Abstract
Previous results from a 16S rRNA gene library analysis showed high diversity within the prokaryotic community of a subterranean radioactive thermal spring, the “Franz-Josef-Quelle” (FJQ) in Bad Gastein, Austria, as well as evidence for ammonia oxidation by crenarchaeota. This study reports further characterization of the community by denaturing gradient gel electrophoresis (DGGE) analysis, fluorescence in situ hybridization (FISH), and semiquantitative nitrification measurements. DGGE bands from three types of samples (filtered water, biofilms on glass slides, and naturally grown biofilms), including samples collected at two distinct times (January 2005 and July 2006), were analyzed. The archaeal community consisted mainly of Crenarchaeota of the soil-subsurface-freshwater group (group 1.1b) and showed a higher diversity than in the previous 16S rRNA gene library analysis, as was also found for crenarchaeal amoA genes. No bacterial amoA genes were detected. FISH analysis of biofilms indicated the presence of archaeal cells with an abundance of 5.3% (±4.5%) in the total 4′,6-diamidino-2-phenylindole (DAPI)-stained community. Microcosm experiments of several weeks in duration showed a decline of ammonium that correlated with an increase of nitrite, the presence of crenarchaeal amoA genes, and the absence of bacterial amoA genes. The data suggested that only ammonia-oxidizing archaea (AOA) perform the first step of nitrification in this 45°C environment. The crenarchaeal amoA gene sequences grouped within a novel cluster of amoA sequences from the database, originating from geothermally influenced environments, for which we propose the designation “thermal spring” cluster and which may be older than most AOA from soils on earth.
The exploration of subsurface environments is a topic of increasing interest in microbiology, because the highest numbers and diversity of microorganisms are found within subterranean and submarine environments as well as in the oceans (51). The total estimated number of prokaryotes in our world is 4 × 1030 to 6 × 1030 cells, wherein the oceans, soils, and oceanic and terrestrial subsurfaces contain 1.2 × 1029, 2.6 × 1029, 3.5 × 1030, and 0.25 × 1030 to 2.5 × 1030 cells, respectively (51). In general, caves are considered to be extreme environments for life and are often resource limited due to the absence of light, which precludes primary production of organic material by phototrophs. On the other hand, such environments provide a variety of redox interfaces, allowing primary growth of chemolithoautotrophic microorganisms (37). The exploration of several subsurface environments, including diverse microbial mats in caves (12, 20, 31, 33), geothermal waters of gold mines or aquifers (18, 24, 47), the Movile cave in Romania (21), and several other cave systems (36, 54), has recently been reported. Many of the dominating groups in these environments belong to the Proteobacteria (e.g., members of Thiobacillus spp. [18, 31]), Bacteroidetes, Acidobacteria, Nitrospirae, and mesophilic Crenarchaeota.
Bad Gastein, a village in the national park Hohe Tauern in the Alps near Salzburg, Austria, and its subsurface radioactive thermal mineral springs provide access to such subsurface realms. The thermal mineral springs in Bad Gastein consist of a cluster of 17 major springs with temperatures up to 47°C, which provide about 4 to 5 million liters of thermal mineral water per day (8, 50). One of the major springs is called “Franz-Josef-Quelle” (FJQ). It consists of a total of 27 single water discharges, issuing directly from rock fissures and delivering 295 m3 water per day. Its average temperature is 45.6°C, and the pH is about 8.0. The microbial community of FJQ was described recently (50). The presence of ammonia-oxidizing Crenarchaeota was shown by the detection of the crenarchaeal amoA gene (50), which encodes the alpha subunit of ammonia monooxygenase, the key enzyme for ammonia oxidation (16, 42, 49). The spring FJQ is thought to represent a quite stable habitat, with nearly no seasonal influences, such as intermixture with cold groundwater or surface water, and no sunlight, as well as stable chemical factors (8).
In this study, denaturing gradient gel electrophoresis (DGGE) and fluorescence in situ hybridization (FISH) analyses were applied to obtain additional information on the taxonomic affiliation of crenarchaeal community members of the subsurface spring. Analysis of DGGE bands from three different types of samples and from two sampling time points (January 2005 and July 2006) was carried out. The DGGE approach was also used to obtain additional information on ammonia-oxidizing microorganisms by searching for bacterial and crenarchaeal amoA genes, since the importance of ammonia-oxidizing archaea (AOA) in the global nitrogen cycle is increasingly being recognized (29, 52). Until now, crenarchaeal amoA genes have been discovered mainly in various soils (23, 29), wastewaters (40), and marine ecosystems and also in the oceans (3, 7, 14, 15, 26, 27, 52), a few geothermally heated environments (like the spring FJQ) (50), a geothermal mine adit (45), and, most recently, in several hot springs (10, 17, 41).
MATERIALS AND METHODS
Sampling and DNA extraction.
Samples were taken from the subsurface water discharges numbered 21, 23, and 24 of the spring FJQ, which are accessible through a tunnel, as described previously (50). All samples were collected under aseptic conditions by using sterile equipment. For chemical analysis (see below), untreated samples, which were taken every few months, were used. Samples for microbiological analyses were collected in January 2005 and in July 2006. Glass slides (75 by 25 by 1 mm) were mounted into a metal rack, which held 10 slides, and were placed into the spring for 14 days. Naturally grown biofilms were removed using sterile spatulas. Slides and biofilm samples were placed after collection into sterile 50-ml reaction tubes. Water samples were collected in 10-liter bottles. In the laboratory, water samples were filtered through a 0.22-μm-pore-size filter and designated F (filtered water). Naturally grown biofilm was designated BF, and biofilm obtained on glass slides was designated OT. The letter -W or -S (for winter or summer, respectively) was added to the sample designations.
