Abstract
Although the route of transmission of Helicobacter pylori remains unknown, drinking water has been considered a possible transmission vector. It has been shown previously that, in water, biofilms are a protective niche for several pathogens, protecting them from stressful conditions, such as low carbon concentration, shear stress, and less-than-optimal temperatures. In this work, the influence of these three parameters on the persistence and cultivability of H. pylori in drinking-water biofilms was studied. Autochthonous biofilm consortia were formed in a two-stage chemostat system and then inoculated with the pathogen. Total numbers of H. pylori cells were determined by microscopy using a specific H. pylori 16S rRNA peptide nucleic acid probe, whereas cultivable cells were assessed by standard plating onto selective H. pylori medium. Cultivable H. pylori could not be detected at any time point, but the ability of H. pylori cells to incorporate, undergo morphological transformations, persist, and even agglomerate in biofilms for at least 31 days without a noticeable decrease in the total cell number (on average, the concentration was between 1.54 × 106 and 2.25 × 106 cells cm−2) or in the intracellular rRNA content may indicate that the loss of cultivability was due to entry into a viable but noncultivable state. Unlike previous results obtained for pure-culture H. pylori biofilms, shear stress did not negatively influence the numbers of H. pylori cells attached, suggesting that the autochthonous aquatic bacteria have an important role in retaining this pathogen in the sessile state, possibly by providing suitable microaerophilic environments or linking biomolecules to which the pathogen adheres. Therefore, biofilms appear to provide not only a safe haven for H. pylori but also a concentration mechanism so that subsequent sloughing releases a concentrated bolus of cells that might be infectious and that could escape routine grab sample microbiological analyses and be a cause of concern for public health.
Helicobacter pylori is one of the most prevalent pathogens in humans, especially in developing countries, where the incidence can be up to 90% of the population (16). Even though most individuals that are infected by this pathogen are asymptomatic, it is now well established that H. pylori infection can lead to the development of peptic and duodenal ulcer disease and gastric mucosa-associated lymphoid tissue lymphoma (8).
The route of transmission of this pathogen is still unknown. Person-to-person transmission seems most likely as the only place where H. pylori has been systematically isolated is the human gastrointestinal tract (3). However, some authors have suggested that water, food, and animals can also be transmission vectors (3, 7, 10, 19, 28, 37, 38). The greatest obstacle to proving that water is a transmission route is the fact that H. pylori has never been cultured from drinking-water distribution systems (DWDS) using standard cultivation techniques (3, 18). Whether this is due to the fastidious nature of the microorganism or to the loss of viability in water is the key question in the transmission debate. Accordingly, some groups have been attempting to develop artificial media to achieve better culture recovery results than those obtained with traditional Columbia blood agar, such as F-12 (36) or the selective medium (HP medium) that has been proposed for recovering H. pylori from water-exposed, heterotrophic microenvironments (15).
In the meantime, molecular techniques, such as PCR, have demonstrated the presence of H. pylori in DWDS, especially in systems with biofilms (10, 28, 29, 40). This shows that H. pylori is present in water, but DNA isolation alone does not provide any indication of the viability of the bacterium.
In recent years another molecular technique, fluorescence in situ hybridization (FISH), has been successfully used to detect this pathogen in DWDS and other bodies of water (9, 30). This technique usually detects rRNA, which implies that not only is it able to detect the presence of H. pylori but it is also able to provide some indication of viability due to the maintenance of a high rRNA content (6, 27, 42).
The aim of this work was to apply both FISH and a selective culture medium to assess the number of H. pylori cells found in an autochthonous complex consortium of drinking-water biofilms formed under different conditions in order to better understand the dynamics of H. pylori populations in real DWDS.
MATERIALS AND METHODS
Biofilm experiments.
