal contacts. Wien. Klin. Wochenschr. 115:125-127. [DOI] [PubMed] [Google Scholar]
- 33.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulaiman, I. M., P. R. Hira, L. Zhou, F. M. Al-Ali, F. A. Al-Shelahi, H. M. Shweiki, J. Iqbal, N. Khalid, and L. Xiao. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanriverdi, S., A. Markovics, O. Arslan, A. Itik, V. Shkap, and G. Widmer. 2006. Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl. Environ. Microbiol. 72:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, H. P., J. S. G. Dooley, J. Kenny, M. McCoy, C. J. Lowery, J. E. Moore, and L. Xiao. 2007. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol. Res. 100:619-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trotz-Williams, L. A., D. S. Martin, W. Gatei, V. Cama, A. S. Peregrine, S. W. Martin, D. V. Nydam, F. Jamieson, and L. Xiao. 2006. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol. Res. 99:346-352. [DOI] [PubMed] [Google Scholar]
- 38.Wu, Z., I. Nagano, T. Boonmars, T. Nakada, and Y. Takahashi. 2003. Intraspecies polymorphism of Cryptosporidium parvum revealed by PCR-restriction fragment polymorphism (RFLP) and RFLP-single-strand conformational polymorphism analyses. Appl. Environ. Microbiol. 69:4720-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao, L., and U. M. Ryan. 2004. Cryptosporidiosis: an update in molecular epidemiology. Curr. Opin. Infect. Dis. 17:483-490. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, L., L. Zhou, M. Santín, W. Yang, and R. Fayer. 2007. Distribution of Cryptosporidium parvum subtypes in calves in eastern United States. Parasitol. Res. 100:701-706. [DOI] [PubMed] [Google Scholar]