Abstract
Campylobacter jejuni and Campylobacter coli are naturally competent, but limited information exists on the impact of environmental conditions on transformation. In this study, we investigated the impact of temperature and microaerobic versus aerobic atmosphere on transformation of C. coli to erythromycin and nalidixic acid resistance. Frequency of transformation was not significantly different between microaerobic (5 to 10% CO2) and aerobic conditions. However, C. coli was transformed to erythromycin resistance at a significantly higher frequency at 42°C than at 25°C (P < 0.05), and few or no transformants were obtained at 25°C. In contrast, transformation to nalidixic acid resistance was highly efficient at both 42°C and 25°C and was similar or, at the most, fourfold higher at 42°C than at 25°C. DNase I treatment experiments suggested that steps both prior and subsequent to internalization of DNA were influenced by temperature in the case of transformation of C. coli to erythromycin resistance. However, the moderately increased (fourfold) frequency of transformation to nalidixic acid resistance at 42°C compared to that at 25°C was exclusively associated with steps prior to DNA internalization. These findings suggest that transformation to erythromycin resistance may be significantly more frequent in the gastrointestinal tract of hosts such as poultry (at 42°C) than in other habitats characterized by lower temperatures, whereas transformation to nalidixic acid resistance may be highly efficient both within and outside the animal hosts.
Campylobacter jejuni and Campylobacter coli are leading bacterial agents of human diarrheal disease in the United States and other industrialized nations (18). Erythromycin and fluoroquinolones are first-line drugs used to treat human infections by Campylobacter in severe cases or immunocompromised people (22), and resistance to these antimicrobial agents can seriously compromise treatment outcomes. Point mutations in 23S rRNA genes and in gyrA are involved in resistance to erythromycin and (fluoro)quinolones, respectively, in Campylobacter (4, 7, 9, 11, 12, 25, 27). For reasons that remain unclear, such resistance is noticeably more common in C. coli than in C. jejuni (2, 3, 7, 26).
Campylobacter is known to be naturally competent (28), and studies with C. jejuni have suggested that transformation (uptake of naked DNA) may make a significant contribution to genetic diversity (1, 5, 21, 29). Transformation may also be important in the dissemination of antibiotic resistance in Campylobacter. In previous studies, we showed that C. coli from animal sources could acquire high-level resistance to erythromycin via transformation, with transformation frequencies being higher among turkey-derived C. coli than among C. coli from swine. Transformants acquired the A2075G mutation in the 23S rRNA gene that was harbored in the 23S rRNA genes of the erythromycin-resistant bacteria employed as sources of donor DNA for the transformations (15).
In other naturally competent bacteria, environmental conditions have been found to strongly affect transformation frequencies. For instance, temperature affected transformation in Bacillus subtilis (14, 31), Streptococcus pneumoniae (17), Haemophilus influenzae (10), and Neisseria gonorrhoeae (24). Length of incubation time with donor DNA as well as type of media and pH also influenced transformation frequencies in some of these species (10, 20, 24, 31).
Surprisingly limited information is currently available on the effects of environmental factors on transformation in Campylobacter spp. The thermophilic species C. jejuni and C. coli are obligate microaerophiles that grow optimally at 42°C and cannot grow below 30°C (19). At the same time, they are among the most frequent zoonotic pathogens, being able to colonize the gastrointestinal tract of many animal species, and frequently experience diverse environmental conditions (e.g., those characteristic of the gastrointestinal tract of different host animals as well as those in the environment, water, and foods). Identification of ecologically relevant environmental influences on transformation will lead to a better understanding of the role of transformation in the generation of genetic diversity and dissemination of antibiotic resistance in Campylobacter spp.
In the case of C. jejuni, CO2 concentration was shown to significantly affect the transformation frequency, with the frequency rate increasing as CO2 concentrations decreased from 10% to 0.7% (30). However, the impact of CO2 concentration on the transformation of C. coli is not known. Furthermore, the possible effects of temperature on transformation have not been characterized for either C. jejuni or C. coli. From the perspective of dissemination of antimicrobial resistance in Campylobacter spp., investigations involving C. coli are clearly needed, considering the propensity of this species to acquire resistance to antibiotics of relevance for treatment of human illness (e.g., erythromycin and fluoroquinolones). In this study, we investigated the effects of temperature (especially at 42°C and 25°C) and microaerobic versus aerobic conditions on the frequency of transformation of C. coli to erythromycin and nalidixic acid resistance.