Biofilms were formed using a two-stage chemostat model system (21). The first stage consisted of a 1-liter vessel (seed vessel), and the second stage consisted of three 1-liter vessels running in parallel but connected in series with the seed vessel. All chemostats were autoclaved and filled with filter-sterilized (0.2-μm-pore-size nylon filter) tap water (1 liter). The seed vessel was then inoculated with a microbial consortium that was obtained from tap water by filtration through a 0.2-μm-pore-size nylon filter (Pall Gelman, United Kingdom). This vessel was maintained in batch mode for 2 days to promote microbial growth and then changed into a continuous mode, in which it was fed filter-sterilized and dechlorinated tap water at a flow rate of 50 ml h−1. The seed vessel was operated at room temperature (approximately 22°C) and stirred at 300 rpm to ensure that the oxygen and nutrient concentrations were homogeneous. The effluent was divided into three parts and used to feed the second-stage chemostats, the biofilm growth vessels. Each biofilm growth vessel was also fed with fresh medium (filter-sterilized tap water) at a flow rate that maintained a dilution rate of 0.2 h−1 to promote typical environmental conditions for biofilm growth. The first vessel was stirred at 300 rpm and contained no additional carbon source (low shear stress and low carbon concentration [LS/LC conditons]); this vessel served as a control. The second vessel was stirred at 1,200 rpm, and no carbon was added (high shear stress and low carbon concentration [HS/LC conditions]). The third vessel was stirred at 300 rpm, and 8.8 mg liter−1 of carbon was added by adding 30 mg liter−1 of sodium acetate to the fresh medium (low shear stress and high carbon concentration [LS/HC conditions]). The temperature was controlled at either 15 or 20°C by a proportional integral derivative unit system (Brighton Systems, United Kingdom). After 10 days, the conditions and total cell numbers in the biofilm growth vessels were stable, and the vessels were inoculated with H. pylori NCTC 11637 at a final concentration of approximately 8 × 105 CFU ml−1 (determined by measuring the absorbance at 640 nm); this was followed by immersion of sterile polyvinylchloride (PVC) coupons (day 0). The coupons were removed after 1, 2, 4, 8, 16, and 31 days, gently rinsed to remove planktonic cells attached to the surface of the biofilm, and scraped to quantify the sessile cells.
Cultivation of H. pylori.
H. pylori NCTC 11637 was maintained in vials frozen at −80°C, and it was recovered by plating onto Columbia blood agar (Oxoid, United Kingdom) supplemented with 5% (vol/vol) defibrinated horse blood (Oxoid, United Kingdom) and incubated for 48 h at 37°C in a variable-atmosphere workstation (MACS VA500; Don Whitley, United Kingdom) set to contain a microaerophilic atmosphere consisting of 10% CO2, 7% H2, 3% O2, and 80% N2. The cultures were subcultured once for 48 h and used to inoculate the second-stage chemostats.
Preparation of coupons.
PVC coupons (1 cm2) were used as supports for biofilms. PVC was chosen for the coupons as it is the material most commonly used in water pipes and also because it has been shown to be one of the less aggressive materials for H. pylori survival (2). Coupons were immersed in water and detergent (Guard professional, United Kingdom) for 5 min, washed with a bottle brush, rinsed twice in distilled water, and air dried. Subsequently, they were washed in 70% (vol/vol) ethanol to remove any organic compounds, attached to the end of a titanium wire, and autoclaved (21).
Quantification of planktonic cells.
Water samples were taken on days 0, 1, 2, 4, 8, 16, and 31 after seeding, and biofilm growth vessels were analyzed to determine the total number of cells, the number of heterotrophic plate count (HPC) cells, and the number of cultivable H. pylori cells. The total cells were quantified using SYTO 9 (Molecular Probes, Invitrogen, United Kingdom). Briefly, 1 ml of an appropriate dilution was mixed with 0.5 μl of SYTO 9 (5 mM solution in dimethyl sulfoxide), incubated in the dark for 15 min, filtered through a 0.2-μm-pore-size polycarbonate black Nuclepore membrane (Whatman, United Kingdom), and allowed to air dry. Then 1 drop of nonfluorescence immersion oil (Fluka, United Kingdom) and a coverslip were added before observation with a Nikon Eclipse E800 episcopic differential interference contrast/epifluorescence microscope (Best Scientific, United Kingdom) (23). As cells were homogeneously distributed, 10 fields of view were randomly chosen, and the cells on each membrane were counted. HPC cells were obtained by plating samples onto R2A agar plates (Oxoid, United Kingdom) and incubating the plates at 22°C for 7 days. Cultivable H. pylori cells were quantified by plating samples in triplicate onto selective HP agar plates as described by Degnan et al. (15), using either calf serum or 5% (vol/vol) defibrinated horse blood (Oxoid, United Kingdom), and adding 0.5 g liter−1 pyruvic acid (Sigma, United Kingdom). The plates were incubated at 37°C in a microaerophilic atmosphere for 7 days. The colonies obtained on HP agar plates were tested using the urease test performed according to the manufacturer's instructions (Oxoid, United Kingdom) and the specific H. pylori peptide nucleic acid (PNA) probe to confirm the identity of H. pylori as described below (20).
Quantification of sessile cells.
