Abstract
A chemotaxis signal transduction pathway (hereafter called Che1) has been previously identified in the alphaproteobacterium Azospirillum brasilense. Previous experiments have demonstrated that although mutants lacking CheB and/or CheR homologs from this pathway are defective in chemotaxis, a mutant in which the entire chemotaxis pathway has been mutated displayed a chemotaxis phenotype mostly similar to that of the parent strain, suggesting that the primary function of this Che1 pathway is not the control of motility behavior. Here, we report that mutants carrying defined mutations in the cheA1 (strain AB101) and the cheY1 (strain AB102) genes and a newly constructed mutant lacking the entire operon [Δ(cheA1-cheR1)::Cm] (strain AB103) were defective, but not null, for chemotaxis and aerotaxis and had a minor defect in swimming pattern. We found that mutations in genes of the Che1 pathway affected the cell length of actively growing cells but not their growth rate. Cells of a mutant lacking functional cheB1 and cheR1 genes (strain BS104) were significantly longer than wild-type cells, whereas cells of mutants impaired in the cheA1 or cheY1 genes, as well as a mutant lacking a functional Che1 pathway, were significantly shorter than wild-type cells. Both the modest chemotaxis defects and the observed differences in cell length could be complemented by expressing the wild-type genes from a plasmid. In addition, under conditions of high aeration, cells of mutants lacking functional cheA1 or cheY1 genes or the Che1 operon formed clumps due to cell-to-cell aggregation, whereas the mutant lacking functional CheB1 and CheR1 (BS104) clumped poorly, if at all. Further analysis suggested that the nature of the exopolysaccharide produced, rather than the amount, may be involved in this behavior. Interestingly, mutants that displayed clumping behavior (lacking cheA1 or cheY1 genes or the Che1 operon) also flocculated earlier and quantitatively more than the wild-type cells, whereas the mutant lacking both CheB1 and CheR1 was delayed in flocculation. We propose that the Che1 chemotaxis-like pathway modulates the cell length as well as clumping behavior, suggesting a link between these two processes. Our data are consistent with a model in which the function of the Che1 pathway in regulating these cellular functions directly affects flocculation, a cellular differentiation process initiated under conditions of nutritional imbalance.
To survive and compete under fluctuating environmental conditions, bacteria must rapidly adjust their behavior to maintain an optimum metabolism. Signal transduction systems enable cells to detect and adapt to these changes by executing appropriate cellular responses, such as regulation of gene expression or modulation of the swimming pattern. Two-component signal transduction systems are found in phylogenetically diverse organisms, but they are most prevalent in bacteria, where they modulate many cellular adaptive responses, including development, virulence, and chemotaxis (38, 41, 44). The best characterized of such systems is the one regulating the run/tumble swimming bias (chemotaxis) in Escherichia coli (for a review, see reference 42). This signal transduction system consists of a set of conserved proteins which includes CheA, CheW, CheY, CheB, and CheR and a set of chemoreceptors (known as methyl-accepting proteins [MCPs]) that perceive environmental cues. Signals sensed by MCPs modulate the activity of the histidine kinase CheA associated with chemoreceptors via CheW. Phosphotransfer from CheA to CheY results in phosphorylated CheY (P∼CheY) that can trigger a switch in the direction of flagellar rotation (tumble). The activity of CheB and CheR modulates the methylation status of MCPs, which affects their sensitivity. CheB, a methylesterase activated through phosphorylation by CheA, removes methyl groups added to MCPs by the constitutive methyltransferase CheR. The analysis of completely sequenced bacterial genomes has revealed that some bacterial species have multiple homologous chemotaxis-like signal transduction pathways (41, 47). Interestingly, Che-like pathways have been recently implicated in controlling cellular functions other than motility, including flagellum biosynthesis, cyst formation, biofilm formation, and initiation of developmental programs (8, 9, 10, 22, 27). In cases where a response regulator (CheY homolog) is present, the mechanism by which the Che-like pathways regulate functions other than the swimming pattern remains unknown. It is also unclear whether cellular responses modulated by parallel chemosensory systems, which function simultaneously in a single organism, may be coordinated.