Filters as well as biofilm samples and glass slides with biofilms were stored at −70°C until further use. DNA was extracted by procedures recently described (50).
Chemical analysis.
Chemical analyses of the spring water were performed by a commercial laboratory (Hydrologische Untersuchungsstelle Salzburg, Austria). The thermal mineral water had the following characteristics (standard deviation from three determinations is given in parentheses): average pH of 7.88 (0.04); cations Na+, 78.33 (3.21) mg/liter; Ca2+, 21.2 (1.37) mg/liter; K+, 3.97 (0.21) mg/liter; Mg2+, 0.51 (0.03) mg/liter; and NH4+, <0.02 mg/liter; anions SO42−, 131 (1.00) mg/liter; HCO3−, 61.63 (2.36) mg/liter; Cl−, 26.00 (1.00) mg/liter, NO3−, <0.5 mg/liter; PO43−, <0.01 mg/liter; and NO2−, <0.003 mg/liter. Additionally, an analysis of trace elements by inductively coupled plasma optical emission spectrometry resulted in the detection of 0.187 mg/liter Ba, 0.4 mg/liter Li, 0.002 mg/liter Mn, 0.025 mg/liter Mo, 0.008 mg/liter Ni, 0.63 mg/liter Sr, 0.043 mg/liter W, and 0.059 mg/liter Zn. The latter analysis was performed by Bühler AG Laboratories, Uzwil, Switzerland.
PCR, DGGE analysis, clone library construction, and grouping of sequences.
Archaeal 16S rRNA genes were amplified with primer pairs A20F (32) and A958R (11) for archaea (PCR conditions were 95°C for 5 min; 35 cycles at 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 10 min) and CTO189f and CTO654r (25) for the β-subdivision of ammonia-oxidizing bacteria (AOB) (PCR conditions were 95°C for 5 min; 30 cycles at 94°C for 30 s, 58°C for 30 s, and 72°C for 20 s; and a final extension step at 72°C for 10 min). Amplification of crenarchaeal and bacterial amoA genes was performed with primer pairs amoA19F (29) and amoA643R (49) for crenarchaeal genes and amoA189F and amoA682R (19) as well as the specific primers amoA-1F and amoA-2R (42) for betaproteobacterial genes (PCR conditions were 94°C for 5 min; followed by 35 cycles at 95°C for 30 s, 55°C [for archaeal amoA genes] or 60°C [for bacterial amoA genes] for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 10 min). For the optimization of PCRs specific for AOB as well as for positive controls, the DNA of Nitrosomonas europaea (a gift of G. Nicol, University of Aberdeen, United Kingdom) was used.
PCRs for DGGE analyses were performed using a nested approach with the primer pairs and PCR programs listed in Table 1. All PCRs were performed using 1.25 U GoTaq polymerase (Promega, Madison, WI). PCR mixtures for DGGE analyses additionally contained 25 mM MgCl2 for a final Mg2+ concentration of 3 mM and 50 ng template DNA of the first PCR product.
TABLE 1.
DGGE primers and PCR cycling programs used in this work
| Primer (reference) | Sequence (5′-3′) | Target | Cycling conditions (source or reference) |
|---|---|---|---|
| amoA189Fb (19) | GGNGACTGGGACTTCTGG | Bacterial amoA gene | 94°C for 5 min; followed by 35 cycles at 95°C |
| amoA682R (19) | GAASGCNGAGAAGAASGC | Bacterial amoA gene | for 30 s, 55°C or 60°Cd for 1 min, and 72°C for 45 s; followed by 72°C for 10 min (this work) |
| ARCHamoAfa (52)c | CNGAYTGGGCHTGGACATC | Crenarchaeal amoA gene | 94°C for 5 min; followed by 35 cycles at 95°C |
| ARCHamoAr (52)c | TTCYKYYTGTHGCCCAGTA | Crenarchaeal amoA gene | for 30 s, 55°C or 60°Cd for 1 min, and 72°C for 45 s; followed by 72°C for 10 min (this work) |
| PARCH340fa (39) | CCCTACGGGGYGCASCAG | Archaeal V3 region | 95°C for 5 min; followed by 5 cycles at 94°C |
| PARCH519r (39) | TTACCGCGGCYGCTG | Archaeal V3 region | for 30 s, 55°C for 1 min, and 72°C for 1 min; followed by 30 cycles at 92°C for 30 s, 55°C for 1 min, and 72°C for 1 min; and finally 72°C for 10 min (35) |
A GC clamp (34) was attached at the 5′ end: TCG CCC GCC GCG CGC GGC GGG CGG GCC GGG GGC ACG GGG GG.
A GC clamp (25) was attached at the 5′ end: CCG CCG CGC GGC GGG CGG GGC GGG GGC ACG GGG GG.
Primers for the crenarchaeal amoA gene were modified to more-universal primers to match clone sequences of the crenarchaeal group 1.1b obtained by Weidler et al. (50).
The annealing temperature for crenarchaeal amoA genes was 55°C and that for bacterial amoA genes was 60°C.
DGGE analysis was performed with a DCodeMutation detection system (Bio-Rad, Hercules, CA) at 60°C and 200 V for 3 h, as described previously (29). Polyacrylamide gels (8%, wt/vol, for 16S rRNA genes and crenarchaeal amoA genes or 6%, wt/vol, for bacterial amoA genes) included a denaturant gradient ranging from 15 to 55% (with 100% denaturant corresponding to 7 M urea and 40% formamide) for archaeal 16S rRNA genes and amoA gene products. About 1 μg of each DGGE PCR product was loaded on the DGGE gel. After electrophoresis, the gels were soaked for 30 min in 1× TAE (40 mM Tris, 20 mM acetic acid, 1 mM EDTA buffer) containing ethidium bromide (1 μg/ml), rinsed for 30 min in 1× TAE, and photographed. All DGGE PCRs and DGGE gel analyses were done in triplicate. Replicate DGGE gels showed identical fingerprint patterns.