MATERIALS AND METHODS
Bacterial strains.
The C. coli strains used as recipients and donors in transformation assays are part of our laboratory's Campylobacter strain collection and were previously described (15). The erythromycin-susceptible turkey-derived C. coli strains 961, 3237, and 1702rnd were used as recipients in transformations to erythromycin resistance. C. coli strains 3237 and 1702rnd were also susceptible to nalidixic acid and were used as recipients in transformations to nalidixic acid resistance. The donor strain, C. coli 1800r, was resistant to several antimicrobial agents, including both erythromycin and nalidixic acid (15). Erythromycin MICs were determined with the agar dilution method as previously described (15). Bacteria were routinely grown on sheep blood agar plates (Remel, Lenexa, KS) and in Mueller-Hinton broth (MHB) (Becton Dickinson, Sparks, MD) at 42°C under microaerobic conditions generated by the CampyPak microaerophilic system (BBL, Sparks, MD).
Transformation assays to study the impact of temperature and aerobic versus microaerobic conditions.
Erythromycin-resistant and erythromycin-susceptible strains were selected as donors and recipients, respectively, for assays of transformation to erythromycin resistance. Similarly, nalidixic acid-resistant and nalidixic acid-susceptible strains were selected as donors and recipients, respectively, for transformation to nalidixic acid resistance. Genomic DNA of erythromycin- or nalidixic acid-resistant donors was extracted using the Qiagen DNeasy tissue kit (Qiagen Inc., Valencia, CA) as described previously (23). Broth transformations followed a previously described protocol (15). Briefly, recipients (strains 961, 3237, and 1702rnd), previously determined to be naturally competent (15), were grown on sheep blood agar plates at 42°C for 36 to 48 h under microaerobic conditions. A single colony was transferred to 5 ml of MHB and incubated at 42°C for 24 h under microaerobic conditions. A portion of the culture (0.1 ml) was added to 50 ml MHB preconditioned at 42°C and incubated for 7 h at 42°C under microaerobic conditions (exponential phase). For each transformation, 1.0 ml of this culture was transferred to a sterile polypropylene round-bottom tube (14 ml; Becton Dickinson), and 15 μl of total genomic DNA (ca. 3 μg) from C. coli 1800r was added. Negative controls were processed identically, except that no genomic DNA was added. Following a 5-h incubation at 42°C or 25°C (or at other specified temperatures) microaerobically and aerobically, 100 μl was plated in triplicate on Mueller-Hinton agar (MHA; MHB with 1.2% agar) plates containing erythromycin (10 μg/ml) and incubated for 48 h at 42°C microaerobically to enumerate erythromycin-resistant transformants. To enumerate nalidixic acid-resistant transformants, the transformation mixture (cells plus DNA) was diluted (10- and 100-fold) in MHB, and the dilutions (100 μl) were plated on MHA plates containing nalidixic acid (16 μg/ml). The plates were incubated for 48 h at 42°C microaerobically, and colonies were enumerated. Dilutions (10−4, 10−6) of the culture at the end of the 5-h transformation period were also plated onto MHA plates and incubated for 48 h at 42°C microaerobically in order to determine the CFU/ml of the recipient. Transformation frequency was indicated as the ratio of the number of transformants/ml to total CFU/ml of the recipient. Agar assays were also used with several strains, as described previously (15).
Effects of temperature on DNA binding versus subsequent steps in transformation.