The alphaproteobacterium Azospirillum brasilense is a soil diazotroph that colonizes the roots of cereals and grasses. A. brasilense cells are motile and, similarly to E. coli, respond to temporal gradients of stimuli by modulating their swimming pattern toward a higher or lower probability of change in swimming direction in response to repellents and attractants, respectively (1). We previously identified a chemotaxis-like signal transduction pathway in this bacterium that is comprised of CheAWYBR homologs (21). An orthologous pathway was shown to exclusively control the motility behavior in a closely related species (24). The ongoing sequencing of the first A. brasilense genome (http://genome.ornl.gov/microbial/abra) indicates that there are three other chemotaxis-like gene clusters in addition to the one identified previously (21), which we now name Che1 (Fig. 1). We demonstrated that the CheB1 and CheR1 proteins from this pathway function as a chemoreceptor-specific methylesterase and methyltransferase, respectively; however, experimental evidence strongly suggested that the primary function of this Che1 pathway is not the control of motility behavior (39). The main line of evidence for this hypothesis is the observation that although mutants lacking CheB and/or CheR functions are impaired in chemoreceptor-specific modifications and are defective in chemotaxis, a mutant in which the entire chemotaxis operon was mutated displays a chemotaxis phenotype mostly similar to that of the parent strain (39). Thus, we hypothesized that this signal transduction system in A. brasilense may control a cellular function(s) other than motility, and we tested this by characterizing the physiology and growth of a set of defined chemotaxis mutants. Here we confirm that this Che pathway contributes to the cell motility pattern, and we also provide evidence that this is not its only or primary function. Our results suggest a function for this chemotaxis pathway in modulating the propensity of cells to clump under conditions of high aeration as well as cell length. Mathematical modeling of the rates of nutrient uptake predicts that shorter cells may have a reduced diffusive nutrient uptake, suggesting that changes in cell length affect cell physiology in order to optimize metabolism. Our data are consistent with a model in which the function of the Che1 pathway in regulating these cellular functions directly affects flocculation, a cellular differentiation process initiated under conditions of nutritional imbalance.
FIG. 1.
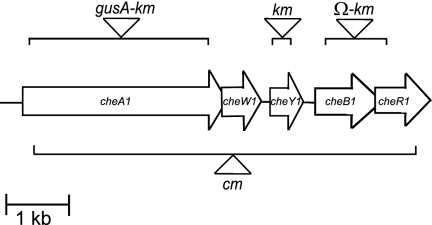
Genetic organization of the Azospirillum brasilense che1 operon and location of mutations. The diagram of the che1 operon is derived from that published previously (21). The bars represent the regions deleted to construct each mutant strain. The triangles represent the insertion of a nonpolar gusA-Km cassette within ΔcheA1 (strain AB101), of a nonpolar Km cassette within ΔcheY1 (strain AB102), of a polar omegon-Km cassette within Δ(cheB1-cheR1) (strain BS104) (39), and of a nonpolar Cm cassette within Δ(cheA1-cheR1) (strain AB103), as described in detail in Materials and Methods. Km, kanamycin resistance; Cm; chloramphenicol resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The wild-type A. brasilense strain Sp7 (ATCC 29145) was used throughout this study. The Che1 pathway and genes therein (cheA1, cheY1, etc.) were previously known as the Che pathway (21, 39). The mutant lacking functional CheB1 and CheR1 [Δ(cheB1-cheR1)::Km] (BS104) (39), the Che1 pathway insertion mutant (BS110) (39), and the IZ15 and IZ21 mutants (21) were described previously. A map detailing the genetic organization of the che1 region and the location of the mutation is shown in Fig. 1. A. brasilense cells were grown in minimal medium (MMAB), supplemented with a carbon source of choice as described previously (21). To visualize Congo red binding by individual colonies, cells to be tested were grown in liquid minimal medium with malate as a carbon source and placed as 5-μl drops on solid TY plates (consisting of [per liter] 10 g Bacto tryptone, 5 g yeast extract, and 15 g Noble agar) containing Congo red (40 μg ml−1). The plates were incubated at 28°C for 1 to 4 days before being photographed. Cell aggregation and flocculation was performed as follows. Overnight cultures in liquid TY medium were normalized to an optical density at 600 nm (OD600) of 1.0, and 100 μl was inoculated into 5 ml of liquid minimal medium containing 8 mM fructose and 0.5 mM NaNO3 and incubated at 28°C with shaking. Flocculation was quantified after 24 and 48 h of incubation as described previously (13). At least three independent cultures were used for each strain. The doubling times of individual strains were determined in at least two independent experiments in minimal medium supplemented with a carbon source of choice during early exponential phase by serial dilution in sterile phosphate-buffered saline (PBS) and plating on minimal medium plates.
Behavioral assays.