DGGE bands were excised from the gels, and the polyacrylamide slices were used directly as templates to reamplify DGGE bands with primers without a GC clamp. Subsequently, PCR products were cloned into a pGEM-T vector II system (Promega) as recommended by the manufacturer and at least five clones of each band were sequenced. 16S rRNA gene sequences were compared with the program Similarity Matrix, version 1.1 (28), available through RDP II, release 8.1, and by a distance matrix generated with the program ClustalX (48), whereas amoA gene sequences were compared only to each other by a distance matrix generated with the ClustalX software. All gene sequences which were ≥99% identical were defined as one operational taxonomic unit (OTU). In the case of archaeal amoA genes, sequences which were ≥95% identical, as suggested by Spear et al. (45), were grouped into OTU groups (designated OTUG#AOA).
Sequencing and phylogenetic analysis.
Sequencing and phylogenetic analysis were performed as recently described by Weidler et al. (50).
FISH and quantitative cell counts.
FISH analysis of biofilms and of glass slides, which had been placed into the spring water for 2 weeks, was performed according to a protocol described previously (1, 6) by using the Cy3-labeled universal probes EUB338 (5′-GCT GCC TCC CGT AGG AGT) (2) and ARCH915 (5′-GTS CTC CCC CGC CAA TTC CT) (46). Cells in biofilms and on glass slides were fixed with 3 volumes of 4% paraformaldehyde (Sigma) for 2 h at room temperature. Prior to fixation, glass slides were placed into 1 volume of filtered and autoclaved spring water mixed with 3 volumes of 4% paraformaldehyde in a 50-ml reaction tube, while biofilm samples were centrifuged at 6,000 × g for 3 min at 4°C, washed with 1× phosphate-buffered saline (PBS), and centrifuged again. Following fixation, glass slides were washed in 1× PBS and subsequently stored in a 50:50 mixture of 100% ethanol and 1× PBS. The pellets of the biofilms were resuspended in 1× PBS, and 1 volume of 100% ethanol was added. Samples were stored at −20°C.
A 10-μl volume of hybridization buffer (0.9 M NaCl, 20 mM Tris-Cl at pH 7.2, 35% formamide for ARCH915 or 25% for EUB338, 0.02% sodium dodecyl sulfate) was spotted in each well of the microscopic slide, and 50 ng of each probe was added. Slides were incubated for 2 h at 46°C in a hybridization chamber of an Eppendorf Comfort thermomixer (Hamburg, Germany). After hybridization, the slides were placed into the washing buffer (20 mM Tris-Cl at pH 7.2, 70 mM NaCl, 5 mM EDTA at pH 8.0, 0.01% sodium dodecyl sulfate) containing 20 μl of an aqueous 4′,6-diamidino-2-phenylindole (DAPI) solution (1 mg/ml; Sigma) at 48°C for 20 min. After being washed, the slides were rinsed with distilled H2O and quickly dried. As positive controls for FISH, cells of Escherichia coli K-12 and Acidianus brierleyi DSM 1651T were used. Quantitative determination of cells was performed manually by counting DAPI-stained cells and ARCH915-labeled cells. A total of approximately 6,000 DAPI-stained particles were evaluated.
Fluorescence microscopy.
Microscopy of the hybridized cells was performed using a Leica fluorescence microscope type DM5000B with a mercury lamp (Hg100W; Leica Microsystems, Wetzlar, Germany) and a digital camera type DFC300FX (Leica Microsystems) combined with Leica image manager IDM50 software, version V5.0 R190.
Microcosm experiments and control PCRs.
Twenty-five liters of thermal mineral water was filtered through a 0.22-μm-pore-size filter (Millipore Corporation, Billerica, MA). The filter was used for the inoculum by placement into 2.5 liters of filtered spring water which was supplemented with components (final concentrations) similar to those described by Simon et al. (44): 2.5 mM (NH4)2SO4, 5 mM Na2HPO4, 5 mM KH2PO4, 2 mM KCl, 1% vitamin solution (9), and 1% trace element solution (43). The batch culture was incubated at 40°C in the dark for at least 38 days. Nitrite and nitrate concentrations were measured three times a week and ammonia was measured once a week by using Merckoquant semiquantitative test strips (Merck, Darmstadt, Germany). The pH was adjusted to 7.5 to 7.7 with 2 N NaOH every second day. A control experiment consisted of medium of the same composition but lacking the inoculum. The presence of archaeal and AOB 16S rRNA and amoA genes in the microcosm was examined after 5 weeks of incubation, with PCRs performed under the same conditions as described above.
Nucleotide sequence accession numbers.
All archaeal small-subunit-rRNA gene sequences as well as the amoA and pmoA gene sequences were deposited in the EMBL/EBI nucleotide sequence database under the following accession numbers: AM749084 to AM749094 for partial 16S rRNA gene sequences, AM749095 to AM749112 for crenarchaeal amoA gene sequences, and AM749113 to AM749117 for bacterial pmoA gene sequences.
RESULTS AND DISCUSSION
DGGE analysis of the prokaryotic community.
