Abstract
Fifty isolates from root nodules of soybean plants sampled in five agricultural-ecological-climatic regions of India were analyzed by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene, the intergenic spacer region between the 16S and 23S rRNA genes (IGS), and the nifH and nodC genes. Eight haplotypes assigned to the Bradyrhizobium genus were identified, and the genetic diversity was conserved across regions. Sequence analyses of the IGS and the dnaK, glnII, recA, and nifH genes revealed three groups. One of them (26% of isolates) was assigned to Bradyrhizobium liaoningense. A second group (36% of isolates) was identified as B. yuanmingense but likely forms a new biovar able to nodulate soybean plants. The third lineage (38% of isolates) was different from all described Bradyrhizobium species but showed the same symbiotic genotype as B. liaoningense and B. japonicum bv. glycinearum.
The soybean plant [Glycine max (L.) Merril] is an important source of high-quality protein and oil and is the most quantitatively important grain legume in the world (>220 Mt in 2006 [http://faostat.fao.org]). Soybeans are a subtropical crop that originated in Southeast Asia but nowadays is cultivated worldwide under various climatic conditions. The high N requirement of the crop is fulfilled mainly by establishing a N2-fixing symbiosis with rhizobia. Soybean inoculation has been used worldwide for a century when effective rhizobia are absent or in insufficient numbers in soil, with millions of hectares inoculated. However, selection of new elite strains adapted to local environmental conditions and to newly bred plant lines stays a topic of interest, fueling research on soybean variety improvement and increasing development of crop cultivation in new areas.
China was the first center of domestication of the soybean plant, about 4,000 years ago, and is the largest producer in Asia. However, although commercial cultivation of soybeans started in India in the 1960s, this crop was probably introduced to this country as soon as it was domesticated in China. Therefore, India is considered a secondary center of domestication of soybeans (2, 10). This country is the fifth producer of soybeans in the world today (8.3 Mt in 2006 [http://faostat.fao.org]). However, the average yield of about 1.1 t ha−1 is low compared to the world average of 1.8 t ha−1, and improving the crop performance is a major challenge for India (6). The effectiveness of symbiotic N2 fixation may be an important factor to take into consideration through successful management of symbiosis between soybean cultivars and native rhizobia.
Soybean-nodulating rhizobia are genetically diverse and are classified into different genera and species, with all the species described so far having been identified in China. The slow growers are distributed in three species of the Bradyrhizobium genus, namely, Bradyrhizobium japonicum (9), Bradyrhizobium liaoningense (30), and Bradyrhizobium elkanii (11). Fast growers belong to Sinorhizobium fredii and S. xinjiangense and also include other unclassified rhizobia (3, 5, 16). Soybean rhizobia with a variable generation time were classified into Mesorhizobium tianshanense (4). However, despite the importance of soybean cultivation in India, little is known about the genetic resources in native rhizobia. In particular, the genetic diversity of soybean rhizobia has not been described so far. Therefore, in this study we examine the genetic diversity of a core collection of 50 isolates from soybean nodules sampled in various fields in different agricultural-ecological-climatic regions of India, including the main cropping areas. Four loci were analyzed by restriction fragment length polymorphism analysis of PCR-amplified DNA fragments (PCR-RFLP), including the 16S rRNA gene, the intergenic spacer (IGS) between the 16S and 23S rRNA genes, and the symbiotic genes nifH and nodC. The IGS and housekeeping genes are currently used as markers for molecular systematics and for estimations of phylogenetic relationships among bradyrhizobia (20, 23, 26-28), with the 16S rRNA gene having few polymorphisms within the Bradyrhizobium genus (23, 27, 28). Genetic relationships of the soybean isolates to other rhizobia were therefore investigated by sequencing the IGS and the housekeeping genes dnaK, glnII, and recA. Phylogenetic analysis of nifH was also performed.
MATERIALS AND METHODS
Bacterial strains.