Coupons were immersed in 2 ml of filter-sterilized tap water containing autoclaved 2-mm-diameter glass beads (Merck, United Kingdom) and vortexed for 1 min to remove the biofilm from the surfaces of the coupons and homogenize the suspension. A previous study in which coupons were observed by using episcopic differential interference contrast microscopy showed that this method completely removes the biofilm. Total cells, HPC cells, and cultivable H. pylori cells were quantified using the methods described above. In addition, total H. pylori cells were quantified using a specific PNA probe (5′-GAGACTAAGCCCTCC-3′) (Eurogentec, Belgium) in a FISH assay (PNA-FISH) (19). PNA-FISH was carried out by first filtering 1 ml of an appropriate dilution through a 0.2-μm Anodisc membrane (Whatman, United Kingdom). This membrane was left to air dry. Then it was covered with 4% (wt/vol) paraformaldehyde for 10 min, followed by 50% (vol/vol) ethanol for 10 min, to fix the cells and finally air dried. The hybridization, washing, and microscopy observation methods were based on the methods described by Guimarães et al. (20). In this case, 20 fields of view were randomly chosen, and the cells on each membrane were counted.
Identification of sessile cells.
All the bacteria isolated on R2A and HP media were identified by 16S rRNA gene sequencing at DNAVision SA (Belgium). Briefly, for each culture DNA was purified, amplified, and sequenced with 16S rRNA primers. The analyzed fragments were about 1,600 bp long. The sequenced fragments were then subjected to a BLAST search using the NCBI public database, and the GenBank accession numbers of the sequences with the highest sequence similarity values were obtained.
Statistical analysis.
The homogeneity of variances of the total number of cells, the total number of H. pylori cells, and the number of HPC cells was checked by the Levene test for equality of variances using a statistical package (SPSS Inc., Chicago, IL). Subsequently, differences were compared by using a one-way analysis of variance, followed by a Bonferroni post hoc test. Differences were considered relevant if the P value was <0.05.
RESULTS AND DISCUSSION
Seed vessel.
The two-stage chemostat system was operated in a continuous mode for 10 days to stabilize the microbial consortia in all of the chemostats. After this period of time, the total number of cells and the number of HPC cells in the seed vessel remained stable during the experiment, and the average concentrations were 3.23 × 106 cells ml−1 and 6.83 × 105 CFU ml−1, respectively. Autochthonous H. pylori cells were not found in the seed vessel using culture recovery or PNA-FISH detection techniques, so the chemostats in the second stage of the system were inoculated directly with H. pylori NCTC 11637, as described above, prior to the immersion of coupons.
Planktonic cells in the biofilm growth vessels.
Initially, the biofilm growth vessels were maintained at 20°C to study the influence of shear stress and carbon concentration. In the planktonic phase an increase in the shear stress did not significantly influence the total number of cells or HPC cells, but in the second-stage chemostat to which acetate carbon was added, the number of HPC cells increased (P < 0.005) (Table 1), indicating that growth was carbon limited. However, a comparison of the percentages of cultivable cells obtained for the three different conditions tested showed that the values were all very similar and that 40 to 45% of the total cells were cultivable. Subsequently, the temperature in the biofilm growth chemostats was decreased to 15°C. In the control vessel (LS/LC conditions) the percentage of cultivable cells was very similar to the value obtained at 20°C; however, the value was higher when either the shear stress (P < 0.005) or the carbon concentration (P < 0.005) was increased (Table 1).
TABLE 1.
Average total numbers of cells and numbers of HPC cells in the planktonic phase and relationship between the number of HPC cells and the total number of cells at 20 and 15°C for all three conditions tested
| Temp (°C) | Conditions | Total no. of cells (cells ml−1) | No. of HPC cells (CFU ml−1) | No. of HPC cells/total no. of cells (%) |
|---|---|---|---|---|
| 20 | LS/LC | 2.74 × 106 | 1.23 × 106 | 45 |
| HS/LC | 2.56 × 106 | 1.01 × 106 | 40 | |
| LS/HC | 6.99 × 106 | 3.17 × 106 | 45 | |
| 15 | LS/LC | 1.74 × 106 | 8.19 × 105 | 47 |
| HS/LC | 1.86 × 106 | 1.18 × 106 | 63 | |
| LS/HC | 1.78 × 107 | 1.08 × 107 | 61 |
For all water samples analyzed it was not possible to recover cultivable H. pylori. Other authors have demonstrated that in pure culture at 15 and 20°C, H. pylori is cultivable for some days both in a water suspension (1, 4, 33) and in biofilms exposed to water (2, 14), which suggests that the difficulty of recovering cultivable H. pylori in this work was due to interactions between this microorganism and other species in the bacterial population present in water, like competition for nutrients or the production of toxic compounds by other microorganisms. However, it is also important to mention that overgrowth of other microorganisms occurred in certain samples. Degnan and colleagues (15) developed HP medium as a way to selectively recover H. pylori from water and have challenged this medium with the most commonly known aquatic microorganisms and with heterotrophic consortia from real water samples, obtaining negative growth in both cases. However, real samples differ from place to place, and in the present study, microorganisms were isolated from the chemostat system on HP medium and identified by 16S rRNA gene sequencing as Brevundimonas spp., Mycobacterium chelonae, and Sphingomonas spp. (GenBank accession numbers EF194089, AM884326, and AY749436, respectively). None of these microorganisms was assessed for growth on HP medium in the original experiments of Degnan et al. (15).