Cultures of the recipient strains (1 ml) were placed in polypropylene round-bottom tubes (14 ml), as described above for broth transformation assays, and mixed with donor DNA (15 μl) from C. coli 1800r. The mixtures were incubated at 42°C and 25°C for 1 h (in experiments involving transformation to erythromycin resistance) or 30 min (in transformations to nalidixic acid resistance). DNase I (Sigma, St. Louis, MO) was added to the mixture at the concentration of 20 Kunitz units/ml, and the tubes were further incubated at 42°C and 25°C for another 4 h (for erythromycin resistance transformations) or 4.5 h (for nalidixic acid resistance transformations). For rapid temperature shifts (42°C → 25°C, 25°C → 42°C), the transformation tubes were placed in a water bath at 42°C or 25°C. All incubations were under aerobic conditions. At the completion of the 5-hour transformation period, erythromycin-resistant and nalidixic acid-resistant transformants were enumerated as described above. DNase I activity was validated by adding DNase I to the transformation mixture immediately after adding the donor DNA and by further incubation of the mixture at 42°C for 5 h.
Statistical analysis.
Transformation frequency data were analyzed with SAS (SAS Institute Inc., Cary, NC) following arc sine transformation of the data. Analysis of variance was used to identify significant differences (P < 0.05).
RESULTS
Effects of temperature on frequency of transformation to erythromycin and nalidixic acid resistance.
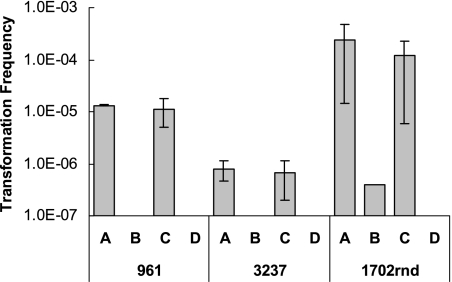
Three turkey-derived C. coli strains (961, 3237, and 1702rnd) previously determined to be naturally competent (15) were investigated in terms of the impact of temperature on transformation to erythromycin resistance. Temperature had a profound effect, with the frequency of transformation being significantly higher at 42°C than at 25°C for all three strains (P < 0.01). For two of the strains (961 and 3237), no erythromycin-resistant transformants were detected following transformations at 25°C (Fig. 1). Testing of several additional C. coli strains by using an agar assay for transformation also indicated that the frequency of transformation to erythromycin resistance was noticeably higher at 42°C than at 25°C, with few or no transformants being obtained at 25°C (data not shown).
FIG. 1.
Effects of temperature and microaerobic versus aerobic atmosphere on frequency of transformation of C. coli strains 961, 3237, and 1702rnd to erythromycin resistance. Transformation frequency was determined as described in Materials and Methods. Conditions employed: A, 42°C, microaerobic atmosphere; B, 25°C, microaerobic atmosphere; C, 42°C, aerobic atmosphere; D, 25°C, aerobic atmosphere.
Transformation to erythromycin resistance at other temperatures was investigated with one strain, C. coli 961. As the temperature decreased from 42°C to 37°C, transformation frequency decreased by 71%, and a 96% decrease was noted at 32°C (data not shown). No erythromycin-resistant colonies were obtained at any of the tested temperatures with negative controls, which lacked donor DNA.
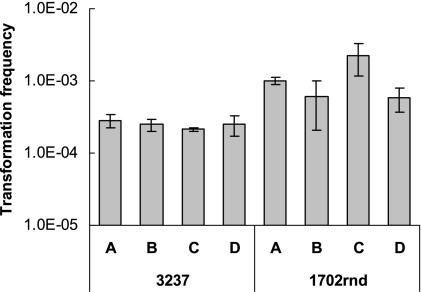
The impact of temperature on transformation to nalidixic acid resistance was evaluated with C. coli strains 3237 and 1702rnd (C. coli 961 was not employed, as it was naturally resistant to nalidixic acid). In contrast to what was observed with transformation to erythromycin resistance, strain 3237 was transformed with similar frequencies to nalidixic acid resistance at 42°C and at 25°C under either aerobic or microaerobic conditions (Fig. 2). The largest observed impact was a fourfold increase in the transformation frequency of strain 1702rnd under aerobic conditions at 42°C compared to that at 25°C (Fig. 2). Negative controls, which lacked donor DNA, yielded either very few (1 to 3 colonies) or, more commonly, no colonies at all (data not shown).
FIG. 2.