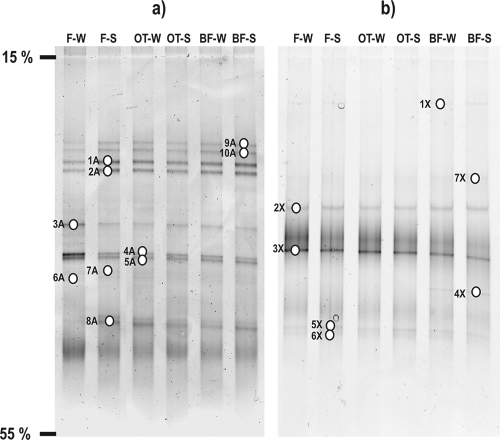
Figure 1a shows the DGGE profiles of the 16S rRNA gene fragments of the archaeal community from the three sample types (filtered spring water, naturally grown biofilms, and biofilms deposited on glass slides), including the two sampling time points January 2005 and July 2006. Figure 1b shows the DGGE gel of the archaeal amoA gene fragments in the same order of samples as for the 16S rRNA genes. All visible bands were excised from the DGGE gels for subsequent analysis of archaeal 16S rRNA and amoA genes. Community DGGE fingerprints of the 16S rRNA gene fragments and subsequent analysis of 10 bands (Fig. 1a) resulted in seven archaeal OTUs with a sequence length of 151 bp; these were designated FJQ-OTU#A (Table 2). Analysis of the amoA gene DGGE bands (Fig. 1b) was performed with seven bands for AOA. A similar DGGE gel obtained with AOB amoA-specific primers contained 16 bands, 9 of which were analyzed. None of them yielded any bacterial amoA gene sequence (not shown; see below). These results corroborated the earlier finding of the lack of beta- and gammaproteobacterial amoA genes (50).
FIG. 1.
DGGE profiles of the spring FJQ. (a) Archaeal 16S rRNA gene fragments and (b) amoA genes of AOA. F-W, filtered water, winter; F-S, filtered water, summer; OT-W, biofilm on glass slides, winter; OT-S, biofilm on glass slides, summer; BF-W, naturally grown biofilm, winter; BF-S, naturally grown biofilm, summer. Numbered open ovals indicate excised and successfully sequenced DGGE bands.
TABLE 2.
Most similar database sequences of crenarchaeal 16S rRNA genes recovered from the subsurface spring FJQ by DGGE analysis
| OTU (phylotype) | Similarity (%) | Nearest database sequence, origin (EMBL/EBI accession no.) | DGGE band(s) |
|---|---|---|---|
| 1A (a) | 98.0 | Unc.a crenarchaeon clone FJQBAA3, subsurface spring (AM039532) | 1A, 3A, 4A, 6A |
| 1A (b) | 98.0 | Unc. soil archaeon clone S223, agricultural soil (AY037656) | 7A, 9A |
| 2A (a) | 96.7 | Unc. crenarchaeon clone FJQBAA5, subsurface spring (AM039530) | 2A |
| 2A (b) | 97.4 | Unc. crenarchaeon clone FJQFA2, subsurface spring (AM039531) | 1A, 8A, 10A |
| 3A | 98.7 | Unc. crenarchaeon clone FJQFA2, subsurface spring (AM039531) | 2A, 9A |
| 4A | 98.0 | Unc. crenarchaeon clone FJQFA2, subsurface spring (AM039531) | 4A, 5A |
| 5A (a) | 98.0 | Unc. crenarchaeon clone FJQFA13, subsurface spring (AM039534) | 7A |
| 5A (b) | 98.0 | Unc. soil archaeon clone S223, agricultural soil (AY037656) | 5A, 7A |
| 6A (a) | 97.4 | Unc. soil archaeon clone S223, agricultural soil (AY037656) | 8A |
| 6A (b) | 96.7 | Unc. soil archaeon clone S223, agricultural soil (AY037656) | 9A |
| 7A | 98.0 | Unc. crenarchaeon clone FJQBAA5, subsurface spring (AM039530) | 3A |
Unc., uncultured.
As reported previously, the archaeal clone library was dominated by clones of the crenarchaeal group 1.1b (50). The seven archaeal OTUs found here also belonged to the crenarchaeal cluster 1.1b, which contains sequences recovered from soil, freshwater, and subsurface environments. Moderately thermophilic Crenarchaeota seemed to be a stable and dominating component in the archaeal/microbial community of the subsurface spring, due to the invariable appearance of DGGE bands of archaeal 16S rRNA and amoA genes among the different samples. This notion was confirmed by the very similar patterns in the DGGE analysis (Fig. 1a) when a comparison between the two time points, January 2005 and July 2006, was carried out.
Archaea of the subsurface thermal spring.
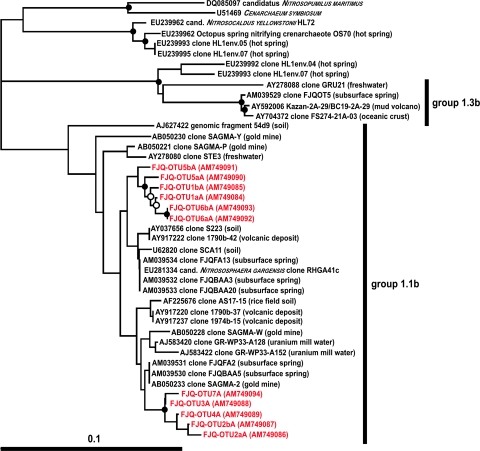
More than 99% of archaeal clones from a 16S rRNA gene analysis were shown recently to belong to two OTUs that represented the crenarchaeal cluster 1.1b (50). Several OTUs (Table 2) represented sequences that were closely related to clones of OTU FJQGA2 (50), which had the nearest database match to a sequence from uranium mill tailings (clone Gitt-GR-39) (Fig. 2) (50), whereas five OTUs grouped together with OTU FJQGA1 (50), whose nearest database match was isolated from a South African gold mine (clone SAGMA-2) (Fig. 2) (47). All analyzed DGGE bands contained sequences of the crenarchaeal group 1.1b (soil-freshwater-subsurface group) (Fig. 2). Members of other crenarchaeal groups or of euryarchaeal origin were not detected in this work. All nearest database sequences represented uncultured Crenarchaeota from the spring FJQ (similarities of 96% to 98%), except one sequence, which was obtained from agricultural soil (similarities of 96% to 99%) (Table 2). Additionally, results of the DGGE approach showed that the diversity within the 1.1b group was larger, as previously shown by the 16S rRNA library approach (50), since a higher number of OTUs was obtained (seven versus two OTUs).