Cell population in the biofilm growth vessels.
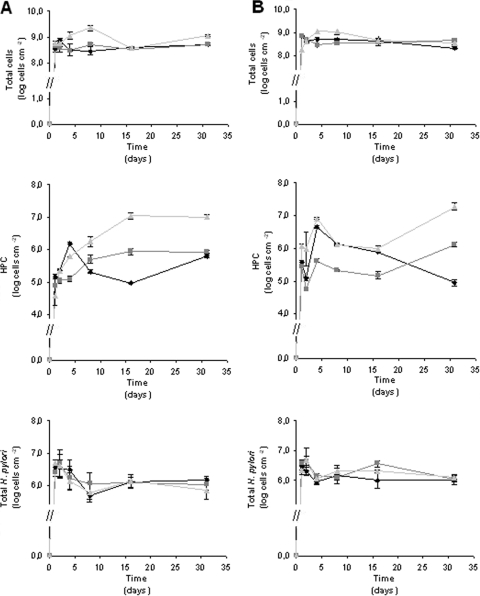
Figure 1A shows the variation in the total number of cells, number of HPC cells, and total number of H. pylori cells in biofilms grown at 20°C for the three conditions tested. As observed for planktonic samples, no cultivable H. pylori was recovered on HP medium for any of the biofilm samples. The biofilm development followed kinetics described by other authors; adhesion of most cells took place on the first day, but no statistically significant change occurred after this (P > 0.05) (13, 32). It should be noted that this pseudo-steady state is actually the result of a dynamic equilibrium typical of biofilms, where parts of biofilms detaching from coupons are balanced by the adherence of new cells (35, 39). This same trend was observed for the experiment carried out at 15°C (P > 0.05) (Fig. 1B).
FIG. 1.
Variation in the total cell number, number of HPC cells, and total number of H. pylori cells in biofilms formed at (A) 20°C and (B) 15°C under the following conditions: low shear stress and low concentration of carbon (⧫), high shear stress and low concentration of carbon (▪), and low shear stress and high concentration of carbon (▴). The error bars indicate standard deviations.
In terms of the total numbers of cells at 20°C, increasing the shear stress did not appear to affect the number, as on average, 4.31 × 108 cells cm−2 was obtained when a biofilm was formed in the LS/LC vessel and 4.14 × 108 cells cm−2 was obtained for the HS/LC vessel. However, addition of a carbon source resulted in a twofold increase in the number (1.01 × 109 cells cm−2), although the difference is not statistically significant (P > 0.05). For HPC cells the differences were even more evident when the carbon concentration was increased (4.00 × 106 CFU cm−2 versus 4.87 × 105 CFU cm−2 for the LS/LC conditions and 4.55 × 105 CFU cm−2 for the HS/LC conditions) (P < 0.1), which is not surprising as carbon is the limiting nutrient in the aquatic environment used. Comparison of the results obtained at the two temperatures suggested that when the concentration of carbon was increased, formation of a biofilm was favored at 20°C (P < 0.005).
Total counts of H. pylori cells in biofilms.
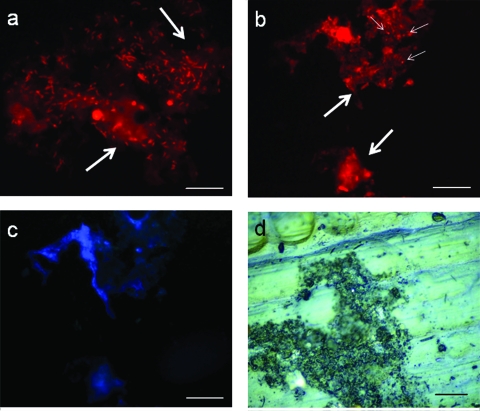
One of the most important breakthroughs in this work was our ability to consistently detect and quantify H. pylori within biofilm structures by use of the H. pylori PNA probe, as previous attempts were hampered by strong autofluorescence of the biofilm stacks in some environments containing autofluorescent contaminants, such as polycyclic aromatic hydrocarbons (6). This problem was overcome by counting the bacteria after removal of the biofilm from the coupons using glass bead agitation and washing. In this way, it was still possible to visualize the matrix structures of biofilm by epifluorescence microscopy (Fig. 2a and b); however, the autofluorescence was not bright, allowing observation of the cells. On the other hand, the number of detected bacteria in the previous study was also considerably lower than the number obtained here, which demonstrates that most of the H. pylori cells in the sessile state are embedded in biofilm structures.
FIG. 2.
Photomicrographs of hybridization with the H. pylori-specific PNA probe in biofilms grown at (a) 20°C and (b) at 15°C using an epifluorescence tetramethyl rhodamine isocyanate filter. The large arrows indicate the autofluorescent matrix of the biofilm, whereas the small arrows indicate coccoid H. pylori cells embedded in the structures. (c) Cells observed using an epifluorescence 4′,6′-diamidino-2-phenylindole (DAPI) filter serving as a control for the autofluorescence of the biofilm stacks and individual cells attached to the substratum. (d) Micrograph of a coupon with a 26-day-old biofilm formed under the LS/LC conditions at 15°C and observed using episcopic differential interference contrast microscopy. Bars = 20 μm.
When the effects of the different conditions tested were compared, the results obtained for the total numbers of H. pylori cells (at 20°C) showed a trend similar to the trend for the results described previously for the total numbers of cells, as in this case the total numbers of H. pylori cells were not statistically different (P > 0.05) for the three conditions tested (on average, 2.25 × 106 cells cm−2 for the LS/LC conditions, 2.12 × 106 cells cm−2 for the HS/LC conditions, and 2.15 × 106 cells cm−2 for the LS/HC conditions) (Fig. 1A). When the values obtained for the total number of H. pylori cells at 15°C were examined, there was no significant difference between the three conditions tested (P > 0.05); on average, the values were 1.54 × 106, 1.89 × 106, and 1.72 × 106 cells cm−2 for the LS/LC, HS/LC, and LS/HC conditions, respectively (Fig. 1B). A comparison of the results obtained at the two temperatures showed that the numbers were also not statistically significantly different (P > 0.05), suggesting that the physicochemical parameters studied in this work did not affect the presence of H. pylori in heterotrophic biofilms, indicating that if a pulse (i.e., a sporadic occurrence) of the pathogen passes through a drinking-water supply, H. pylori is included in biofilms regardless of the DWDS characteristics.
The lack of an effect of temperature on H. pylori adhesion has been demonstrated previously by Azevedo et al. (5); however, contrary to what was observed in this work, the study of Azevedo et al. demonstrated that shear stress hinders biofilm formation. Even though the systems used to generate biofilms were different (the system used in the previous study was operated in batch mode, with planktonic H. pylori cells during the entire experiment), the altered behavior might be more logically explained by the heterotrophic nature of the biofilms in the current work. Heterotrophic DWDS-associated biofilms are known to create a safe haven that protects microorganisms from external stresses, such as temperature, shear, oxygen, and nutrient concentration stresses (24, 31, 41), and might help retain H. pylori attached to surfaces.
In general, the total numbers of H. pylori cells at both temperatures decreased during the first week (P < 0.05) and were more stable after this, whereas the total number of bacteria in the biofilm remained constant throughout the experiment (Fig. 1A and B). This might be explained by the fact that the vessels were pulsed with H. pylori only at the beginning of the experiment rather than being continually challenged. Consequently, after the initial decline in the number of cells of the pathogen in the chemostats (which occurred in approximately 5 h), H. pylori cells that had only loosely adhered detached from the biofilms and could not be replenished. After 1 week, the remaining H. pylori cells were well protected within the biofilms and by any extra layers of heterotrophic cells that attached on top of them, and hence the total number of H. pylori bacteria stabilized under most conditions. Figures 2a and 2b show that most of the H. pylori cells in the sessile state were embedded in biofilm structures, which supported this hypothesis.
H. pylori morphology and location within biofilms.
The major difference between the experiments carried out at different temperatures was the difference in the shape of the cells detected by PNA-FISH. Spiral and coccoid cells were observed at both temperatures; however, at 20°C there was a larger proportion of spiral cells than there was at 15°C (Fig. 2a and 2b). This is extremely relevant as the morphology of H. pylori cells has been intimately connected with viability and infection capacity. Although it has been shown previously that coccoid H. pylori cells might be dead (17, 26), recent reports on the behavior of H. pylori in water have shown that coccoid cells are the manifestation of an environmentally robust type of cells that might be considered to be in the viable but noncultivable (VBNC) state (2, 11, 12). For H. pylori, cells in this morphological condition appear to be able to become cultivable again and cause infections when they are inoculated into mice (34).
An ecological explanation for the presence of H. pylori in biofilms is provided by the microaerophilic nature of this pathogen. In fact, other microorganisms, such as Legionella pneumophila, Campylobacter jejuni, and even Escherichia coli, have been shown to prefer the microaerophilic environments demonstrated to be present in biofilm stacks (21, 22, 31) due to their intricate structure (Fig. 2d). A recent study (25) confirmed that even at high shear stresses, the oxygen concentration remains quite low in fronds or stacks, which is certainly beneficial to H. pylori and might explain why the number of H. pylori cells is not affected at high shear stresses. However, oxygen availability is certainly dependent on the conditions under which biofilms are formed, and thus, it is not yet possible to ascertain that all biofilms contain microaerophilic microniches.
Cultivability of H. pylori in water and water-associated biofilms and implications for transmission.
Adams et al. (1) have shown that in pure culture H. pylori cells remain cultivable longer at 15°C than at 20°C. In the current study it was not possible to recover cultivable H. pylori from water samples and biofilms. However, considering the shape of the cells detected by PNA-FISH and considering that cultivable cells are spiral shaped, while coccoid cells are VBNC and therefore likely to be noncultivable, the expectation is that there should be more cultivable H. pylori cells at 20°C, demonstrating that the behavior of this pathogen in heterotrophic biofilms might be completely different than its behavior in pure culture. Additionally, the PNA probe used in this work targets sites on the 16S rRNA molecule, and it is known that the RNA content of a cell can be indicative of viability (6), which suggests that the cells detected were still viable. It is shown above that the concentration of all H. pylori cells in the biofilms formed in this work is either higher than or very similar to the concentrations found when pure-culture biofilms were formed (5). In addition, the detection of H. pylori embedded in biofilms suggests that there is a close association with other bacteria present in the biofilms. These two factors, together with the persistence of a bright PNA-FISH signal, which is indicative of a high rRNA content, suggest that the heterotrophic bacteria present in the biofilms formed in this study were not a negative influence on H. pylori but only induced its transformation to the more robust coccoid morphology (2).
The mode of transmission of H. pylori is not well established, and although there is considerable evidence that water is a strong candidate, several authors are skeptical about accepting this route of transmission. This work provides new evidence about the survival of H. pylori in drinking-water biofilms and shows that this pathogen, although it is fastidious and loses cultivability easily and rapidly, can still remain viable in the environmentally robust coccoid VBNC state for long periods of time. The fact that this work, in contrast to the pure-culture studies described previously, was done using natural, heterotrophic microbial consortia shows the capacity of H. pylori to adapt to stress situations by “taking advantage” of the presence of other microorganisms.
Conclusions.
The results of this study are in agreement with the data obtained for the aquatic environment, in which H. pylori can be detected by molecular techniques but not by plating methods (9). In fact, it is shown here that even in artificially inoculated systems, H. pylori recovery and growth on agar culture plates remain elusive despite the abundance of the bacterium in biofilms as assessed by PNA-FISH. On the other hand, previous experiments have shown that H. pylori NCTC 11637 exposed to water remains cultivable in pure-culture biofilms for at least 24 h (3).
The high numbers of H. pylori cells in biofilms and the maintenance of high levels of rRNA in the cells for at least 31 days strongly suggest that, far from being deleterious, interactions indeed protect this pathogen by providing a stable, possibly microaerophilic environment in which this pathogen can subsist. This indicates that H. pylori might be found in biofilms in a VBNC state, confirming that standard cultivation methods are not the best approach to assess the safety of drinking water with respect to this pathogen and that while improved recovery methods are not available, it is important to utilize PNA-FISH as a monitoring method. This work shows that even when cultivable H. pylori is not detected by standard methods, this pathogen persists in biofilms under most conditions found in aquatic environments, suggesting that water biofilms might have a role in H. pylori transmission.
Acknowledgments
We thank Nuno Guimarães for his technical assistance with the PNA-FISH method.
This work was supported by the Portuguese Institute Fundação para a Ciência e Tecnologia (Ph.D. grant SFRH/BD/17088/2004 and postdoctoral grant SFRH/BPD/20484/2004).