Effects of temperature and microaerobic versus aerobic atmosphere on frequency of transformation of C. coli strains 3237 and 1702rnd to nalidixic acid resistance. Transformation frequency was determined as described in Materials and Methods. Conditions employed: A, 42°C, microaerobic atmosphere; B, 25°C, microaerobic atmosphere; C, 42°C, aerobic atmosphere; D, 25°C, aerobic atmosphere.
Lack of difference in transformation frequency between microaerobic and aerobic conditions.
No significant differences (P > 0.05) in frequency of transformation to either erythromycin or nalidixic acid resistance were observed between microaerobic (5 to 10% CO2) and aerobic conditions (P > 0.05) (Fig. 1 and 2). The 5-h incubation period under aerobic conditions did not affect the viability of the recipient, based on CFU determinations immediately prior to and at the completion of this period (data not shown). At 42°C, the frequencies of transformation to erythromycin resistance were similar under microaerobic and aerobic conditions with the agar assay as well (data not shown). Transformants had high-level erythromycin resistance (MIC, >256 μg/ml), regardless of whether they were obtained under microaerobic or aerobic conditions. Erythromycin MICs for these transformants were similar to those for C. coli 1800r (>256 μg/ml), used as the source of the donor DNA.
Steps both prior and subsequent to internalization of DNA are susceptible to temperature in transformation to erythromycin resistance.
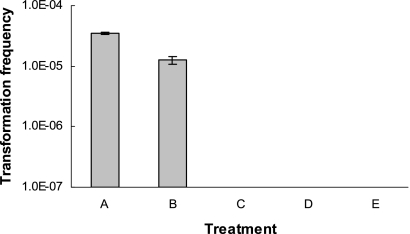
The addition of DNase I immediately after adding the donor DNA to the recipient cells abolished transformation to either erythromycin or nalidixic acid resistance (data not shown), confirming DNase I activity. The addition of DNase I after 30 min reduced the efficiency of transformation to erythromycin resistance by approximately eightfold (data not shown). This could result in numbers of transformants falling below detection level, especially for strain 3237, which had a relatively low frequency of transformation to erythromycin resistance (Fig. 1). When DNase I was added to the transformation mixture at 42°C 1 hour after addition of DNA and the tubes were further incubated at 42°C for 4 h, the frequency of transformation to erythromycin resistance was 10−5, about threefold lower than that in the untreated control (Fig. 3). Thus, the addition of DNase I 1 hour after incubation of cells with DNA was employed to investigate the impact of temperature on internalization of DNA.
FIG. 3.
Effect of temperature on transformation of C. coli 961 to erythromycin resistance. Temperature shifts (42°C versus 25°C) and DNase I treatments to degrade noninternalized DNA were done as described in Materials and Methods. Transformation frequency was determined as described in Materials and Methods. Treatments: A, standard transformation conditions (42°C for 5 h, without addition of DNase); B, positive-control transformation with DNase added after 1 h of incubation at 42°C and subsequent incubation of the mixture at 42°C for an additional 4 h; C, DNase added after 1 h at 25°C and subsequent incubation of the mixture at 42°C for an additional 4 h; D, DNase added after 1 h at 42°C and subsequent incubation of the mixture at 25°C for an additional 4 h; E, DNase added after 1 h at 25°C and subsequent incubation of the mixture at 25°C for an additional 4 h.
When DNase I was added to the mixture of cells and donor DNA following an hour of incubation at 25°C, no transformants could be obtained regardless of whether the mixture was maintained at 25°C or shifted to 42°C for four additional hours (Fig. 3), suggesting that at 25°C, DNA failed to become internalized during 1 hour of incubation and, thus, was susceptible to DNase I. Transformants also failed to be produced when DNase I was added after one hour of incubation at 42°C and the mixture was then shifted to 25°C for four additional hours (Fig. 3), suggesting that steps subsequent to DNA internalization (i.e., after the first hour of transformation) were inhibited at 25°C as well. As would be expected from these findings, conducting the entire transformation at 25°C failed to yield any erythromycin-resistant transformants (Fig. 3).