Nodules were collected from soybean plants grown in various fields in different agricultural-ecological-climatic regions of India (Table 1). The sampled sites had a history of intensive soybean cultivation of nearly 40 years, except for the Chengalpattu site, where soybeans had only recently been cultivated (for 5 years before sampling), and had no known history of inoculation of soybeans. The distance between sampled sites was at least 60 km, and the distance between individual sampled plants was at least 200 m at each site. A single nodule per plant was excised. The nodules were surface sterilized, and crushed nodules were streaked on yeast-mannitol agar plates according to the method of Vincent (25). Single colonies from each nodule isolate were inoculated onto soybean cultivars JS335 and Bragg. Five weeks after inoculation, one nodule per plant was excised and rhizobia were reisolated as described above. Single colonies were grown on yeast-mannitol agar slants at 28 ± 2°C, and cultures were transferred to fresh slants monthly.
TABLE 1.
Origins of soybean rhizobial isolates and agricultural-ecological-climatic characteristics of sampled regions
| Site | Designation of isolates | No. of fields sampled | No. of isolates | Previous cropa | Agricultural-ecological-climatic region (region and soil characteristics)b |
|---|---|---|---|---|---|
| Ujjain (Madhya Pradesh) | SR66-70 | 2 | 5 | Wheat | A (hot and semiarid, medium and deep black soils, coarse to fine loamy, highly alkaline, moderate to gentle slope) |
| Indore (Madhya Pradesh) | SR71-75 | 1 | 5 | Chickpea | A |
| Khandwa (Madhya Pradesh) | SR76-79 | 2 | 4 | Fallow | A |
| Raipur (Chattisgarh) | SR80-91 | 5 | 12 | Wheat | B (hot and subhumid, red and yellow soils, moderate clay to deep clayey, slightly acidic to slightly alkaline, moderate to gentle slope) |
| Jabalpur (Madhya Pradesh) | SR92-95 | 2 | 4 | Black gram | C (hot and subhumid, red and black soils, deep loamy, neutral to slightly acidic, moderate to gentle slope) |
| Sehore (Madhya Pradesh) | SR96-97 | 1 | 2 | Red gram | C |
| Chengalpattu (Tamil Nadu) | SR98-100 | 1 | 3 | Green gram-red gram intercropping | D (hot and subhumid to semiarid, coastal alluvium-derived soils, clayey, slightly alkaline to moderately alkaline, level to very gentle slope) |
| Gwalior (Madhya Pradesh) | SR101-108 | 3 | 8 | Wheat | E (hot and semiarid, alluvium-derived soils, loamy to clayey, slightly alkaline, gentle to very gentle slope) |
| Mirzapur (Uttar Pradesh) | SR109-115 | 4 | 7 | Wheat | E |
Crop grown in the previous season.
From the National Bureau of Soil Survey and Land Use Planning, India, and Indian Council of Agricultural Research, New Delhi, India. Hot and semiarid, mean annual rainfall of 500 to 1,100 mm, moisture index of −33.3 to −66.7, temperature of >22°C, and length of growing period (LGP) of 90 to 150 days; hot and subhumid, mean annual rainfall of 1,000 to 1,500 mm, moisture index of +20 to −33.3, temperature of 15 to 22°C, and LGP of 150 to 180 days; hot and subhumid to semiarid, mean annual rainfall of 900 to 1,600 mm, moisture index of −66.7 to 0, temperature of >15 to 22°C, and LGP of 90 to 210 days. Soil pH descriptions: neutral, 6.6 to 7.5; slightly acidic, 6.1 to 6.5; slightly alkaline, 8.0 to 8.5; moderately alkaline, 8.6 to 9.0; and highly alkaline, >9.0. Soil topology (slope) descriptions: level, <1 to 3%; very gentle, 1 to 3%; gentle, 3 to 8%; moderate, 8 to 15%.
Characterization of rhizobial isolates by PCR fingerprinting.
The isolates were characterized by PCR-RFLP with the various restriction enzymes listed in Table 2, as previously described for the 16S rRNA gene (13), the IGS (12), and the nifH and nodC genes (14).
TABLE 2.
Distribution of rhizobia isolated from root nodules of soybean plants into genotypes
| Genotype | RFLP pattern of 16S rRNA genea | 16S rRNA type | IGS RFLP patterna | IGS type | nifH RFLP patterna | nif type | nodC RFLP patterna | nod type | % of isolates |
|---|---|---|---|---|---|---|---|---|---|
| I | AAAAAAA | I | AAA | I | AAA | I | AA | I | 14 |
| II | AAAAAAA | I | AAA | I | AAA | I | AB | II | 2 |
| III | AAAAAAA | I | AAA | I | ABB | II | BC | III | 12 |
| IV | AAAAAAA | I | ABA | II | ABB | II | BC | III | 2 |
| V | AAAAAAA | I | BAB | III | AAA | I | AA | I | 6 |
| VI | AAABBAB | II | CCC | IV | BCC | III | CD | IV | 38 |
| VII | AAABBAB | II | DDD | V | BCC | III | CD | IV | 20 |
| VIII | AAABBAB | II | DED | VI | BCC | III | CD | IV | 6 |
The letters identify RFLP patterns obtained with CfoI, DdeII, HaeIII, HinfI, MspI, NdeII, and RsaI for the 16S rRNA gene; with AluI, CfoI, and HaeIII for the IGS; with CfoI, HaeIII, and MspI for nifH; and with HaeIII and MspI for nodC.
Sequencing and phylogenetic analysis.
Fragments of dnaK, glnII, and recA genes were amplified by PCR, using the primer pairs described by Vinuesa et al. (26) and/or Stêpkowski et al. (20). PCR products from the IGS, nifH, dnaK, glnII, and recA genes were purified using PCR purification kits and sequenced using the PCR primers. The nucleotide sequences were determined using either a CEQ 8000 XL sequencer (Beckman Coulter) or an ABI Prism 310 DNA sequencer (Applied Biosystems) with sequencing reaction kits recommended by the manufacturer, or sequencing was carried out by MacroGen Inc. (Seoul, South Korea). Multiple sequence alignment was performed with Clustal W, version 1.8 (22), and manually corrected using GeneDoc software (version 2.6.002 [http://www.psc.edu/biomed/genedoc]). The phylogenetic analyses were performed on the Phylogeny.fr platform by maximum likelihood (ML) analyses with the GTR substitution model (7). Shimodaira-Hasegawa tests of congruence of tree topologies were performed using PAUP, version 4.0b10 (21).
Nucleotide sequence accession numbers.
All sequences from soybean isolates have been deposited in the GenBank database under accession numbers EU357926 through EU357931 for the IGS, EU357921 through EU357925 for nifH, EU818928 through EU818931 for dnaK, EU818932 through EU818935 for glnII, and EU818936 through EU818939 for recA. The nifH sequences of the B. liaoningense, Bradyrhizobium canariense, and Bradyrhizobium yuanmingense type strains have also been deposited, under accession numbers EU818925, EU818926, and EU818927, respectively.
RESULTS
Genetic diversity of isolates.
All nodule isolates were slow growers, with generation times varying from 8 to 23 h (data not shown). The data from PCR-RFLP analysis of each isolate at different loci are shown in Table 2. Two 16S rRNA haplotypes were identified. The first type (36% of isolates) corresponded to that of the B. yuanmingense type strain, as predicted by mapping of restriction sites from the 16S rRNA gene sequence (GenBank accession number AF193818). The second type was identical to that of B. japonicum and B. liaoningense (13). Each 16S rRNA type was subdivided into three IGS haplotypes, and the isolates were distributed in four symbiotic (nif-nod) genotypes. 16S rRNA type II was associated with only one symbiotic genotype (III-IV). The combined data from the multilocus RFLP analysis resulted in the characterization of eight distinct genotypes, with four of them containing 14 to 38% of isolates each.
Geographical distribution of genotypes.
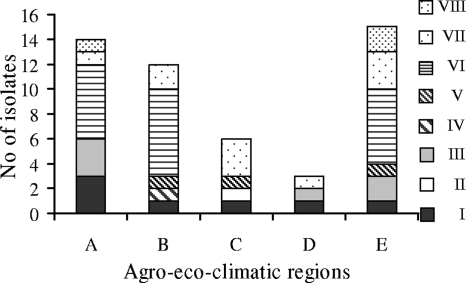
Three to six genotypes were identified in each region, and genotypes I and VII were detected in all geographic areas (Fig. 1). An exact test of sample differentiation based on haplotype frequencies according to geographic region was performed using Arlequin software (18). No differentiation was found (P = 0.2) according to the global test as well as in testing differentiation between all pairs of samples, except between the populations from regions A and C (P = 0.02). One genotype (VI) was predominant in three populations (40 to 60% of isolates). Because the sampling in each region was very small, diversity was presumably underestimated. However, it should be noticed that the three isolates sampled at the Chengalpattu site (region D) were classified into three predominant genotypes, I, III, and VII. This result suggests that the diversity of soybean bradyrhizobia might be especially high in this geographic area, which is not a traditional site for soybean cultivation, in contrast to the other sampled regions, which are considered soybean hubs of India.
FIG. 1.
Distribution of bradyrhizobial genotypes (I to VIII) defined by the combined data from PCR-RFLP analysis of the 16S rRNA gene, IGS, nifH, and nodC among agricultural-ecological-climatic regions of India.
Phylogenetic analysis of the IGS.
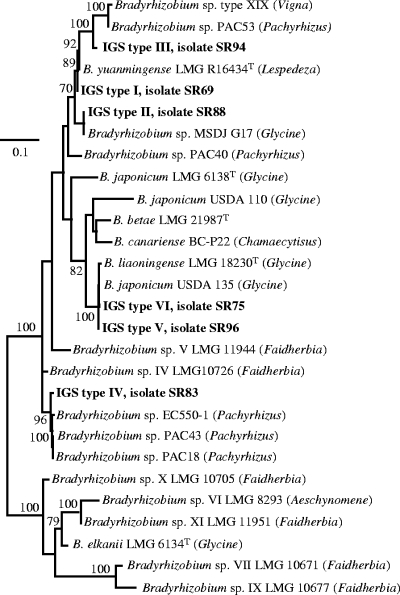
The sequences of six strains representative of each IGS type were determined and compared to those available in databases. The phylogenetic tree shown in Fig. 2 includes six unnamed genospecies of bradyrhizobia defined by Willems et al. (29). The six IGS types were distributed in three different clades supported by bootstrap values of ≥70%. The three representatives of IGS types I, II, and III formed a cluster (97 to 100% similarity between the aligned parts of the sequences) with B. yuanmingense and other unclassified bradyrhizobia, including strain MSDJ G17, isolated from a soybean plant in Argentina (A. Hartmann, personal communication). The IGS type V and VI sequences were closely related (99 to 100% similarity) to the type strain of B. liaoningense and to strain USDA 135 from soybean, which was formerly assigned to B. japonicum prior to the description of B. liaoningense as a novel species. The IGS type IV sequence (strain SR83) and unclassified Bradyrhizobium strains isolated from Pachyrhizus erosus nodules (17) formed a tight cluster that could not be related to any described species (only 76 to 80% similarity between the IGS sequences) but which might have a closer relationship with Bradyrhizobium genospecies IV (93 to 94% similarity). The sequence of strain SR83 did not show high percent identities with those of the various B. japonicum serotypes and other bradyrhizobial accessions of the USDA ARS National Rhizobium Germplasm Collection (23, 24). Therefore, our results show that 74% of isolates (IGS types I to IV) could not be related to a species (or genospecies) that was described so far as including soybean symbionts.
FIG. 2.
Phylogenetic ML tree based on 938-bp alignment of nucleotide sequences of the IGS between the 16S and 23S rRNA genes. Only bootstrap probability values of ≥70% (over 100 replicates) are indicated at the branching points. The scale bar indicates the number of substitutions per site. Soybean isolates are shown in bold, with their IGS haplotypes indicated. The letter “T” indicates the type strain of the species. Accession numbers of sequences extracted from GenBank are as follows: PAC53, AY628088; type XIX, AY493859; LMG R16434T, AJ534605; MSDJ G17, AF338851; PAC40, AY628087; LMG 6138T, AJ279264; BC-P22, AY386706; LMG 21987T, AJ631967; USDA 110, BA000040; LMG 18230T, AJ279301; USDA 135, AF208511; LMG 11944, AJ279287; LMG 10726, AJ279281; EC550-1, AY628085; PAC43, AY628089; PAC18, AY628093; LMG 10677, AJ534597; LMG 10671, AJ279272; LMG 10705, AJ534592; LMG 6134T, AJ279308; LMG 11951, AJ534594; and LMG 8293, AJ279311.
Phylogenetic analysis of housekeeping genes.
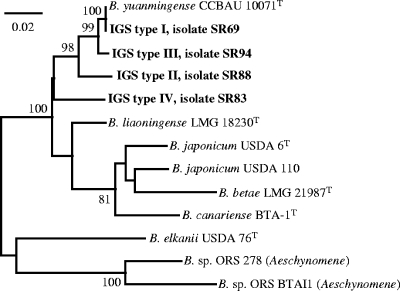
To confirm the taxonomic status of the strains showing IGS types I to IV, the partial sequences of dnaK, glnII, and recA were determined for four representative strains. Based on nucleotide sequence comparisons (analyses in GenBank, updated on June 2008), the dnaK, glnII, and recA sequences of representatives of IGS types I to III showed the highest similarities (96 to 100%) to those of B. yuanmingense strains from various hosts, including recently released glnII and recA sequences of soybean bradyrhizobia isolated in China (e.g., GenBank accession numbers EU152364 and EU152382). The recA and glnII sequences of IGS type IV (strain SR83) were also identical to those of two Bradyrhizobium sp. strains isolated from soybeans in China (GenBank accession numbers EU419721, EU419725, EU419736, and EU419744). Otherwise, the SR83 sequences did not show more than 94 to 96% similarity with the other sequences available in the databases. We analyzed the phylogeny for each locus, and we also constructed a tree derived from the three pooled concatenated sequence alignments (Fig. 3). This tree did not show significant differences in topology from those derived from individual markers, based on Shimodaira-Hasegawa tests (P > 0.2). This multilocus analysis confirmed the conclusions drawn from IGS sequence analysis, grouping representatives of IGS types I, II, and III with B. yuanmingense, while the representative of IGS type IV formed a lineage that did not show a significant relationship with the bradyrhizobial species described so far.
FIG. 3.
Phylogenetic ML tree based on 1,493-bp alignment of concatenated nucleotide sequences of dnaK (489 bp), glnII (519 bp), and recA (482 bp). Only bootstrap probability values of ≥70% (over 100 replicates) are indicated at the branching points. The scale bar indicates the number of substitutions per site. Soybean isolates are shown in bold, with their IGS haplotypes indicated. The letter “T” indicates the type strain of the species. Accession numbers of sequences extracted from GenBank are as follows: CCBAU 10071T, AY923039 (dnaK), AY386780 (glnII), and AY591566 (recA); LMG 18230T, AY923041 (dnaK), AY386775 (glnII), and AY591564 (recA); USDA 6T, AM168362 (dnaK), AF169582 (glnII), and AM168341 (recA); USDA 110, BA000040 (complete genome); LMG 21987T, AY923046 (dnaK), AB353733 (glnII), and AB353734 (recA); BTA-1T, AY923047 (dnaK), AY386765 (glnII), and AY591553 (recA); USDA 76T, AM168363 (dnaK), AY599117 (glnII), and AM168342 (recA); ORS 278, CU234118 (complete genome); and BTAI1, CP000495 (complete genome).
Analysis of nifH and nodC sequences.
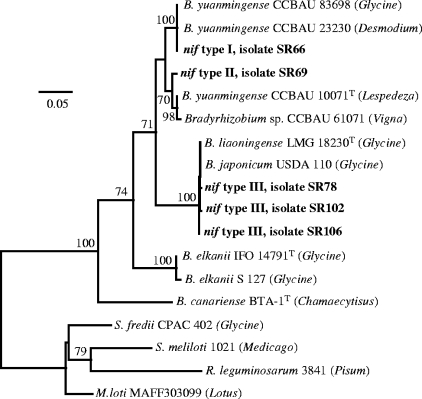
The sequences (∼700 bp) of representatives of the three nifH haplotypes and of the type strains of B. liaoningense, B. canariense, and B. yuanmingense were determined. The three nifH haplotypes were placed in two distinct clusters (Fig. 4). nif types I and II were grouped with B. yuanmingense reference strains, including Chinese soybean isolates (e.g., GenBank accession number EU146010). As expected, nif type III sequences were very similar to those of the B. japonicum bv. glycinearum and B. liaoningense reference strains.
FIG. 4.
Phylogenetic ML tree based on 612-bp alignment of nucleotide sequences of the nifH gene. Only bootstrap probability values of ≥70% (over 100 replicates) are indicated at the branching points. The scale bar indicates the number of substitutions per site. Soybean isolates are shown in bold, with their nifH haplotypes indicated. The letter “T” indicates the type strain of the species. Accession numbers of sequences extracted from GenBank are as follows: CCBAU 83698, EU146010; CCBAU 23230, AY934871; CCBAU 61071, EU113228; USDA 110, BA000040; S 127, DQ485701; IFO 14791T, AB094963; CPAC 402, DQ485714; MAFF303099, BA000012; 3841, AM236084; and 1021, AE007235.
Mapped restriction site analysis of nodC fragments from the available sequences of the B. yuanmingense strains (GenBank accession numbers AB354633 and DQ010033) and from the previously published RFLP data for nodC (14) confirmed the results of nifH phylogeny analysis. nod types I and III were identical to those of B. yuanmingense strain CCBAU 23230 and of the B. yuanmingense type strain, respectively, and differed by only one restriction site out of nine from nod type II. nod type IV was identical to that of B. japonicum bv. glycinearum and B. liaoningense and differed by 6 or 7 of 12 restriction sites from nodC of B. yuanmingense reference strains.
DISCUSSION
The 50 soybean isolates collected in India were classified in the Bradyrhizobium genus and were distributed in three IGS clusters within the clade including all valid bradyrhizobial species described so far, except for B. elkanii, which is most distantly related (23, 29). This result was corroborated by phylogenetic analysis of the housekeeping genes dnaK, glnII, and recA.
Only 26% of the Indian soybean isolates (genotypes VII and VIII) could be assigned to one valid species known to include soybean symbionts, i.e., B. liaoningense. Our current knowledge of the genetic diversity of rhizobia associated with soybeans refers to various studies in different continents. However, all of the species described so far could be found in China (32), which is considered the center of origin of the soybean plant. Therefore, assignment of a significant proportion of Indian soybean isolates to B. liaoningense was not surprising, since this species is widespread in Asia (19, 30, 32).
The finding that 36% of isolates (genotypes I to V) were closely related to B. yuanmingense was firstly unexpected, since spontaneous nodulation of soybean plants by this species has not been reported. However, in searching for sequence similarities in databases, we found highly similar sequences suggesting that soybean bradyrhizobia closely related to B. yuanmingense also exist in China and South America. The analysis of nifH and nodC corroborated the close relationship of these isolates to B. yuanmingense. This result argues for vertical acquisition of the symbiotic genes rather than relatively recent horizontal gene transfer of symbiotic genes from B. liaoningense or B. japonicum bv. glycinearum. The B. yuanmingense species was originally created based on the description of nodule isolates from Lespedeza spp. (33). Since then, several studies have reported nodulation of various legumes by B. yuanmingense-related rhizobia (8, 15, 17, 30). Studies of host specificity showed that these B. yuanmingense isolates were not capable of effective nodulation of soybean plants (8, 17, 31, 33). Two representative B. yuanmingense soybean isolates were evaluated on soybean plants and were effective in symbiotic N2 fixation (data not shown). Therefore, our results suggest the occurrence of different biovars within the B. yuanmingense species.
The third genomic group (38% of isolates; IGS type IV, genotype VI) characterized in this study could not be assigned to previously described Bradyrhizobium species but harbors the same symbiotic genetic information as B. japonicum bv. glycinearum and B. liaoningense, which might result from acquisition by horizontal transfer of a B. liaoningense or B. japonicum symbiotic island. Such an event was recently reported to have occurred in Brazilian soybean fields (1). IGS type IV showed significant similarity with unclassified bradyrhizobia from Pachyrhizus erosus plants native to Central and South America, but these were not able to nodulate soybean plants (17). We also evaluated one representative of IGS type IV on soybean plants and found that the strain was fully effective for N2 fixation (data not shown). These results again suggest the occurrence of different biovars of the same genomic species. Further phylogenic and taxonomic analyses are in progress to clarify the classification of soybean isolates grouped in genotype VI.
In conclusion, the present study provides the first analysis of the genetic diversity of soybean-nodulating Bradyrhizobium isolates from India, a major soybean producer. The results show that the diversity is wider than expected based on previous studies in various geographic areas and on the current taxonomy of soybean rhizobia. Notably, the diversity of the soybean symbionts appears to be conserved across the agricultural-ecological-climatic regions sampled. Further studies should investigate the link between genetic diversity and functional variability of the soybean-rhizobium symbiosis.
Acknowledgments
We thank G. S. Chauhan, National Research Centre for Soybean, Indore (M.P.), India, for kindly providing pure seeds of the soybean cultivar. We are also grateful to Lionel Moulin for helpful advice on phylogenetic analyses and to Geraldine Depret and David Bru for technical assistance.
Chinnaswamy Appunu was supported by a research fellowship from the Indian Council of Scientific and Industrial Research—University Grant Commission and by a short-term training fellowship from the European Molecular Biology Organization. This work was supported by grant 45 from the French governmental organization Bureau des Ressources Génétiques.
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Barcellos, F. G., P. Menna, J. S. da Silva Batista, and M. Hungria. 2007. Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian Savannah soil. Appl. Environ. Microbiol. 73:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chauhan, G. S., and O. P. Joshi. 2005. Soybean (Glycine max)—the 21st century crop. Indian J. Agric. Sci. 75:461-469. [Google Scholar]
- 3.Chen, L. S., A. Figueredo, F. O. Pedrosa, and M. Hungria. 2000. Genetic characterization of soybean rhizobia in Paraguay. Appl. Environ. Microbiol. 66:5099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W., E. Wang, S. Wang, Y. Li, and X. Chen. 1995. Characteristics of Rhizobium tianshanense sp. nov., a moderately and slowly growing root nodule bacterium isolated from an arid saline environment in Xinjiang, People's Republic of China. Int. J. Syst. Bacteriol. 45:153-159. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W. X., G. H. Yan, and J. L. Li. 1988. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Bacteriol. 38:392-397. [Google Scholar]
- 6.Deosthali, V., A. Akmanchi, and C. Salunke. 2005. Soybean agriculture in India, a spatial analysis. Trans. Inst. Indian Geogr. 27:13-30. [Google Scholar]
- 7.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J.-F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J.-M. Claverie, and O. Gascuel. 19 April 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed]
- 8.Gu, J., E. T. Wang, and W. X. Chen. 2007. Genetic diversity of rhizobia associated with Desmodium species grown in China. Lett. Appl. Microbiol. 44:286-292. [DOI] [PubMed] [Google Scholar]
- 9.Jordan, D. C. 1982. Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing root nodule bacteria from leguminous plants. Int. J. Syst. Bacteriol. 32:136-139. [Google Scholar]
- 10.Khoshoo, T. N. 1995. Census of India's biodiversity; tasks ahead. Curr. Sci. 69:14-17. [Google Scholar]
- 11.Kuykendall, L. D., B. Saxena, E. E. Devine, and S. E. Udell. 1992. Genetic diversity in Bradyrhizobium japonicum Jordan 1982, and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 38:501-505. [Google Scholar]
- 12.Laguerre, G., P. Mavingui, M.-R. Allard, M.-P. Charnay, P. Louvrier, S. I. Mazurier, L. Rigottier-Gois, and N. Amarger. 1996. Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl. Environ. Microbiol. 62:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laguerre, G., P. van Berkum, N. Amarger, and D. Prevost. 1997. Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl. Environ. Microbiol. 63:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laguerre, G., S. M. Nour, V. Macheret, J. Sanjuan, P. Drouin, and N. Amarger. 2001. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981-993. [DOI] [PubMed] [Google Scholar]
- 15.Ormeño-Orrillo, E., P. Vinuesa, D. Zúñiga-Dávila, and E. Martínez-Romero. 2006. Molecular diversity of native bradyrhizobia isolated from lima bean (Phaseolus lunatus L.) in Peru. Syst. Appl. Microbiol. 29:253-262. [DOI] [PubMed] [Google Scholar]
- 16.Peng, G. X., Z. Y. Tan, E. T. Wang, B. Reinhold-Hurek, W. F. Chen, and W. X. Chen. 2002. Identification of isolates from soybean nodules in Xinjiang region as Sinorhizobium xinjiangense and genetic differentiation of S. xinjiangense from Sinorhizobium fredii. Int. J. Syst. Evol. Microbiol. 52:457-462. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Navarro, D. N., M. Camacho, E. O. Leidi, R. Rivas, and E. Velazquez. 2004. Phenotypic and genotypic characterization of rhizobia from diverse geographical origin that nodulate Pachyrhizus species. Syst. Appl. Microbiol. 27:737-745. [DOI] [PubMed] [Google Scholar]
- 18.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin ver. 2000: a software for population data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- 19.Shutsrirung, A., T. Yokoyama, K. Senoo, S. Tajima, K. Minamisawa, R. Sameshima, A. Bhromsiri, and M. Hisamatsu. 2003. Genetic diversity of native Bradyrhizobium populations in soybean growing areas of Northern Thailand. Soil Sci. Plant Nutr. 49:255-265. [Google Scholar]
- 20.Stêpkowski, T., C. E. Hughes, I. J. Law, £. Markiewicz, D. Gurda, A. Chlebicka, and L. Moulin. 2007. Diversification of lupine Bradyrhizobium strains: evidence from nodulation gene trees. Appl. Environ. Microbiol. 73:3254-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford, D. L. 1998. PAUP. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 22.Thompson, J. D., T. J. Gibson. F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Berkum, P., and J. J. Fuhrmann. 2000. Evolutionary relationships among the soybean bradyrhizobia reconstructed from 16S rRNA gene and internally transcribed spacer region sequence divergence. Int. J. Syst. Evol. Microbiol. 50:2165-2172. [DOI] [PubMed] [Google Scholar]
- 24.van Berkum, P., and J. J. Fuhrmann. 2001. Characterization of soybean bradyrhizobia for which serogroup affinities have not been identified. Can. J. Microbiol. 47:519-525. [DOI] [PubMed] [Google Scholar]
- 25.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. IBP handbook no. 15. Blackwell Scientific Publications, Oxford, United Kingdom.
- 26.Vinuesa, P., C. Silva, D. Werner, and E. Martinez-Romero. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34:29-54. [DOI] [PubMed] [Google Scholar]
- 27.Willems, A., A. Munive, P. de Lajudie, and M. Gillis. 2003. In most Bradyrhizobium groups sequence comparison of 16S-23S rDNA internal transcribed spacer regions corroborates DNA-DNA hybridizations. Syst. Appl. Microbiol. 26:203-210. [DOI] [PubMed] [Google Scholar]
- 28.Willems, A., R. Coopman, and M. Gillis. 2001. Comparison of sequence analysis of 16S-23S rDNA spacer regions, AFLP analysis and DNA-DNA hybridizations in Bradyrhizobium. Int. J. Syst. Evol. Microbiol. 51:623-632. [DOI] [PubMed] [Google Scholar]
- 29.Willems, A., R. Coopman, and M. Gillis. 2001. Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Syst. Appl. Microbiol. 51:111-117. [DOI] [PubMed] [Google Scholar]
- 30.Xu, L. M., C. Ge, Z. Cui, J. Li, and H. Fan. 1995. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45:706-711. [DOI] [PubMed] [Google Scholar]
- 31.Yang, J. K., and J. C. Zhou. 2008. Diversity, phylogeny and host specificity of soybean and peanut bradyrhizobia. Biol. Fertil. Soils 44:843-851. [Google Scholar]
- 32.Yang, J. K., W. T. Zhang, T. Y. Yuan, and J. C. Zhou. 2006. Genotypic characteristics of the rrn operon and genome of indigenous soybean bradyrhizobia in cropping zones of China. Can. J. Microbiol. 52:968-976. [DOI] [PubMed] [Google Scholar]
- 33.Yao, Z. Y., F. L. Kan, E. T. Wang, G. H. Wei, and W. X. Chen. 2002. Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int. J. Syst. Evol. Microbiol. 52:2219-2230. [DOI] [PubMed] [Google Scholar]