To compare chemotaxis responses to different carbon sources, minimal medium plates containing 0.3% agar and lacking a carbon source were supplemented with 10 mM malate, 10 mM succinate, or 10 mM fructose. These plates were then inoculated with wild-type A. brasilense strain Sp7 or the mutant derivatives AB101 [Δ(cheA1)::gusA-Km], AB102 [Δ(cheY1)::Km], AB103 [Δ(cheA1-cheR1)::Cm], BS104 [Δ(cheB1-cheR1)::Km, IZ15, and IZ21 as described previously (39). Measurements of chemotactic ring diameter were performed after 48 h. Aerotaxis assays using the capillary method were performed as described previously (39). In order to determine the reversal frequency (probability of change in the swimming direction) of cells under steady-state conditions, free-swimming cells were first grown in MMAB supplemented with malate or fructose (final concentrations of 10 mM) to early exponential phase, observed with a dark-field microscope, and videotaped. Malate and fructose are the strongest chemoattractants for A. brasilense (1), and wild-type cells of A. brasilense grown with malate have a different swimming motility pattern under steady-state conditions than when grown with fructose (39). The reversal frequency under steady-state conditions was measured by determining the number of reversals of a single cell during a 4-s time frame, corresponding to the average time that a single cell can be tracked, as described previously (39). The reversal frequencies of at least 90 cells (from three independent cultures) were measured in each experiment.
Construction and characterization of mutants.
To construct a mutant lacking a functional cheA1 gene, pGA115 containing a Δ(cheA1)::gusA-Km deletion-insertion was introduced into A. brasilense by triparental mating (38). Double-homologous recombinants carrying a deletion-insertion in cheA1 were selected. Southern hybridization and PCR were used to verify the correct insertion and the loss of the recombinant plasmid, and one such mutant strain was named AB101 (Fig. 1).
The mutant lacking a functional cheY1 gene [Δ(cheY1)::Km was constructed by deletion of an internal 320-bp fragment in frame and insertion of a nonpolar kanamycin cassette from pCM184 (29). A 531-bp fragment directly upstream of cheY1 and containing the start codon was amplified from the pFAJ451 cosmid (21) using the primers pair cheY1UF (5′GCCACCACCAAGAAGTCCAAG) and cheY1UR (5′CATGGTGAACGGGCCTCAGGC). This fragment was cloned in the pCR2.1TOPO vector (Invitrogen). A 554-bp fragment directly downstream of cheY1 and encompassing the last 43 bp at the 3′ end of the gene was amplified from pFAJ451 using the primers pair cheY1DF (5′CGACATCATCCAGACCAAGTTC) and cheY1DR (5′CTTGACCTTCGACACCAGCTC) and cloned into pGEM-T (Promega). Individual fragments were verified by sequencing and subsequently isolated by digestion with NcoI and NotI (upstream fragment) and ApaI and SacI (downstream fragment) before being cloned into corresponding sites in the pCM184 vector. In the resulting construct, the kanamycin cassette from pCM184 is inserted in frame between the two cloned fragments, yielding a Δ(cheY1)::Km construct. The entire Δ(cheY1)::Km construct was amplified using the primers cheY1UF and cheY1DR and blunt cloned into the pSUP202 suicide vector (digested with EcoRI, followed by end repair for bluntness), which was used for allelic exchange (39). Correct allelic replacement was verified by PCR, and one such mutant strain was named AB102 (Fig. 1).
We also constructed a second mutant deleted for most of the genes encoded within Che1. A 614-bp region immediately upstream of the che1 gene cluster and including 222 bp of the first gene in the operon (cheA1) was PCR amplified with the primer pair CheA1TMupFwd (5′-CCCAAGCTTCAGCGCGATGAACTGGTTG), including a 5′ HindIII site (engineered restriction sites are underlined), and CheA1TMupRev (5′-CTTGAGCAGGGACATGTTGTAGGCGGC). An 831-bp region beginning 12 bp from the end of the last gene in the operon, cheR1, and extending downstream of the operon was PCR amplified with the primer pair CheR1dwnFwd (5′-AACATGTCCCTGCTCAAGCAGCGTTCC) and CheR1dwnRev (5′-CGG GGTACCTTAGACGGCCGCCCGGAG), including a 5′ KpnI site. DNA fragments with the cheA1-to-cheR1 deletion were generated by a two-step, overlap PCR procedure (36) by using DNA polymerase (Epicentre Biotechnologies, Inc.). The fragments with appropriate deletions were excised with the restriction enzymes HindIII and KpnI and inserted into the vector pUC19 at the appropriate restriction sites to generate the new construct pUCAR. A cassette encoding resistance to chloramphenicol was excised as a SmaI fragment from the p34SCm vector (15) and cloned into pUCAR to create plasmid pUCARCm. A 2,363-bp region encompassing the Δ(cheA1-cheR1)::Cm construct was then amplified by PCR using the primer pair CheA1TMupFwd and CheR1dwnRev (a 5′ XbaI site replaced the KpnI site). The PCR fragments were excised with the restriction enzymes HindIII and XbaI and inserted into the suicide vector pKGmobGII (25) at the appropriate restriction sites to generate the deletion construct pKGmobGARCm. The deletion construct was transferred into A. brasilense wild-type strain Sp7 by biparental conjugation for allelic exchange (39). PCR was used to verify the correct replacement in the Δ(cheA1-cheR1)::Cm mutants obtained, and one such mutant strain was named AB103 (Fig. 1). All intermediate constructs were verified by sequencing prior to being transferred into A. brasilense.