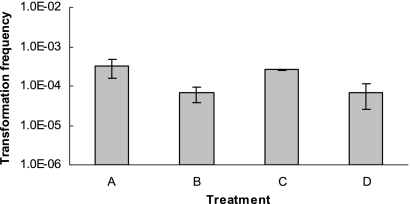
DNase treatment experiments were also employed to determine whether steps prior or subsequent to DNA internalization were affected by temperature in the observed modest (fourfold) increase in frequency of transformation to nalidixic acid resistance at 42°C versus 25°C observed with C. coli 1702rnd under aerobic conditions (Fig. 2). In transformation to nalidixic acid resistance, DNase I treatment after 30 min of incubation reduced the transformation frequency by about sixfold (data not shown). Considering the relatively high efficiencies of transformation of strains 3237 and 1702rnd to nalidixic acid resistance (Fig. 2), DNase I treatment at 30 min was chosen, because the frequency of transformation remained sufficiently high to permit the evaluation of the DNase I treatments. Transformation frequency was about fourfold higher when the mixture of cells and donor DNA was incubated at 42°C for the first 30 min prior to the addition of DNase I than it was at 25°C. However, the incubation temperature (42°C versus 25°C) of the transformation mixture after the first 30 min did not affect transformation frequencies (Fig. 4), suggesting that the step(s) subsequent to the internalization of DNA were not affected by temperature (42°C versus 25°C) in transformation of C. coli 1702rnd to nalidixic acid resistance.
FIG. 4.
Effect of temperature on transformation of C. coli 1702rnd to nalidixic acid resistance. Temperature shifts (42°C versus 25°C) and DNase I treatments to degrade noninternalized DNA were done as described in Materials and Methods. Transformation frequency was determined as described in Materials and Methods. Treatments: A, positive-control transformation with DNase added after the first 30 min of incubation at 42°C and subsequent incubation of the mixture at 42°C for an additional 4.5 h; B, DNase added after 30 min at 25°C and subsequent incubation of the mixture at 42°C for an additional 4.5 h; C, DNase added after 30 min at 42°C and subsequent incubation of the mixture at 25°C for an additional 4.5 h; D, DNase added after 30 min at 25°C and subsequent incubation of the mixture at 25°C for an additional 4.5 h.
DISCUSSION
In this study, temperature was found to be an environmental factor with important influence on transformation of C. coli to erythromycin resistance. Our findings suggest that 42°C may be the optimal temperature for transformation to erythromycin resistance since a significant drop in transformation frequency was noticed even at 37°C. Thus, transformation to erythromycin resistance may take place much more efficiently in the gastrointestinal tract of poultry, whose body temperature is 42 to 43°C, than in other habitats where temperature may be lower, such as surface water, soil, excreted fecal samples, milk, and meat (e.g., refrigerated raw meat or poultry). The noticeable decrease in transformation frequency for erythromycin resistance at 37°C also suggests a relatively low likelihood for such transformation within the gastrointestinal tract of animals with body temperatures such as those seen in humans (37°C) and in swine (38 to 39°C). However, we cannot exclude the possibility that other factors in the environment, or in the gastrointestinal tracts of animals such as swine and humans, may alleviate the effect of reduced temperatures and enable frequent transformation to erythromycin resistance within these habitats and hosts. Further experimental studies are needed to address these possibilities and to further define the environmental conditions that may limit transformation to erythromycin resistance within animal hosts and in the environment.
It is of interest that transformation to nalidixic acid resistance was either not significantly affected or affected to only a modest (fourfold) extent by temperature (25°C versus 42°C). Nalidixic acid-resistant C. coli isolates are typically also resistant to ciprofloxacin and other fluoroquinolones (6, 8). The results from transformation to nalidixic acid resistance suggest that acquisition of fluoroquinolone resistance through transformation may occur frequently and consistently in a wide range of CO2 concentrations and temperatures, both in animal hosts and in the environment. Thus, acquisition of fluoroquinolone resistance in Campylobacter through transformation may occur frequently in various niches, whereas transformation-mediated acquisition of high-level resistance to erythromycin may be much more common inside avian hosts than in animals with lower body temperatures or in the environment.