FIG. 2.
Archaeal phylogenetic tree based on 16S rRNA gene sequences of the Crenarchaeota (groups 1.1b and 1.3b), including various sequences obtained from the DGGE gels of the spring FJQ. The tree was inferred by a neighbor-joining analysis. The scale bar represents a 10% nucleotide sequence difference. “Cultivated” Crenarchaeota constitute the outgroup, and strains depicted in uppercase letters represent cultivated species. Accession numbers are from the EMBL/EBI database. Symbols on the branches indicate bootstrap confidence values, as follows: •, >90%; ○, >75%.
Chemical analysis for crenarchaeal amoA and bacterial pmoA genes.
Bock and Wagner (4) pointed out that ammonia or ammonium is the most frequently found form of nitrogen compounds in the biosphere. In contrast, nitrite is usually found in trace amounts in aerobic habitats. Under oxic conditions and in the presence of nitrifiers, ammonia and nitrite are converted to nitrate. Chemical analysis of the FJQ spring agreed with these notions, since the concentration of ammonium/ammonia was low (<0.001 mM), which could indicate microbial oxidation of released organic nitrogen compounds, and the nitrite concentration in the spring was very low (<0.065 μM). Thus, the oxidation to nitrate by nitrifying organisms (e.g., Nitrospira spp., which were found in FJQ [50]) might explain the higher nitrate concentration in the spring water (<0.008 mM).
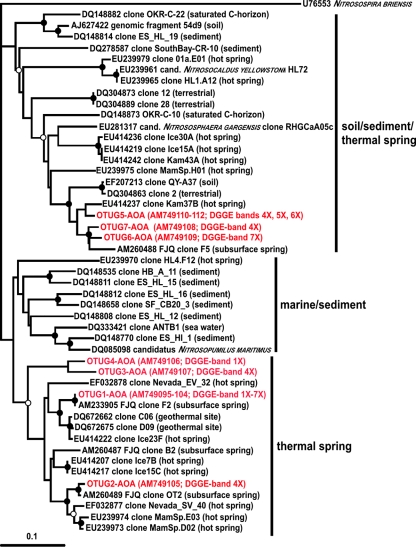
The visible bands of crenarchaeal amoA genes which were recovered from the DGGE gel (Fig. 1b) were 256 bp long. The grouping of crenarchaeal amoA gene sequences resulted in 16 phylotypes (designated OTU#AOA), which were further grouped into seven OTUs with a cutoff of 5% sequence similarity (designated OTUG#AOA) (45). Fourteen phylotypes had similarities to the amoA gene sequences obtained from recently performed experiments ranging from 94% to 100% (50). Two phylotypes exhibited only 84% and 68% similarity to clone F2 (EMBL/EBI accession number AM233905) recovered from the subsurface spring FJQ (50). Five OTUs of the analysis described here grouped together in the phylogenetic tree with sequences of amoA genes from the soil/sediment/thermal spring cluster (Fig. 3). These OTUs were similar to clone F5 of the spring FJQ (50) (91% to 94% similarity) and to a terrestrial clone from Australia (clone 2, EMBL/EBI accession number DQ304863), as well as to clones recovered from hot springs from Kamchatka, Iceland (41), and Yellowstone National Park (10). All other OTUs also grouped with sequences of geothermally influenced sites (hot springs and a geothermal mine adit) (10, 41, 45) (Fig. 3). Therefore, we propose to designate this cluster of amoA gene sequences the “thermal spring” cluster (Fig. 3). An affiliation of the amoA genes to moderately thermal environments, including subsurface caves or springs, was also recently discussed in a review by Francis et al. (13).
FIG. 3.
Phylogenetic tree of amoA genes obtained from the DGGE fingerprinting analysis of the thermal spring FJQ and closely related crenarchaeal genes from the database. The tree was inferred by a neighbor-joining analysis. The scale bar represents a 10% nucleotide sequence difference. The amoA gene of Nitrosospira briensis was used as the outgroup. The designation of origin as soil/sediment or water column/sediment was suggested by Francis et al. (14), and the new designation of the “thermal spring” cluster is proposed in this work. Accession numbers are from the EMBL/EBI database. Symbols on the branches indicate bootstrap confidence values, as follows: •, >90%; ○, >75%.
Comparison of the AMO peptide sequences showed high similarities (between 98% and 100%) to clones recovered from the spring FJQ (clones F2, OT2, and F5 [50]), whereas phylotypes FJQ-OTU3AOA (AM749098) and OTU9AOA (AM749104) were 100% similar to clones E06 (ABI21670) and A04 (ABI21625), respectively, from a geothermal mine adit (45). All remaining phylotypes, FJQ-OTU12AOA (AM749107), OTU13AOA (AM749108), OTU15aAOA (AM749111), OTU15bAOA (AM749110), and OTU16AOA (AM749112), had similarities of 98% to 100% to AMO sequences from a sandy-ecosystem soil (29).
Putative bacterial amoA genes only could be amplified with the primer pair amoA189F and amoA682R (42), which also amplified the very closely related pmoA genes. The use of the specific primers amoA-1F and amoA-2R (42) did not yield any PCR products. Additionally, the amplification of the 16S rRNA gene of the β-subdivision of AOB with the specific primers CTO189f and CTO654r (25) did not result in a PCR product. Gammaproteobacterial nitrifiers, e.g., Nitrosococcus spp., were not detected in this study, although the primers used in this work would bind to gammaproteobacterial amoA genes. No gammaproteobacteria had been found in a recently performed 16S rRNA gene library analysis (50). Bacterial DGGE sequence analysis of possible amoA genes was performed with nine (from a total of 16 visible) representative DGGE bands. None of the nine examined DGGE bands contained bacterial amoA gene sequences, but three contained sequences of pmoA genes, with a length of 531 bp, which were successfully sequenced.
FISH analysis and cell counts.
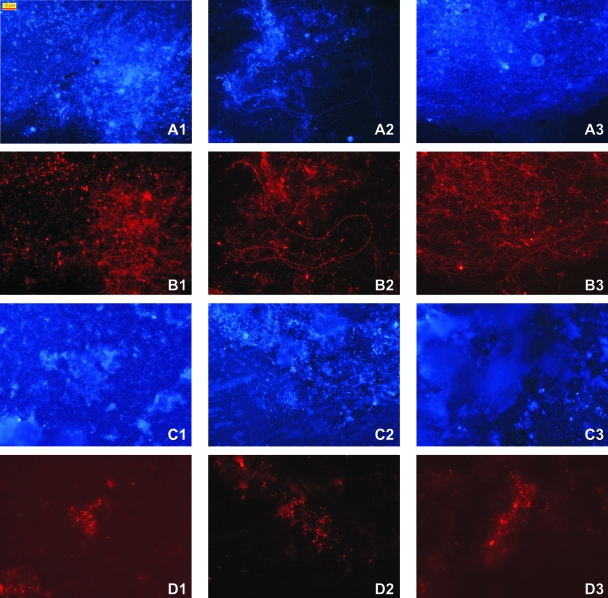
FISH analysis of naturally grown biofilms and artificial biofilms on glass slides showed highly diverse morphologies within the bacterial community (Fig. 4A and B1 to 3), which consisted of long rods (1.5 to 2 μm in length) that formed chains (Fig. 4B2) or long thin filamentous cells that formed networks (Fig. 4B3), as well as thick rods or cocci (1 to 2 μm in diameter) (Fig. 4B1) and several other morphologies like small rods or cocci. In contrast, the archaeal cells always seemed to be very small rods (about 0.8 to 1.5 μm in length), which sometimes looked like very small cocci, a morphology which was also reported by Macalady et al. (30), Hatzenpichler et al. (17), and de la Torre et al. (10). Archaea were detected with the universal probe ARCH915 on glass slides and in biofilms of the spring and showed similar morphologies. The labeled cells probably constituted members of the crenarchaeal group 1.1b, because of their dominance in the 16S rRNA gene library (>99.8% of about 300 clones [50]) and their similarity to the recently enriched moderately thermophilic Crenarchaeota (17). Other archaeal probes, such as CREN512 (22), were tried but without success. A quantitative measurement of the abundance of archaeal cells compared to that of bacterial cells was obtained by counting DAPI-stained cells and FISH-labeled cells. The archaeal cells constituted about 5.3% (standard deviation of ±4.5%) of the DAPI-stained population on glass slides and the surrounding biofilms of the spring FJQ.
FIG. 4.
FISH analysis of naturally grown biofilms and glass slides, which were placed into the spring water for 2 weeks. Panels A1 to 3 show DAPI-stained naturally grown biofilms, and panels B1 to 3 show the same sections in the microscope labeled with probe EUB338 to show the high morphological diversity within the bacterial spring community. Panel C2 shows a DAPI-stained biofilm, and panels C1 and 3 show DAPI-stained cells from a glass slide; panels D1 to 3 constitute their counterparts with ARCH915-labeled cells. Bar, 10 μm.
Microcosm experiments.
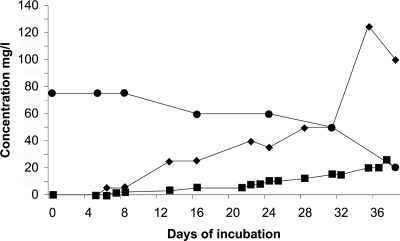
A microcosm consisting of 2.5 liters of filtered spring water, which was supplemented with a vitamin solution, trace elements, and 2.5 mM ammonium sulfate, was inoculated with the deposit of cells on a 0.22-μm filter, following filtration of 25 liters of spring water. During incubation at 40°C, the ammonium concentration decreased, while nitrite and nitrate concentrations concomitantly increased (Fig. 5); starting from 4.22 mM ammonium and finishing (day 37) with about 0.4 mM NO2−, 2.1 mM NO3−, and residual 0.8 mM NH4+ (sum of 3.3 mM), the conversion was almost stoichiometric. No conversion of ammonium was observed with uninoculated controls. This suggested the occurrence of autotrophic nitrification in the microcosm. The formation of nitrate may be due to the presence of Nitrospira sp. in the spring (50). An estimation of the activity yielded about 5.4 fmol converted ammonium per cell per day (assuming a density of approximately 2 × 107 cells/ml) (F. Gerbl, unpublished results), which compares well with about 4 fmol of NH3 per cell per day for “Candidatus Nitrosopumilus maritimus” at 28°C (26) (calculation by Wuchter et al. [52]).
FIG. 5.
Oxidation of ammonium and formation of nitrite and nitrate in a microcosm containing spring water enriched with vitamins and trace elements. The inoculum consisted of the prokaryotic planktonic community of the FJQ, which was concentrated by filtration. Incubation was done in the dark at 40°C for a period of 38 days. •, ammonium; ▪, nitrite; ⧫, nitrate.
The microcosm fluid was tested for the presence of archaeal 16S rRNA and amoA genes and also for betaproteobacterial amoA and 16S rRNA genes with the primers mentioned above following 5 weeks of incubation. The microcosm samples were positive for the archaeal 16S rRNA and amoA genes, but amplification of their betaproteobacterial counterparts did not yield any PCR products, while procedural controls with Nitrosomonas europaea DNA showed the expected PCR products for both genes. The data presented here suggested the absence of AOB and the presence of AOA, while ammonium was converted to nitrite; therefore, we argue that the first step of nitrification in the spring FJQ is probably carried out only by AOA. The lack of AOB, which was deduced previously from the 16S rRNA gene library analysis (50) and corroborated in this work, as well as the absence of bacterial amoA genes supports this notion.
Conclusions.
Prokaryotic ammonia oxidation had long been thought to be performed only by chemolithoautotrophic bacteria, but increasing evidence for the involvement of perhaps vast numbers of archaea in this process has been obtained (17, 26, 27, 29, 52). Our previous study (50) suggested strongly the occurrence of crenarchaeal ammonium oxidation at elevated temperatures (46°C); this notion has been corroborated and demonstrated recently with an enrichment culture from a Siberian hot spring (17) and two further hot spring environments (10, 41). While several lines of evidence support the finding of crenarchaeal ammonium oxidation, as shown here for alpine thermal springs, it should be noted that general difficulties with cultivation of both AOB and AOA and the lack of detailed knowledge of the influence of environmental parameters on community structure or niche differentiation underscore the need for broad explorations in this rapidly growing field and the necessity of applying numerous techniques, e.g., quantitative PCR, stable isotope probing, etc.
Nitrification apparently takes place at higher temperatures than thought before, for example, at 45 to 47°C in the spring FJQ (reference 50 and this work) or the Siberian Garga spring (17), as well as hot springs in Iceland and Kamchatka (temperatures from 38 to 97°C) (41) or Yellowstone National Park (temperatures up to 74°C) (10), and at 50°C in a geothermal mine adit (45) as well as probably in Nevada hot springs (EF032878 and EF032877) (Fig. 3). Therefore, we suggest moving the new cluster of amoA genes from the “marine/sediment” cluster to a new one named the “thermal spring” cluster (Fig. 3).
The question of a potential thermophilic origin of ammonia oxidation is of interest (17). Most of the soil on the earth's surface, except some paleosoils, is from the Pleistocene Age (1.8 million to about 11,000 years ago) or younger (5). The folding of the Alps started about 250 million years ago upon the breakup of Pangaea (38), and geothermal environments, including hot springs, were formed. It can be speculated that the massive outflow of warm spring waters from deep subsurface locations contained large numbers of thermophiles, which adapted gradually to cooler temperatures in soils. This scenario would be consistent with a thermophilic origin of ammonia oxidation (17, 41). However, whether a thermophilic ancestry of the processes for biogeochemical nitrogen cycling can be traced before the tectonic activities 250 million years ago is much less certain now than it seemed in recent years, since many arguments which do not support the notion of thermophilic ancestral cell lines have accumulated (compiled, e.g., in reference 53).
Acknowledgments
We thank the personnel of the water works of the community of the village Bad Gastein, especially Johann Knoll, the technical manager of the thermal springs in Bad Gastein, for help in obtaining samples, as well as Gerhard Steinbauer, the mayor of Bad Gastein, for the permission of sampling.
This work was funded by FWF project P19250-B17 of the Austrian Science Foundation.
Footnotes
Published ahead of print on 22 August 2008.
REFERENCES
- 1.Amann, R. I. 1995. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes, section 3.3.6., p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beman, M. J., and C. A. Francis. 2006. Diversity of ammonia-oxidizing Archaea and Bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, E., and M. Wagner. 2006. Oxidation of inorganic nitrogen compounds as an energy source, p. 457-495. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 2. Springer Science and Business Media, LLC, Berlin, Germany. [Google Scholar]
- 5.Buol, S. W., R. J. Southard, R. C. Graham, and P. A. McDaniel. 2003. Soil genesis and classification, 5th ed. Iowa State Press, Ames.
- 6.Burggraf, S., T. Mayer, R. Amann, S. Schadhauser, C. R. Woese, and K. O. Stetter. 1994. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 60:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coolen, M. J. L., B. Abbas, J. van Bleijswijk, E. C. Hopmans, M. M. M. Kuypers, S. G. Wakeham, and J. S. Sinninghe Damste. 2007. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 9:1001-1016. [DOI] [PubMed] [Google Scholar]
- 8.Cudrigh, S. F. 2002. Die Wasserwegigkeit des Gasteiner Thermalwassers. Diploma thesis. University of Salzburg, Salzburg, Austria.
- 9.Daniels, L., N. Belay, and B. S. Rajagopal. 1986. Assimilatory reduction of sulfate and sulfite by methanogenic bacteria. Appl. Environ. Microbiol. 51:703-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Torre, J. R., C. B. Walker, A. E. Ingalls, M. Konneke, and D. A. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel, A. S., M. L. Porter, L. A. Stern, S. Quinlan, and P. C. Bennett. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria.” FEMS Microbiol. Ecol. 51:31-53. [DOI] [PubMed] [Google Scholar]
- 13.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 14.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing Archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallam, S. J., K. T. Konstantinidis, N. Putnam, C. Schleper, Y.-I. Watanabe, J. Sugahara, C. Preston, J. de la Torre, P. M. Richardson, and E. F. DeLong. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. USA 103:18296-18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, P. M. Richardson, and E. F. DeLong. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzenpichler, R., E. V. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. USA 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirayama, H., K. Takai, F. Inagaki, Y. Yamato, M. Suzuki, K. H. Nealson, and K. Horikoshi. 2005. Bacterial community shift along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles 9:169-184. [DOI] [PubMed] [Google Scholar]
- 19.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, A. J., N. A. Tujula, M. Holley, A. Contos, J. M. James, P. Rogers, and M. R. Gillings. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256-264. [DOI] [PubMed] [Google Scholar]
- 21.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 22.Jurgens, G., F.-O. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 23.Kemnitz, D., S. Kolb, and R. Conrad. 2007. High abundance of Crenarchaeota in a temperate acidic forest soil. FEMS Microbiol. Ecol. 60:442-448. [DOI] [PubMed] [Google Scholar]
- 24.Kimura, H., M. Sugihara, H. Yamamoto, B. K. C. Patel, K. Kato, and S. Hanada. 2005. Microbial community in a geothermal aquifer associated with the subsurface of the Great Artesian Basin, Australia. Extremophiles 9:407-414. [DOI] [PubMed] [Google Scholar]
- 25.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 26.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, M. T. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam, P., M. M. Jensen, G. Lavik, D. F. McGinnis, B. Muller, C. J. Schubert, R. Amann, B. Thamdrup, and M. M. M. Kuypers. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. USA 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen, N., G. J. Olsen, B. L. Maidak, M. J. McCaughey, R. Overbeek, T. J. Macke, T. L. Marsh, and C. R. Woese. 1993. The ribosomal data base project. Nucleic Acids Res. 21:3021-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 30.Macalady, J. L., D. S. Jones, and E. H. Lyon. 2007. Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ. Microbiol. 9:1402-1414. [DOI] [PubMed] [Google Scholar]
- 31.Macalady, J. L., E. H. Lyon, B. Koffman, L. K. Albertson, K. Meyer, S. Galdenzi, and S. Mariani. 2006. Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72:5596-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meisinger, D. B., J. Zimmermann, W. Ludwig, K.-H. Schleifer, G. Wanner, M. Schmid, P. C. Bennett, A. S. Engel, and N. M. Lee. 2007. In situ detection of novel Acidobacteria in microbial mats from a chemolithoautotrophically based cave ecosystem (Lower Kane Cave, WY, USA). Environ. Microbiol. 9:1523-1534. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicol, G. W., L. A. Glover, and J. I. Prosser. 2003. The impact of grassland management on archaeal community structure in upland pasture rhizosphere soil. Environ. Microbiol. 5:152-162. [DOI] [PubMed] [Google Scholar]
- 36.Northup, D. E., S. M. Barns, L. E. Yu, M. N. Spilde, R. T. Schelble, K. E. Dano, L. J. Crossey, C. A. Connolly, P. J. Bosten, D. O. Natvig, and C. N. Dahm. 2003. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider caves. Environ. Microbiol. 5:1071-1086. [DOI] [PubMed] [Google Scholar]
- 37.Northup, D. E., and K. H. Lavoie. 2001. Geomicrobiology of caves: a review. Geomicrobiol. J. 18:199-222. [Google Scholar]
- 38.Ollier, C., and C. Pain. 2000. The origin of mountains. Routledge, London, United Kingdom.
- 39.Øvreås, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, H.-D., G. F. Wells, H. Bae, C. S. Criddle, and C. A. Francis. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reigstad, L. J., A. Richter, H. Daims, T. Urich, L. Schwark, and C. Schleper. 2008. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol. Ecol. 64:167-174. [DOI] [PubMed] [Google Scholar]
- 42.Rotthauwe, J.-H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer, D. M., C. L. Davis, and M. P. Bryant. 1980. Ammonia saturation constants for predominant species of rumen bacteria. J. Dairy Sci. 63:1248-1263. [DOI] [PubMed] [Google Scholar]
- 44.Simon, H. M., C. E. Jahn, L. T. Bergerud, M. K. Sliwinski, P. J. Weimer, D. K. Willis, and R. M. Goodman. 2005. Cultivation of mesophilic soil crenarchaeotes in enrichment cultures from plant roots. Appl. Environ. Microbiol. 71:4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spear, J. R., H. A. Barton, C. E. Robertson, C. A. Francis, and N. R. Pace. 2007. Microbial community biofabrics in a geothermal mine adit. Appl. Environ. Microbiol. 73:6172-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl, D. A., and R. I. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley & Sons, Chichester, England.
- 47.Takai, K., D. P. Moser, M. DeFlaun, T. C. Onstott, and J. K. Fredrickson. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67:5750-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H.-P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic Crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 50.Weidler, G. W., M. Dornmayr-Pfaffenhuemer, F. W. Gerbl, W. Heinen, and H. Stan-Lotter. 2007. Communities of Archaea and Bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl. Environ. Microbiol. 73:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. Sinninghe Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, Y., and N. Glansdorff. 2007. Lessons from extremophiles: early evolution and border conditions of life, p. 409-421. In C. Gerday and N. Glansdorff (ed.), Physiology and biochemistry of extremophiles. ASM Press, Washington, DC.
- 54.Zhou, J., Y. Gu, C. Zou, and M. Mo. 2007. Phylogenetic diversity of bacteria in an earth-cave in Guizhou province, southwest of China. J. Microbiol. 45:105-112. [PubMed] [Google Scholar]