Chemotaxis mutant complementation.
Functional complementation of the AB101 strain carrying the Δ(cheA1)::gusA-Km mutation was performed by using a wild-type gene expressed from a broad-host-range plasmid. First, PCR amplification of cheA1 and 545 bp of DNA sequence upstream of the chemotaxis operon was performed by using the previously described pGA111 vector (39), a pBluescript (Stratagene) derivative, as a template. The cheA1 region was isolated by EcoRV-XhoI digestion and cloned into the SmaI-XhoI-linearized medium-copy vector pBBR-MCS3 (28) to generate pBBR-cheA1. This cheA1 DNA region was also cloned into the low-copy plasmid pRK415 (26) by purifying an XbaI-KpnI fragment from pBBR-cheA1, yielding pRK415-cheA1. Both constructs were then introduced into the A. brasilense cheA1 mutant using triparental mating with pRK2013 as a helper (20), as described previously (21). As a control, the empty pRK415 and pBBR-MCS3 vectors were introduced into the A. brasilense wild-type strain (Sp7) and the AB101 strain carrying the Δ(cheA1)::gusA-km mutation. Complementation of the BS104 and AB102 strains was performed essentially as described previously, with the exception that a ribosome-binding site (5′-AGGAGGA-3′) was engineered at the 5′ end of each sequence to facilitate expression from the plasmid promoters (16). In order to complement the BS104 strain carrying a mutation deleting a region encompassing the cheB1-cheR1 genes, the cheB1-cheR1 genes were amplified by PCR from genomic DNA using the CheB1R1 forward primer (5′-CCCAAGCTTTAAGGAGAGGCCCGTATGTCCGATGGTTTCGGCAGAC-3) and CheB1R1 reverse primer (5′-GCTCTAGATTAGACGGCCGCCCGGAGC-3′) (the restriction sites are underlined and the ribosome-binding site is in bold). The PCR product was cloned into the pCR2.1TOPO (Invitrogen) vector and verified by sequencing. The cheB1-cheR1 region was then digested with the restriction enzymes HindIII and XbaI, prior to cloning into pRK415 (25), yielding pRK415-cheB1R1. The wild-type cheY1 gene was cloned in a similar manner using the CheY1 forward primer (5′-CCCAAGCTTTAAGGAGAGGCCCGTATGAAAGTTTGTCTGGTC-3′) and the CheY1 reverse primer (5′-CGGATTTCTCACAGCAGCCCGACCTCGAAC-3′) into the pCR2.1TOPO vector, and the construct was verified by sequencing. Next, cheY1 was isolated by digestion with HindIII and XbaI and cloned into pRK415, yielding pRK415-cheY1 (26). The pRK415-cheB1R1 and pRK415-cheY1 constructs were then introduced into the A. brasilense BS104 and AB102 strains, respectively, using triparental mating, as described previously. As a control, an empty pRK415 (and a pBBR1 MCS3 vector for cheA1 complementation) was introduced into A. brasilense wild-type (Sp7), AB101, BS104, and AB102 strains using triparental mating. The BS104 strain carrying the pRK415-cheB1R1 vector did not require IPTG (isopropyl-β-d-thiogalactopyranoside) for complementation (perhaps due to “leaky” expression), whereas the AB102 strain carrying the pRK415-cheY1 vector was complemented using 1 mM IPTG. We also complemented the previously described IZ15 and IZ21 mutants (21) using the pRK415 and pBBR-MCS3 vector derivatives containing the cheA1, cheB1-cheR1, or cheY1 gene as described above. The transconjugants from each mating were inoculated en masse as a straight line in semisolid (0.3% [wt/vol] agar) MMAB medium (lacking nitrogen) and incubated at 28°C for 48 h before being photographed. Complemented strains (but not controls) are expected to move away from the inoculation sites under these conditions. All constructs were verified by sequencing prior to being transferred into A. brasilense strains.