The DNase treatment experiments suggested that in the case of transformation to erythromycin resistance, steps both prior to internalization of DNA (e.g., DNA binding) and subsequent to internalization required high temperatures (42°C) and were severely impaired at 25°C. The underlying mechanisms are not clear at this time, and the biochemical aspects of transformation in Campylobacter are currently poorly understood. Alternative approaches such as experiments employing labeled DNA would need to be included in future studies aiming to accurately monitor internalization and the subsequent fate of transforming DNA. Nonetheless, the fact that temperature affected transformation to erythromycin resistance much more profoundly than it did to nalidixic acid resistance suggests that these mechanisms may be specific to the target sequences. In the case of nalidixic acid resistance, the sequences correspond to the fragment of gyrA that harbors the base substitution associated with fluoroquinolone resistance (27). On the other hand, in the case of erythromycin resistance, the target DNA involves fragments of the 23S rRNA genes (9, 12, 25), and single-stranded DNA fragments with these sequences would be expected to harbor extensive secondary structure. In other competent bacteria, donor DNA is rendered single stranded in the process of uptake for transformation (16), and secondary structure implications would therefore be relevant for such donor DNA. It is tempting to speculate that uptake and integration via homologous recombination of the 23S rRNA gene fragments harboring the mutation are impaired at 25°C, due to limitations on the secondary structure of the target molecules at that temperature. Secondary-structure-related impact of temperature on single-stranded nucleic acid fragments has been demonstrated in other bacterial systems. For instance, in Listeria monocytogenes, mRNA secondary structure of the transcript of prfA (encoding a transcription factor for several virulence genes) is profoundly affected in response to temperature (37°C versus 30°C), resulting in the well-known temperature-regulated expression of key virulence determinants in this pathogen (13). It would be of interest to determine whether transformation of mutations in rRNA genes may be impacted by temperature in other competent bacteria besides C. coli.
Wilson et al. found that the efficiency of transformation of C. jejuni to chloramphenicol resistance increased by 10- to 100-fold as CO2 concentration declined from 10% to 0.7% (30). In our study, we showed that transformation was equally effective under microaerobic (5 to 10% CO2) and aerobic (0.03% CO2) conditions, suggesting that these differences in CO2 concentrations did not significantly affect transformation. Differences in the Campylobacter species and strains that were used, the antibiotic resistance markers that were transformed, and the experimental conditions, including the CO2 concentration range, may account for the difference in findings between the study of Wilson et al. (30) and this study.
The findings from the current study clearly indicate that the elucidation of environmental factors influencing transformation in Campylobacter will be important for further characterization of the ecological constraints on the dissemination of antimicrobial resistance in this organism. In addition, these findings imply that the use of one marker to study environmental factors affecting transformation frequency in Campylobacter may lead to pitfalls. For example, temperature would be mistakenly considered as a relatively insignificant factor in transformation, if only transformation to nalidixic acid resistance were studied. Further studies are needed to determine the extent to which environmental factors such as temperature may affect transformation of other genetic determinants, including those involved in pathogenesis and other adaptations of this pathogen.
Acknowledgments
We thank Roger Thompson (Food Science Research Unit, USDA-ARS, Raleigh, NC) and Dennis Boos (Department of Statistics, North Carolina State University) for assistance in statistical analysis of the data. We are grateful to Robin Siletzky for technical assistance.
Partial support for this project was provided by USDA grant NRI 2003-0299.
Footnotes
Published ahead of print on 15 August 2008.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. Significance of duplicated flagellin genes in Campylobacter. J. Mol. Biol. 230:359-363. [DOI] [PubMed] [Google Scholar]
- 2.Avrain, L., F. Humbert, R. L'Hospitalier, P. Sanders, C. Vernozy-Rozand, and I. Kempf. 2003. Antimicrobial resistance in Campylobacter from broilers: association with production type and antimicrobial use. Vet. Microbiol. 96:267-276. [DOI] [PubMed] [Google Scholar]
- 3.Bae, W., K. N. Kaya, D. D. Hancock, D. R. Call, Y. H. Park, and T. E. Besser. 2005. Prevalence and antimicrobial resistance of thermophilic Campylobacter spp. from cattle farms in Washington state. Appl. Environ. Microbiol. 71:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carattoli, A., A. M. Dionisi, and I. Luzzi. 2002. Use of a LightCycler gyrA mutation assay for identification of ciprofloxacin-resistant Campylobacter coli. FEMS Microbiol. Lett. 214:87-93. [DOI] [PubMed] [Google Scholar]
- 5.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. M. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 6.Delsol, A. A., J. Sunderland, M. J. Woodward, L. Pumbwe, L. J. V. Piddock, and J. M. Roe. 2004. Emergence of fluoroquinolone resistance in the native Campylobacter coli population of pigs exposed to enrofloxacin. J. Antimicrob. Chemother. 53:872-874. [DOI] [PubMed] [Google Scholar]
- 7.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudreau, C., and H. Gilbert. 1997. Comparison of disc diffusion and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni subsp. jejuni and Campylobacter coli. J. Antimicrob. Chemother. 39:707-712. [DOI] [PubMed] [Google Scholar]
- 9.Gibreel, A., V. N. Kos, M. Keelan, C. A. Trieber, S. Levesque, S. Michaud, and D. E. Taylor. 2005. Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype. Antimicrob. Agents Chemother. 49:2753-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodgal, S. H., and R. M. Herriott. 1961. Studies on transformations of Hemophilus influenzae. I. Competence. J. Gen. Physiol. 44:1201-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrow, S. A., B. J. Gilpin, and J. D. Klena. 2004. Characterization of erythromycin resistance in Campylobacter coli and Campylobacter jejuni isolated from pig offal in New Zealand. J. Appl. Microbiol. 97:141-148. [DOI] [PubMed] [Google Scholar]
- 12.Jensen, L. B., and F. M. Aarestrup. 2001. Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 14.Khanna, M., and G. Stotzky. 1992. Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl. Environ. Microbiol. 58:1930-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J., D. K. Carver, and S. Kathariou. 2006. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 72:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacks, S. 1962. Molecular fate of DNA in genetic transformation of Pneumococcus. J. Mol. Biol. 5:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Lacks, S. A., S. Ayalew, A. G. de la Campa, and B. Greenberg. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: expression of dpnA, a late competence gene encoding a DNA methyltransferase of the DpnII restriction system. Mol. Microbiol. 35:1089-1098. [DOI] [PubMed] [Google Scholar]
- 18.Nachamkin, I., J. Engberg, and F. M. Aarestrup. 2000. Diagnosis and antimicrobial susceptibility of Campylobacter species, p. 45-66. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 19.Park, S. F. 2000. Environmental regulatory genes, p. 423-440. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 20.Perry, D., and H. D. Slade. 1963. Optimal conditions for the transformation of streptococci. J. Bacteriol. 85:636-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phongsisay, V., V. N. Perera, and B. N. Fry. 2006. Exchange of lipooligosaccharide synthesis genes creates potential Guillain-Barré syndrome-inducible strains of Campylobacter jejuni. Infect. Immun. 74:1368-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, DC.
- 23.Smith, K., N. Reimers, H. J. Barnes, B. C. Lee, R. Siletzky, and S. Kathariou. 2004. Campylobacter colonization of sibling turkey flocks reared under different management conditions. J. Food Prot. 67:1463-1468. [DOI] [PubMed] [Google Scholar]
- 24.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vacher, S., A. Ménard, E. Bernard, and F. Mégraud. 2003. PCR-restriction fragment length polymorphism analysis for detection of point mutations associated with macrolide resistance in Campylobacter spp. Antimicrob. Agents Chemother. 47:1125-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Looveren, M., G. Daube, L. De Zutter, J. M. Dumont, C. Lammens, M. Wijdooghe, P. Vandamme, M. Jouret, M. Cornelis, and H. Goossens. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J. Antimicrob. Chemother. 48:235-240. [DOI] [PubMed] [Google Scholar]
- 27.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wassenaar, T. M., B. N. Fry, and B. A. M. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, D. L., J. A. Bell, V. B. Young, S. R. Wilder, L. S. Mansfield, and J. E. Linz. 2003. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149:3603-3615. [DOI] [PubMed] [Google Scholar]
- 31.Young, F. E., and J. Spizizen. 1963. Incorporation of deoxyribonucleic acid in the Bacillus subtilis transformation system. J. Bacteriol. 86:392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]