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Adams, B. L., T. C. Bates, and J. D. Oliver. 2003. Survival of Helicobacter pylori in a natural freshwater environment. Appl. Environ. Microbiol. 69:7462-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azevedo, N. F., C. Almeida, L. Cerqueira, S. Dias, C. W. Keevil, and M. J. Vieira. 2007. Coccoid form of Helicobacter pylori as a morphological manifestation of cell adaptation to the environment. Appl. Environ. Microbiol. 73:3423-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azevedo, N. F., N. Guimaraes, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. A new model for the transmission of Helicobacter pylori: role of environmental reservoirs as gene pools to increase strain diversity. Crit. Rev. Microbiol. 33:157-169. [DOI] [PubMed] [Google Scholar]
- 4.Azevedo, N. F., A. P. Pacheco, C. W. Keevil, and M. J. Vieira. 2004. Nutrient shock and incubation atmosphere influence recovery of culturable Helicobacter pylori from water. Appl. Environ. Microbiol. 70:490-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azevedo, N. F., A. R. Pinto, N. M. Reis, M. J. Vieira, and C. W. Keevil. 2006. Shear stress, temperature, and inoculation concentration influence the adhesion of water-stressed Helicobacter pylori to stainless steel 304 and polypropylene. Appl. Environ. Microbiol. 72:2936-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azevedo, N. F., M. J. Vieira, and C. W. Keevil. 2003. Establishment of a continuous model system to study Helicobacter pylori survival in potable water biofilms. Water Sci. Technol. 47:155-160. [PubMed] [Google Scholar]
- 7.Bellack, N. R., M. W. Koehoorn, Y. C. Macnab, and M. G. Morshed. 2006. A conceptual model of water's role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiol. Infect. 134:439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bragança, S. M., N. F. Azevedo, L. C. Simões, C. W. Keevil, and M. J. Vieira. 2007. Use of fluorescent in situ hybridisation for the visualisation of Helicobacter pylori in real drinking water biofilms. Water Sci. Technol. 55:387-393. [DOI] [PubMed] [Google Scholar]
- 10.Bunn, J. E. G., W. G. Mackay, J. E. Thomas, D. C. Reid, and L. T. Weaver. 2002. Detection of DNA in drinking water biofilms: Helicobacter pylori implications for transmission in early life. Lett. Appl. Microbiol. 34:450-454. [DOI] [PubMed] [Google Scholar]
- 11.Cellini, L., R. Grande, E. Di Campli, S. Di Bartolomeo, D. Di Lorio, and S. Caputi. 2005. Biofilm formation and modulation of luxS and rpoD expression by Helicobacter pylori. Biofilms 2:1-9. [Google Scholar]
- 12.Chaput, C., C. Ecobichon, N. Cayet, S. E. Girardin, C. Werts, S. Guadagnini, M. C. Prevost, D. Mengin-Lecreulx, A. Labigne, and I. G. Boneca. 2006. Role of amiA in the morphological transition of Helicobacter pylori and in immune escape. PLoS Pathog. 2:844-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Codony, F., J. Morato, F. Ribas, and J. D. Mas. 2002. Effect of chlorine, biodegradable dissolved organic carbon and suspended bacteria on biofilm development in drinking water systems. J. Basic Microbiol. 42:311-319. [DOI] [PubMed] [Google Scholar]
- 14.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degnan, A. J., W. C. Sonzogni, and J. H. Standridge. 2003. Development of a plating medium for selection of Helicobacter pylori from water samples. Appl. Environ. Microbiol. 69:2914-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eaton, K. A., C. E. Catrenich, K. M. Makin, and S. Krakowka. 1995. Virulence of coccoid and bacillary forms of Helicobacter pylori in gnotobiotic piglets. J. Infect. Dis. 171:459-462. [DOI] [PubMed] [Google Scholar]
- 18.Engstrand, L. 2001. Helicobacter in water and waterborne routes of transmission. J. Appl. Microbiol. 90:80S-84S. [DOI] [PubMed] [Google Scholar]
- 19.Gomes, B. C., and E. C. P. De Martinis. 2004. The significance of Helicobacter pylori in water, food and environmental samples. Food Control 15:397-403. [Google Scholar]
- 20.Guimarães, N., N. F. Azevedo, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 45:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keevil, C. W. 2001. Continuous culture models to study pathogens in biofilms. Methods Enzymol. 337:104-122. [DOI] [PubMed] [Google Scholar]
- 22.Keevil, C. W. 2002. Pathogens in environmental biofilms, p. 2339-2356. In G. Bitton (ed.), The encyclopedia of environmental microbiology. Wiley, New York, NY.
- 23.Keevil, C. W. 2003. Rapid detection of biofilms and adherent pathogens using scanning confocal laser microscopy and episcopic differential interference contrast microscopy. Water Sci. Technol. 47:105-116. [PubMed] [Google Scholar]
- 24.Keevil, C. W., J. Rogers, and J. T. Walker. 1995. Potable water biofilms. Microbiol. Eur. 3:10-14. [Google Scholar]
- 25.Kühl, M., L. F. Rickelt, and R. Thar. 2007. Combined imaging of bacteria and oxygen in biofilms. Appl. Environ. Microbiol. 73:6289-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kusters, J. G., M. M. Gerrits, J. A. Van Strijp, and C. M. Vandenbroucke-Grauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehtola, M. J., E. Torvinen, L. T. Miettinen, and C. W. Keevil. 2006. Fluorescence in situ hybridization using peptide nucleic acid probes for rapid detection of Mycobacterium avium subsp avium and Mycobacterium avium subsp paratuberculosis in potable-water biofilms. Appl. Environ. Microbiol. 72:848-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackay, W. G., L. T. Gribbon, M. R. Barer, and D. C. Reid. 1998. Biofilms in drinking water systems—a possible reservoir for Helicobacter pylori. Water Sci. Technol. 38:181-185. [DOI] [PubMed] [Google Scholar]
- 29.Park, S. R., W. G. Mackay, and D. C. Reid. 2001. Helicobacter sp. recovered from drinking water biofilm sampled from a water distribution system. Water Res. 35:1624-1626. [DOI] [PubMed] [Google Scholar]
- 30.Queralt, N., R. Bartolome, and R. Araujo. 2005. Detection of Helicobacter pylori DNA in human faeces and water with different levels of faecal pollution in the north-east of Spain. J. Appl. Microbiol. 98:889-895. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, P. J., J. T. Walker, C. W. Keevil, and J. Cole. 1995. Reporter genes and fluorescent-probes for studying the colonization of biofilms in a drinking-water supply line by enteric bacteria. FEMS Microbiol. Lett. 129:183-188. [DOI] [PubMed] [Google Scholar]
- 32.Rogers, J., A. B. Dowsett, P. J. Dennis, J. V. Lee, and C. W. Keevil. 1994. Influence of temperature and plumbing material selection on biofilm formation and growth of Legionella pneumophila in a model potable water system containing complex microbial flora. Appl. Environ. Microbiol. 60:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shahamat, M., U. Mai, C. Paszko-Kolva, M. Kessel, and R. R. Colwell. 1993. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl. Environ. Microbiol. 59:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.She, F. F., J. Y. Lin, J. Y. Liu, C. Huang, and D. H. Su. 2003. Virulence of water-induced coccoid Helicobacter pylori and its experimental infection in mice. World J. Gastroenterol. 9:516-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 36.Testerman, T. L., D. J. McGee, and H. L. T. Mobley. 2001. Helicobacter pylori growth and urease detection in the chemically defined medium Ham's F-12 nutrient mixture. J. Clin. Microbiol. 39:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Duynhoven, Y. T., and R. de Jonge. 2001. Transmission of Helicobacter pylori: a role for food? Bull. W. H. O. 79:455-460. [PMC free article] [PubMed] [Google Scholar]
- 38.Velazquez, M., and J. M. Feirtag. 1999. Helicobacter pylori: characteristics, pathogenicity, detection methods and mode of transmission implicating foods and water. Int. J. Food Microbiol. 53:95-104. [DOI] [PubMed] [Google Scholar]
- 39.Vieira, M. J., L. F. Melo, and M. M. Pinheiro. 1993. Biofilm formation: hydrodynamic effects on internal diffusion and structure. Biofouling 7:67-80. [Google Scholar]
- 40.Voytek, M. A., J. B. Ashen, L. R. Fogarty, J. D. Kirshtein, and E. R. Landa. 2005. Detection of Helicobacter pylori and fecal indicator bacteria in five North American rivers. J. Water Health 3:405-422. [DOI] [PubMed] [Google Scholar]
- 41.Walker, J. T., C. W. Mackerness, J. Rogers, and C. W. Keevil. 1995. Heterogenous mosaic biofilm—a haven for waterborne pathogens, p. 196-204. In H. M. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 42.Wilks, S. A., and C. W. Keevil. 2006. Targeting species-specific low-affinity 16S rRNA binding sites by using peptide nucleic acids for detection of legionellae in biofilms. Appl. Environ. Microbiol. 72:5453-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]