Measurements of cell size parameters.
Transmission electron micrographs were taken of cells grown to mid-exponential phase (OD600 of <0.5) in the medium indicated, washed twice in sterile PBS, spotted on Formvar-coated nickel grids, and negatively stained with 2% uranyl acetate or 0.75% uranyl formate. The transmission electron microscopy images were taken at random within a particular grid. Several grids were collected for each experiment. For cell length measurements, all cells within the field of view were measured from one cell pole to the other at the longest axis, provided that both poles of the cells were clearly visible. The only cells not measured were those for which both poles were not clearly seen, either because of cell-to-cell overlap or because they were not entirely within the image. Both the images of the cells and the measurements of cell length were taken blindly to avoid bias in the collection of data; i.e., the particular mutant studied at a given time was identified only by grid number until all data (both images and measurements) were collected. Cell size data were collected from at least three independent experiments for the wild type and each mutant under all growth conditions.
We also measured cell length using fluorescence microscopy of actively growing cells stained with the live fluorescent dye FM4-64 (Invitrogen Molecular Probes) in order to ensure that the differences detected did not result from the fixation protocol used for preparing samples for transmission electron microscopy. Briefly, aliquots of the cultures were stained with 1 μM FM4-64 for 5 min (Invitrogen Molecular Probes) at room temperature. Stained cells were immobilized on slides using molten 1% agarose in PBS. Pictures were taken using a Nikon Eclipse 80i fluorescence microscope equipped with a Nikon CoolSnap HQ2 cooled charge-coupled device camera, and cell lengths were measured using the Nikon NIS-Elements BR program for a minimum of 50 cells per sample taken from at least four different fields of view.
EPS isolation and quantitation.
Extraction and quantitation of exopolysaccharides (EPS) was performed as described by Enos-Berlage and McCarter (18) with the following modifications. Cells were grown overnight at room temperature with shaking in 250 ml of minimal medium supplemented with 5 mM malate and 5 mM fructose. Cells were harvested via centrifugation at 8,000 rpm for 10 min in a Sorvall GSA rotor. Cells were resuspended in PBS (20 mM sodium phosphate [pH 7.3], 100 mM NaCl) and then vortexed at high speed for 1 min. Cells were then incubated at 30°C on a gyratory shaker for 1.5 h. The vortexing and incubation steps were repeated once. Cells were then pelleted at 8,000 rpm for 15 min in a PTI F18S-12 × 50 rotor. An aliquot of the cell pellet was set aside for protein quantitation according to the method of Bradford (12). The supernatant was collected, treated with proteinase K to a final concentration of 200 μg/ml, and incubated on a gyratory shaker overnight at 37°C. The samples were then extracted using phenol-chloroform and precipitated with 95% ethanol. The pellets were washed once with 70% ethanol before being resuspended in water. Quantitation of EPS production was then determined using a 5% phenol-sulfuric acid solution according to the method described by Dubois et al. (17).
Mathematical analysis of nutrient uptake by diffusion.
We have used the model described by Berg and Purcell (7) and modified by Wagner et al. (43) for comparing the diffusion rates of nutrient uptake in each of the mutants relative to the wild-type strain. We calculated the rate of nutrient uptake for each mutant using a model for cells with prolate spheroid shapes (43), assuming a constant concentration of the nutrient and the same diffusion constant for each cell type. In addition, we estimated the number of receptors to be around 50,000 per cell (7), assuming the same density of receptors on the surface for cells used on the reference (2) and assuming the diameter of each receptor to be 1 nm. Changing the values of these assumptions for all strains did not affect the results.
Statistical analysis.
First, one-way analysis-of-variance tests (alpha level of 0.05) were performed to compare cell length between all strains and all growth conditions. Significant differences were found in cell length (for cells grown on malate, P = 3.38e−41; for cells grown on malate and fructose, P = 2.85e−51) among the strains in the analysis-of-variance tests. Similar differences were also found for septating cells (for cells grown in malate, P = 2.45e−8; for cells grown in malate and fructose, P = 2.61e−22). