Abstract
Despite the importance of salmonellae as one of the major causes of food-borne infections worldwide, data regarding the presence of these organisms in the environment are limited. We investigated the presence of Salmonella spp. in Bahia de Todos Santos (Baja California, Mexico) and evaluated the environmental factors that affect the occurrence of Salmonella spp. in this arid region. A total of 1,331 samples collected from 21 sites along the coast during a period of 3 years were analyzed for Salmonella spp. Geographical and seasonal distribution of Salmonella spp. was evaluated in association with environmental parameters and with human infections in the area. The incidence of Salmonella bacteria throughout the study was 4.8%, with the highest incidence detected in wastewater (16.2%), followed by stream water (10.6%), mollusks (7.4%), and seawater (2.3%). Twenty different serotypes were identified among the 64 Salmonella isolates. The dominant serotype was Typhimurium (23.4%), followed by Vejle (6.2%). The presence of Salmonella spp. in coastal areas was mostly confined to rainy periods and areas of stream discharges, and runoff was identified as the predominant factor influencing the transport of Salmonella bacteria from source points to the sea via streams. Isolation of Salmonella spp. was negatively and significantly associated with temperature, probably because of the effect of solar radiation in the decline of permanence of Salmonella bacteria. Conversely, human infections prevailed during the warmest months and were negatively correlated with the presence of Salmonella spp. in the marine environment.
Salmonellae are one of the major causes of food-borne infections worldwide, associated with the ingestion of contaminated water or food of animal origin. Salmonella comprises more than 2,500 different serotypes included in the Kauffmann-White scheme (27), most of them belonging to Salmonella enterica subspecies enterica. Although all the serotypes are considered potentially pathogenic, only 50 serotypes are dominantly isolated from humans or animals, while the pathogenicity of most of the other infrequent serotypes isolated from the environmental or from clinical sources remains undefined (28). The results of large numbers of studies covering all the biological and epidemiological aspects of the dominant serotypes isolated from humans or animals also contrast with the limited data available regarding the distribution and characteristics of the serotypes present in the environment. This information is essential in assessing the roles of nonhost habitats in the transmission of Salmonella enterica and as natural reservoirs for this organism. The capacity of Salmonella bacteria to survive in the environment has been suggested as a critical step in the Salmonella life cycle, increasing the probability of colonizing new hosts (31).
Salmonella bacteria have been detected in a wide range of environments, such as water (1, 26), sewage effluents (21), and soil, where they can survive for long periods (6). The marine environment has traditionally been considered an adverse habitat for the survival of Salmonella spp., according to the low risk of food-borne illness for this pathogen in association with marine products (24). Nevertheless, Salmonella spp. are commonly isolated from seawater and seafood (1, 3, 10, 11, 19, 26, 30). Detection of enteric bacteria in coastal waters has been linked to periods of sporadic torrential rain discharge, whereby the contamination is transported from the source points to the sea via river water (1, 8, 20, 25). This relationship has been well established in temperate and tropical regions with climates characterized by long periods of heavy rainfall (23). However, there is a complete lack of information about the occurrence of Salmonella bacteria in the coastal areas of dry regions of the world with scarce rains and warm temperatures and where other variables may modulate the presence and permanence of this pathogen in the sea.
In order to assess the dynamics of contamination of Salmonella spp. in arid regions, we conducted a study of the presence of these organisms along the coast of Bahia de Todos Santos (Todos Santos Bay) in Mexico, a country with high rates of reported cases of Salmonella infections (9). Bahia de Todos Santos is located on the Pacific coast of Baja California and is influenced by cold seawater currents that contrast with the elevated temperatures inland. The climatic pattern is notably different from other previously investigated warm areas of the world, such as tropical regions, where the presence of rain and warm seawater provide different environmental conditions for the presence of Salmonella spp. The aim of the present study was to evaluate the environmental factors that influence the pattern of contamination of Salmonella bacteria in the marine environment in a region with high temperatures and scarce rainfall and characterized by the presence of cold seawater, evaluating additionally the association between the pattern of contamination and the rates and dynamics of human infections in the area.
MATERIALS AND METHODS
Study area.
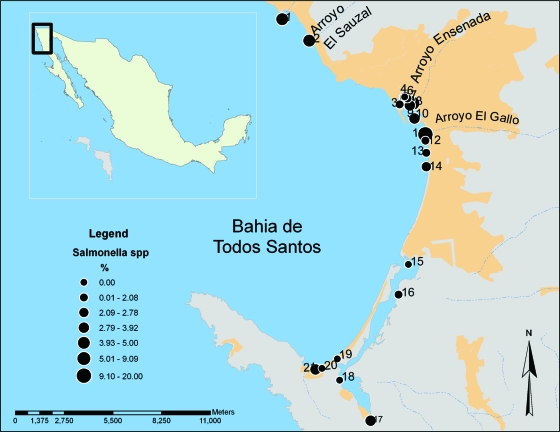
Bahia de Todos Santos (Todos Santos Bay) is located on the Pacific coast of Baja California (Mexico) and covers an area of approximately 180 km2 (Fig. 1). The central area of the bay is occupied by the municipality of Ensenada (a population of 413,481 in 2005 [12]). The climate in Bahia de Todos Santos is characteristically temperate and dry, with an average annual temperature of 18°C (a minimum 12.3°C in March and a maximum of 23.8°C in July) and low annual rainfall values of 250 mm (a monthly average of 10 mm). The rainy season is highly variable, although rains are most frequent in the last weeks of fall and during the winter months. Streams flowing into Bahia de Todos Santos are characteristically temporary, with water present only during the rainy periods. However, the El Gallo stream flows throughout the year, owing to a permanent influx of sewage discharge effluents from one of Esenada's primary treatment plants.
FIG. 1.
Area of study and locations of the sampling stations in Bahia de Todos Santos and spatial distribution of presence of Salmonella species throughout the study period (2004 to 2006) in the different sampling sites according to GIS results.
Sampling program.
A total of 1,331 samples were collected between October 2004 and July 2006 for analysis of Salmonella spp. Samples were collected from 21 fixed sites located along the coast of Bahia de Todos Santos (Fig. 1). Sample stations were selected according to their proximities to streams and sewage treatment discharges and the concentration of humans and livestock in the area. Additionally, random samples of mollusks, water discharges, puddles, and wastewater were collected throughout the area to complement the information obtained from the study of the fixed sites.
Analysis for Salmonella isolates.
Samples were collected in sterile containers and transported to the laboratory under refrigeration. The presence of Salmonella spp. was determined according to the ISO 6579:1993 standard method (13). For liquid samples, 100 ml was filtered through 0.45-μm sterile filters (Millipore Corporation, Bedford, MA); filters were added to 225 ml of buffered peptone water (BPW) (Merck, Darmstadt, Germany). For solid samples, 25 g of sample product was added to 225 ml of BPW (Merck), and mixed for 90 s with a Stomacher homogenizer. Inoculated BPW broths were incubated at 37°C for 20 h. Ten milliliters of preenriched cultures was then transferred to 100 ml of selenite cystine broth (Difco, Detroit, MI), 0.1 ml was transferred to 10 ml of RV10 Rappaport-Vassiliadis broth (Difco), and they were incubated at 37°C and 42°C, respectively, for 24 h. Selective enrichments were streaked onto Hektoen enteric agar (Difco), phenol red-brilliant green agar (Difco), and bismuth sulfite agar (Difco) and incubated at 37°C for 24 h (if only slight growth was observed, the plates were reincubated for an additional 24 h). Typical colonies were selected and streaked onto nutrient agar and subjected to initial biochemical screening in triple sugar iron agar (Difco). Cultures showing a reaction typical of Salmonella bacteria—an alkaline slant and acid butt, with or without production of H2S—were confirmed by biochemical tests on an API-20E strip (bioMérieux, Marcy-l'Etoile, France) and PCR analysis by the amplification of 284 bp of the invA gene according to the protocol of Malorny et al. (16).
Salmonella serotyping.
All Salmonella isolates were serotyped by seroagglutination with commercial antisera (Statens Serum Institut, Copenhagen, Denmark). Polyvalent Salmonella O and H antisera were used to obtain a presumptive diagnosis, and the definitive antigenic designation was then determined with monovalent antisera.
Environmental parameters.
The environmental parameters considered in the study were as follows: temperature, wind, hours of sunshine per day, atmospheric pressure, relative humidity, solar radiation, and rainfall. The daily ambient temperature was taken as the minimum, maximum, and average of the temperature registered in a day. Wind direction was measured as the time in hours that the wind blew in each of the four prevailing quadrants (northwest, northeast, southwest, and southeast) or was measured as no wind (calm). Wind speed was expressed as kilometers per day. Atmospheric pressure was measured in pascals, while relative humidity was expressed as a percentage and solar radiation as Watts per square meter (W/m2). Rainfall was measured as millimeters of precipitation per day. All the variable data except rainfall were obtained from the weather station of the Instituto de Investigaciones Oceanlogicas of the University Autonoma of Baja California, located in Ensenada (116°39′58″W and 31°51′46″N). Rainfall data were provided by the Comision Nacional de Agua and were collected from the weather station of Emilio Lopez Zamora. (116°36′12″W and 31°53′29″N).
Additionally, data for discharges from the sewage treatment plants (liters/s) provided by the Comision Estatal de Servicios Publicos de Ensenada were obtained from the facility of Naranjo-Gallo. This facility discharges its effluents in close proximity to the sea through the El Gallo stream.
Human infections.
Information about the number of cases of human nontyphoid and typhoid Salmonella spp. infections recorded in Baja California was inferred from the records reported in the weekly Epidemiological Bulletin published by the Sistema Unico de Información para la Vigilancia Epidemiológica of the Secretaría de Salud Publica in Mexico (Public Health Department) (5).
Spatial analysis.
Results of the analyses were processed with the Geographical Information System (GIS) software, ArcGIS version 9.1, and the extension Spatial Analyst by ESRI. The formats of the data were Shapefile (vector data) and GRID (raster data) by ESRI, and the vector data source was the Instituto Nacional de Estadistica Geografica e Informatica of Mexico (INEGI).
Statistical analysis.
The differences in the frequencies of Salmonella spp. present at different sites and periods were evaluated by the chi-square and Fisher's exact tests.
The associations between environmental factors and presence of Salmonella spp. were initially analyzed by Pearson correlation coefficients with weekly average values of incidence. Relationships between the presence of Salmonella spp. and each of the environmental parameters included in the study were initially surveyed by simple logistic regression analysis. Once the significant variables at an individual level were selected, a multiple logistic regression model was then conducted. Predicted probability and odds ratios were estimated by logistic regression analysis. The odds ratio is defined as the predicted change in odds for a unit increase in the corresponding independent variable.
All statistical analyses were carried out with SPSS version 14.0.1 (SPSS Inc.), and the level of significance was set at a P value of <0.05.
RESULTS
The overall incidence of Salmonella bacteria in Bahia de Todos Santos throughout the period of study (2004 to 2006) was 4.8%, with a total of 64 positive samples of the 1,331 analyzed (Table 1). The highest incidence of Salmonella bacteria was detected in wastewater, with 11 positive samples of the 68 analyzed (16.2%); followed by stream water, with 23 positive samples (10.6%); mollusks, with 5 positive samples (7.4%); and seawater, with 22 positive samples (2.3%). The analysis of a small number of samples of puddle water revealed the presence of Salmonella bacteria in 17.6% of the samples. Puddle water, wastewater, and stream water showed a significantly higher incidence of Salmonella bacteria than the other groups analyzed (P < 0.05).
TABLE 1.
Number of samples and incidence of Salmonella spp. throughout the period of study per type of sample
| Yr | Mo | No. of samples (% positive for Salmonella spp.)
|
Total (%) | ||||
|---|---|---|---|---|---|---|---|
| Stream water | Seawater | Sewage water | Mollusks | Puddles | |||
| 2004 | October | 0 | 1 (0) | 14 (0) | 0 | 0 | 15 (0) |
| November | 24 (8.3) | 69 (0) | 4 (0) | 11 (0) | 0 | 108 (1.8) | |
| December | 8 (0) | 32 (0) | 0 | 8 (0) | 0 | 48 (0) | |
| Total | 32 (6.3) | 102 (0) | 18 (0) | 19 (0) | 0 | 171 (1.2) | |
| 2005 | January | 25 (20) | 78 (7.7) | 3 (60) | 6 (16.7) | 0 | 114 (13.2) |
| February | 12 (0) | 31 (0) | 3 (0) | 2 (0) | 0 | 48 (0) | |
| March | 0 | 0 | 0 | 0 | 0 | 0 | |
| April | 12 (8.3) | 46 (4.3) | 1 (100) | 0 | 0 | 59 (14.8) | |
| May | 3 (0) | 16 (0) | 0 | 0 | 0 | 19 (0) | |
| June | 20 (5) | 90 (2.2) | 4 (25) | 0 | 0 | 114 (3.5) | |
| July | 8 (0) | 39 (0) | 1 (100) | 0 | 1 (0) | 49 (2) | |
| August | 9 (11.1) | 60 (3.3) | 2 (0) | 0 | 0 | 71 (4.2) | |
| September | 11 (0) | 79 (0) | 1 (0) | 0 | 0 | 91 (0) | |
| October | 13 (7.7) | 73 (0) | 1 (100) | 0 | 0 | 87 (2.3) | |
| November | 8 (12.5) | 34 (0) | 5 (20) | 0 | 0 | 47 (4.2) | |
| December | 4 (0) | 18 (0) | 0 | 4 (0) | 0 | 26 (0) | |
| Total | 125 (8) | 564 (2.1) | 23 (34.8) | 12 (8.3) | 1 (0) | 725 (4.3) | |
| 2006 | January | 8 (50) | 33 (0) | 4 (25) | 0 | 0 | 45 (11.1) |
| February | 14 (7.15) | 52 (0) | 10 (20) | 11 (0) | 11 (9.1) | 98 (3.1) | |
| March | 12 (16.6) | 63 (12.7) | 5 (0) | 18 (22.2) | 2 (0) | 100 (14) | |
| April | 9 (22.2) | 33 (6) | 2 (50) | 6 (0) | 3 (66.7) | 53 (13.2) | |
| May | 7 (28.6) | 52 (0) | 1 (0) | 2 (0) | 0 | 62 (3.2) | |
| June | 6 (0) | 46 (0) | 2 (0) | 0 | 0 | 54 (0) | |
| July | 3 (0) | 17 (0) | 3 (0) | 0 | 0 | 23 (0) | |
| Total | 59 (18.6) | 296 (3.4) | 27 (11.1) | 37 (10.8) | 16 (18.75) | 435 (7.1) | |
| Total | 216 (10.6) | 962 (2.3) | 68 (16.2) | 68 (7.4) | 17 (17.6) | 1,331 (4.8) | |
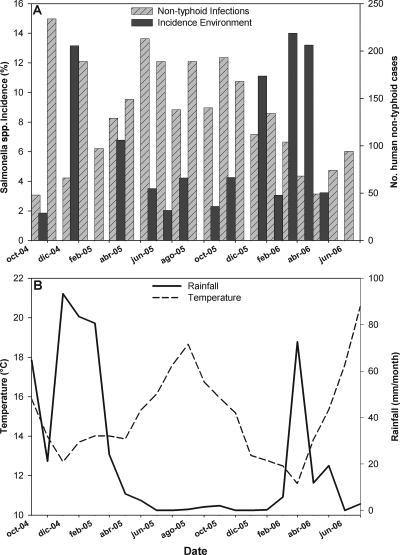
The incidence of Salmonella bacteria in the 171 samples investigated between October and December 2004 was 1.2% (Table 1), which was significantly lower than that of the other years of study. In 2005, Salmonella bacteria were detected in 31 samples of the 725 investigated (4.3%), whereas the incidence of Salmonella bacteria in 2006 was significantly higher than in other years, with 31 positive samples from a total of 435 samples (7.1%) obtained between January and July. The month with the highest incidence of Salmonella bacteria was April 2005 (Table 1; Fig. 2) when 8 of 59 samples (14.8%) tested positive. Significantly greater differences (P < 0.05) were also observed in January 2005 (13.2%) and March 2006 (14%). A high presence of Salmonella bacteria (13.2%) was also observed in samples in April 2006, although it was not significantly higher than at other sampling times.
FIG. 2.
Monthly distribution of incidence of Salmonella species in Bahia de Todos los Santos and human infections of Salmonella reported in Baja California throughout the 3-year study (A) and variations in rainfall and temperature throughout the same period (B). oct, October; dic, December; feb, February; abr, April; jun, June; ago, August.
Results obtained from the 21 fixed sample stations located along the coast of Bahia de Todos Santos (Table 2) over the period of study showed a significantly higher incidence (P < 0.05) of Salmonella bacteria at sites 11 (Arroyo El Gallo) and 8 (Arroyo Ensenada), with 11 (20%) and 5 (9.3%) positive samples, respectively. The site with the second-highest presence of Salmonella bacteria was Arroyo Sauzal (sampling station 2), with an incidence of 5% in the 40 samples investigated. An association between the presence of Salmonella spp. and areas of stream discharge was also observed by GIS analysis (Fig. 1). Salmonella bacteria were detected mostly in close proximity to streams, and the incidence decreased gradually as the distance from the point increased.
TABLE 2.
Number of samples analyzed and Salmonella incidence per year and sampling site
| Sampling site | No. of samples (% positive for Salmonella spp.)
|
Total
|
|||
|---|---|---|---|---|---|
| 2004 | 2005 | 2006 | No. of samples | No. of samples positive for Salmonella spp. (%) | |
| 1 | 7 (0) | 30 (6.7) | 10 (0) | 47 | 2 (4.3) |
| 2 | 12 (0) | 22 (0) | 6 (33.3) | 40 | 2 (5) |
| 3 | 5 (0) | 32 (3.1) | 18 (0) | 55 | 1 (1.8) |
| 4 | 7 (0) | 31 (0) | 17 (0) | 55 | 0 (0) |
| 5 | 7 (0) | 32 (3.1) | 18 (5.6) | 57 | 2 (3.5) |
| 6 | 7 (0) | 23 (0) | 18 (5.6) | 48 | 1 (2.1) |
| 7 | 3 (0) | 33 (6.1) | 18 (0) | 54 | 2 (3.7) |
| 8 | 7 (14.3) | 32 (9.4) | 16 (6.3) | 55 | 5 (9.1) |
| 9 | 5 (0) | 32 (0) | 17 (11.8) | 54 | 2 (3.7) |
| 10 | 5 (0) | 32 (6.3) | 18 (0) | 55 | 2 (3.6) |
| 11 | 7 (14.3) | 31 (16.1) | 17 (29.4) | 55 | 11 (20) |
| 12 | 7 (0) | 31 (3.2) | 17 (0) | 55 | 1 (1.8) |
| 13 | 7 (0) | 32 (0) | 17 (6) | 56 | 1 (1.8) |
| 14 | 19 (0) | 17 (6) | 36 | 1 (2.8) | |
| 15 | 7 (0) | 32 (0) | 17 (0) | 56 | 0 (0) |
| 16 | 6 (0) | 31 (3.2) | 16 (0) | 53 | 1 (1.9) |
| 17 | 7 (0) | 27 (0) | 17 (11.8) | 51 | 2 (4) |
| 18 | 7 (0) | 6 (0) | 1 (0) | 14 | 0 (0) |
| 19 | 23 (0) | 18 (0) | 41 | 0 (0) | |
| 20 | 8 (0) | 25 (0) | 17 (0) | 50 | 0 (0) |
| 21 | 7 (0) | 32 (3.1) | 17 (6) | 56 | 2 (3.6) |
| Other | 43 (0) | 137 (8.8) | 108 (13) | 288 | 26 (9.1) |
| Total | 171 (1.2) | 725 (4.3) | 435 (7.1) | 1,331 | 64 (4.8) |
Only 45 of the 64 Salmonella isolates obtained throughout the study were able to be characterized by serotyping (Table 3). Twenty different serotypes were identified among the 45 isolates. The dominant serotype was Salmonella enterica serotype Typhimurium, identified in 15 isolates (23.4%) obtained primarily from stream water samples. The remaining isolates showed highly diverse serovar profiles, with 19 different serotypes determined in 30 isolates. The serotypes Vejle (four isolates), Suberu (three isolates), and Urbana (three isolates) were the most abundant in this group. The 19 untyped isolates showed a rugose phenotype and originated mainly from marine environments.
TABLE 3.
Distribution of serotypes among the Salmonella isolates obtained throughout the study per type of sample
| Serotype | No. of isolates per sample type
|
Total (%) | ||||
|---|---|---|---|---|---|---|
| Stream water | Seawater | Sewage water | Mollusks | Puddles | ||
| Typhimurium | 9 | 4 | 2 | 0 | 0 | 15 (23.4) |
| Vejle | 2 | 1 | 1 | 0 | 0 | 4 (6.2) |
| Suberu | 2 | 1 | 0 | 0 | 0 | 3 (4.7) |
| Urbana | 3 | 0 | 0 | 0 | 0 | 3 (4.7) |
| Galiema | 0 | 2 | 0 | 0 | 0 | 2 (3.1) |
| Othmarschem | 1 | 1 | 0 | 0 | 0 | 2 (3.1) |
| Soerenga | 1 | 0 | 1 | 0 | 0 | 2 (3.1) |
| Tonev | 0 | 1 | 0 | 1 | 0 | 2 (3.1) |
| Amherstiana | 0 | 0 | 0 | 0 | 1 | 1 (1.6) |
| Augustenborg | 1 | 0 | 0 | 0 | 0 | 1 (1.6) |
| Breda | 1 | 0 | 0 | 0 | 0 | 1 (1.6) |
| Bulovka | 0 | 1 | 0 | 0 | 0 | 1 (1.6) |
| Coeln | 1 | 0 | 0 | 0 | 0 | 1 (1.6) |
| Corvallis | 0 | 0 | 1 | 0 | 0 | 1 (1.6) |
| Djugu | 1 | 0 | 0 | 0 | 0 | 1 (1.6) |
| Nchanga | 0 | 0 | 1 | 0 | 0 | 1 (1.6) |
| Nitra | 0 | 0 | 1 | 0 | 0 | 1 (1.6) |
| Stanley | 0 | 1 | 0 | 0 | 0 | 1 (1.6) |
| Stanleyville | 0 | 0 | 1 | 0 | 0 | 1 (1.6) |
| Winnipeg | 0 | 0 | 0 | 1 | 0 | 1 (1.6) |
| Salmonella spp. | 1 | 10 | 2 | 3 | 3 | 19 (29.7) |
| Total | 23 | 22 | 10 | 5 | 4 | 64 |
The rain pattern in Bahia de Todos Santos proved to be highly irregular over the period of study. Rainfall was significantly higher (P < 0.05) in 2004 than in 2005 or 2006. The heaviest rains were recorded in October and December 2004, January and February 2005, and March 2006, although the difference was only statistically significant in October 2004. The greatest number of Salmonella isolates coincided with the months of highest rainfall (Fig. 2). Temperatures in 2004 were significantly lower than those in 2005 and 2006. The warmest year was 2005, although the temperatures were only significantly higher than those in 2004. The warmest periods occurred between May and October 2005 and in June and July 2006.
Interactions between the daily incidence of Salmonella bacteria in Bahia de Todos Santos and the environmental factors were initially examined by correlation analysis (Table 4). The presence of Salmonella spp. was positively and significantly correlated (P < 0.05) with rainfall 1 and 2 days before sample collection and with the accumulated rainfall during the 3 days prior to sample collection. There was a negative significant association with registered temperatures on the day of collection and the day before the sample collection, whether the maximum, minimum, or averaged values of temperature were considered, along with the relative humidity on the day of the sample collection. The highest significant association was observed with rainfall the day before the sampling day, with a correlation coefficient of 0.349 (P < 0.01).
TABLE 4.
Correlation coefficient values for the presence of Salmonella spp. in Bahia de Todos Santos and different environmental parameters
| Environmental parameter | Correlation coefficienta
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| Wind speed | 0.194 | 0.107 | 0.105 | −0.035 |
| Wind direction | 0.025 | 0.067 | −0.052 | 0.009 |
| Temp | −0.315** | −0.310** | −0.287* | −0.164 |
| Maximum temp | −0.309** | −0.332** | −0.278* | −0.172 |
| Minimum temp | −0.306** | −0.268* | −0.163 | −0.103 |
| Rainfall | 0.167 | 0.349** | 0.279* | 0.004 |
| Atmospheric pressure | 0.210 | 0.177 | 0.023 | 0.176 |
| Relative humidity | −0.308** | −0.131 | −0.076 | −0.118 |
| Radiation | 0.122 | 0.091 | −0.031 | 0.035 |
| Sun exposure | −0.036 | −0.047 | −0.050 | −0.053 |
| Sewage flow | 0.097 | 0.207 | 0.175 | 0.119 |
0, day of sample collection; 1, 1 day before sampling date; 2, 2 days before sampling date; 3, 3 days before sampling date; *, significant correlation at a P value of <0.05; **, significant correlation at a P value of <0.01.
The model obtained by simple logistic regression analysis showed a Nagelkerke r-square value of 0.145 and identified the presence of rain during the 2 days prior to sample collection as the dominant factor in the model, with a highly significant positive effect on the presence of Salmonella spp. in Bahia de Todos Santos (P < 0.00001, 1 day before; P < 0.001, 2 days before). The model also revealed a positive significant association (P < 0.05) with winds on the day of the sample collection and a negative relationship with the average temperature on the day of the sample collection.
The comparison of the data obtained in this study with the number of cases of human nontyphoid and typhoid Salmonella infections recorded in Baja California over the same period showed an inverse relationship between the presence of Salmonella spp. in coastal areas and the dynamics of infections in the region (Fig. 2). The weekly incidence of Salmonella bacteria in coastal environments was negatively and significantly associated (P < 0.05) with the weekly number of nontyphoid and typhoid cases, with correlation coefficients of −0.254 and −0.261, respectively. Human infections prevailed in the warmest months of the year, whereas the presence of Salmonella bacteria in the environment was governed primarily by rainy periods during the coldest periods.
DISCUSSION
The presence of Salmonella bacteria is an important public health issue in Mexico, with high rates of infection reported annually. The number of recorded cases varies greatly from one year to the next among different states in the country. In 1994, a total of 100,342 infections were reported in Mexico (111.21 cases per 100,000 people), whereas nearly twice the number of cases were recorded in 1998 (215,155 cases; a rate of 223.53 per 100,000 people) (9). Similar values were obtained between 2003 and 2005, with the number of infections reaching 106,000 cases, an average rate of 102.7 cases per 100,000 people (5). These values are significantly higher than those reported for other parts of the world, such as the United States with 17.7 cases per 100,000 people in 1999 (15), the European Union with 35 cases per 100,000 people in 2006 (7), and New Zealand with 32.3 cases per 100,000 people in 2006 (22). During the same period, the number of infections reported in Baja California was lower than the national rate, although it remained elevated, with a mean value of 70 cases per 100,000 people, which represented 1.8% of all cases reported for the entire country. The low number of cases reported in Baja California, a warm location with scarce rainfall, contrasts with the number of infections recorded in other Mexican states with extreme climates and high temperatures, such as Chihuahua, where incidences of Salmonella infection reached 850 cases per 100,000 people, which represents 14% of the total number of cases reported in Mexico (5).
The results obtained in the present study reflect a low incidence of Salmonella bacteria in the coastal environments of Bahia Todos los Santos, with an overall value of 4.8% throughout the 22 sampling months. Similar low levels of Salmonella presence were obtained in marine samples from regions with similar oceanographic conditions to Baja California and temperate seawater temperatures, such as Galicia (northwest Spain), the United States, and the United Kingdom, which have incidences of 2.4%, 7.4%, and 8%, respectively (2, 20, 30), far from the values obtained in tropical or warm seawater areas where Salmonella incidence in seafood can reach up to 20% (10, 11, 19).
Isolation of Salmonella spp. in the coastal environments of Bahia Todos los Santos was significantly and negatively associated with atmospheric temperature. Conversely, atmospheric temperature was the parameter that modulated human infections during the same period in this area, with cases peaking during the warmer months. The presence of Salmonella spp. in this study was associated primarily with rainy periods and confined to areas close to stream discharges. Similar associations between storm-generated flows, torrential rains, and the monsoon season have been reported in previous studies in temperate and tropical regions of the world with frequent rainy periods (1, 2, 10, 14, 19, 25, 29), signaling the washing effect of torrential rains as one of the principal environmental drivers of Salmonella contamination in coastal areas (19). According to the pattern observed in Bahia Todos los Santos, the arrival of Salmonella spp. into marine environments was predominantly governed by the presence of rains persistent enough to transport the contamination from the original source points to the sea via streams, whereas the permanence of Salmonella contamination in coastal areas appears to have been modulated by a combination of oceanographic characteristics and atmospheric conditions related primarily to the effects of sunlight. Bahia Todos los Santos has a semiarid climate with warm temperatures accompanied by rains restricted to only a few days throughout the year that restore water flow along dry streams for short periods. According to the results of the present study, Salmonella spp. were only detected if rain occurred on the days prior to sampling and the presence of Salmonella spp. was especially marked if rainy conditions prevailed for several consecutive days. Alternatively, the occurrence of rain on the day of the sample collection alone did not have any effect on the detection of Salmonella species.
Once the contamination reached the sea, the presence of Salmonella spp. was affected mainly by atmospheric conditions on the day of the sample collection, primarily recorded temperatures and winds. In contrast to observations for human infection, high temperatures in the Bahia Todos los Santos had a negative effect on the occurrence of Salmonella spp., which may be linked to the effect of sunlight on the survival of bacteria. In Mexico, solar radiation reaches maximum values in the northwestern states, with annual average values for Baja California ranging from 6 to 7 kWh/m2/day. The incoming fresh water from streams characteristically spreads to the sea surface because of its lower density, thereby exposing the bacteria trapped in the water to the direct deleterious effect of solar radiation. A similar association may be suggested for the favorable effect of the wind on the same sampling day, since the presence of strong winds causes turbulence and waves on the sea surface that reduce the penetration of sunlight and its bactericidal effect. Additionally, seawater temperature may play a secondary role in the long-term survival rate of Salmonella bacteria. The presence of cold waters may reduce the permanence of Salmonella spp. in the marine environment, while warm waters together with high levels of organic matter—i.e., typical conditions in tropical coastal areas—may contribute to a more appropriate habitat for an increased survival of bacteria, as reflected in the disparate incidence of Salmonella spp. described in diverse studies in temperate and tropical regions (2, 10, 11, 20, 30).
Previous studies of the presence of Salmonella spp. in marine environments characteristically identified a maximum of 20 different serovars, independent of the number of samples processed or strains isolated (18). Serovar Typhimurium has been shown to be the most common clinically significant serovar isolated from marine samples in different parts of the world (1, 3, 20, 26, 30), probably because of its enhanced capacity for adaptation and survival in saline environments (1). A similar pattern was also observed in the present investigation, with 20 serovars identified among the Salmonella isolates and a clear dominance of serovar Typhimurium among the strains recovered. Serovar Typhimurium is the most common serovar isolated from human sources in Mexico (9, 32) and also predominates in animals and in meat products (32). The different seasonal relationship between human infections and Salmonella contamination observed in Bahia Todos los Santos may suggest a nonhuman origin for the Salmonella serovar Typhimurium contamination detected in coastal areas. There is significant livestock production in the area surrounding Bahia Todos los Santos, and there are several ranches located in the vicinity. The rainfall that occurs after long periods of drought probably carries animal waste from source points to streams and finally to the sea. These results contrast with data from previous studies carried out in the United States and Spain (2, 20), in which a different serotype dominance was reported in marine environments and humans. Whereas the serovars Typhimurium and Enteritidis are the major serotypes isolated from humans in both countries (4, 20), serovar Newport prevailed in oysters collected in the United States, and serovar Senftenberg prevailed in shellfish from northwest Spain. This distinctive pattern may be related to the relative epidemiological or zoonotic importance of these serovars in the regions investigated and may have biased the serotype distribution present in the sea (17).
In conclusion, rain has shown to be the distinctive factor influencing the transport of Salmonella contamination from source points to the sea in the arid region of Baja California. A similar pattern has been described for both rainy temperate and tropical regions of the world, signaling runoff as a universal environmental driver for the presence of Salmonella spp. in the marine environment. Furthermore, the intense sunlight prevailing in Bahia Todos los Santos emerges as a critical variable for the drastic reduction of the permanence of Salmonella spp. in the sea. The integration of the major environmental factors governing the dynamics of contamination of Salmonella spp. in the sea in the surveillance programs of coastal areas and shellfish may greatly contribute to the development of improved and more operative risk management systems.
Acknowledgments
We are grateful to M. Victoria Orozco-Borbon for hosting us at the Instituto de Investigaciones Oceanologicas in the Universidad Autonoma de Baja California during the undertaking of the present study. We also thank Silvia Carles Gonzalez and Isabel Mayan Barreiro (Insitituto de Acuicultura, University of Santiago de Compostela, Spain) for technical assistance in serotyping the Salmonella strains, Emmanuel Alvarez (Comision Nacional del Agua), Alfonso Da Silveira Mascarenhas, and Eduardo Gil Silva (Instituto de Investigaciones Oceanologicas, UABC) for providing the weather data included in the study, the Comision Estatal de Servicios Publicos de Ensenada for data of discharges from the sewage treatment plants, and Marilu Morales Fraga for assistance with GIS analysis.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Baudart, J., K. Lemarchand, A. Brisabois, and P. Lebaron. 2000. Diversity of Salmonella strains isolated from the aquatic environment as determined by serotyping and amplification of the ribosomal DNA spacer regions. Appl. Environ. Microbiol. 66:1544-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brands, D. A., A. E. Inman, C. P. Gerba, C. J. Maré, S. J. Billington, L. A. Saif, J. F. Levine, and L. A. Joens. 2005. Prevalence of Salmonella spp. in oysters in the United States. Appl. Environ. Microbiol. 71:893-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catalao Dionisio, L. P., M. Joao, V. S. Ferreiro, M. L. Fidalgo, M. E. García Rosado, and J. J. Borrego. 2000. Occurrence of Salmonella spp. in estuarine and coastal waters of Portugal. Antonie van Leeuwenhoek 78:99-106. [DOI] [PubMed] [Google Scholar]
- 4.CDC. 2007. Salmonella surveillance: annual summary, 2005. Department of Health and Human Services, CDC, Atlanta, GA. http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2005/SalmonellaAnnualSummary2005.pdf.
- 5.Comité Nacional de Vigilancia Epidemiológica. 2006. Manual de procedimientos de la notificación semanal de casos nuevos: casos sujetos a notificación obligatoria, primera edición. Sistema Nacional de Vigilancia Epidemiológica, Secretaría de Salud, Mexico. http://www.dgepi.salud.gob.mx.
- 6.Davies, R. H., and C. Wray. 1996. Seasonal variations in the isolation of Salmonella typhimurium, Salmonella enteritidis, Bacillus cereus and Clostridium perfringens from environmental samples. J. Vet. Med. Ser. B 43:119-127. [DOI] [PubMed] [Google Scholar]
- 7.European Food Safety Authority. 2007. The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European Union in 2006. European Food Safety Authority, Parma, Italy. http://www.efsa.europa.eu/EFSA/DocumentSet/Zoon_report_2006_en.pdf.
- 8.Goyal, S. M., C. P. Gerba, and J. L. Melnick. 1977. Occurrence and distribution of bacterial indicators and pathogens in canal communities along the Texas coast. Appl. Environ. Microbiol. 34:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez-Cogco, L., E. Montiel-Vazquez, P. Aguilera-Perez, and M. C. Gonzalez-Andrade. 2000. Serotipos de Salmonella identificados en los servicios de salud de México. Salud Pública Mex. 42:490-495. [PubMed] [Google Scholar]
- 10.Hatha, A. A. M., and P. Lakshmanaperumalsamy. 1997. Prevalence of Salmonella in fish and crustaceans from markets in Coimbatore, South India. Food Microbiol. 14:111-116. [Google Scholar]
- 11.Heinitz, M. L., R. D. Ruble, D. E. Wagner, and S. R. Tatini. 2000. Incidence of Salmonella in fish and seafood. J. Food Prot. 63:579-592. [DOI] [PubMed] [Google Scholar]
- 12.Instituto Nacional de Estadística, Geografía e Informática de México. 2006. Conteo de población y vivienda 2005. Identidad Federativa, Baja California, Mexico. http://www.inegi.gob.mx.
- 13.International Organization for Standardization. 1993. Microbiology: general guidance on methods for the detection of Salmonella, 3rd ed. International standard method ISO 6579:1993. International Organization for Standardization, Geneva, Switzerland.
- 14.Kaper, J. B., G. S. Sayler, M. M. Baldini, and R. R. Colwell. 1977. Ambient-temperature primary nonselective enrichment for isolation of Salmonella spp. from an estuarine environment. Appl. Environ. Microbiol. 33:829-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy, M., F. J. Angulo, and the FoodNet Working Group. 2000. Incidence of foodborne illnesses: 1999 data from FoodNet. Irish J. Agric. Food Res. 39:295-300. [Google Scholar]
- 16.Malorny, B., J. Hoorfar, C. Bunge, and R. Helmuth. 2003. Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Urtaza, J., and E. Liebana. 2005. Investigation of clonal distribution and persistence of Salmonella Senftenberg in the marine environment and identification of potential sources of contamination. FEMS Microbiol. Ecol. 52:255-263. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Urtaza, J., E. Liebana, L. Garcia-Migura, P. Perez-Piñeiro, and M. Saco. 2004. Characterization of Salmonella enterica serovar Typhimurium from marine environments in coastal waters of Galicia (Spain). Appl. Environ. Microbiol. 70:4030-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Urtaza, J., M. Saco, G. Hernandez-Cordova, A. Lozano, O. Garcia-Martin, and J. Espinosa. 2003. Identification of Salmonella serovars isolated from live molluscan shellfish and their significance in the marine environment. J. Food Prot. 66:226-232. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Urtaza, J., M. Saco, J. de Novoa, P. Perez-Pineiro, J. Peiteado, A. Lozano-Leon, and O. Garcia-Martin. 2004. Influence of environmental factors and human activity on the presence of Salmonella serovars in a marine environment. Appl. Environ. Microbiol. 70:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mezrioui, N., B. Baleux, and M. Trousselier. 1995. A microcosm study of the survival of Escherichia coli and Salmonella typhimurium in brackish water. Water Res. 29:459-465. [Google Scholar]
- 22.Ministry of Health. 2006. Notifiable and other diseases in New Zealand, annual report. New Zealand Ministry of Health, Wellington, New Zealand. http://www.moh.govt.nz/foodborne.html#salmonella.
- 23.Mohandass, C., and P. A. Loka Bharathi. 2003. Representation, dispersion, and variation of bacterial indicators in the coastal waters of Nagore (east coast of India). Water Environ. Res. 75:66-72. [DOI] [PubMed] [Google Scholar]
- 24.National Advisory Committee on Microbiological Criteria for Foods. 1992. Microbiological criteria for raw molluscan shellfish. J. Food Prot. 55:463-480. [DOI] [PubMed] [Google Scholar]
- 25.O'Shea, M. L., and R. Field. 1992. Detection and disinfection of pathogens in storm-generated flows. Can. J. Microbiol. 38:267-276. [DOI] [PubMed] [Google Scholar]
- 26.Polo, F., M. J. Figueras, I. Inza, J. Sala, J. M. Fleisher, and J. Guarro. 1999. Prevalence of Salmonella serotypes in environmental waters and their relationships with indicator organisms. Antonie van Leeuwenhoek 75:285-292. [DOI] [PubMed] [Google Scholar]
- 27.Popoff, M. Y., J. Bockemuhl, and L. L. Gheesling. 2004. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155:568-570. [DOI] [PubMed] [Google Scholar]
- 28.Uzzau, S., D. J. Brown, T. Wallis, S. Rubino, G. Leori, S. Bernard, J. Casadesús, D. J. Platt, and J. E. Olsen. 2000. Host adapted serotypes of Salmonella enterica. Epidemiol. Infect. 125:229-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkateswaran, K., T. Takai, I. M. Navarro, H. Nakano, H. Hashimoto, and R. J. Siebeling. 1989. Ecology of Vibrio cholera non-O1 and Salmonella spp. and role of zooplankton in their seasonal distribution in Fukuyama coastal waters, Japan. Appl. Environ. Microbiol. 55:1591-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson, I. G., and J. E. Moore. 1996. Presence of Salmonella spp. and Campylobacter spp. in shellfish. Epidemiol. Infect. 116:147-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaidi, M. B., J. J. Calva, M. T. Estrada-Garcia, V. León, G. Vazquez, G. Figueroa, E. Lopez, J. Contreras, J. Abbott, S. Zhao, P. MacDermott, and L. Tollefson. 2008. Integrated food chain surveillance system for Salmonella spp. in Mexico. Emerg. Infect. Dis. 14:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]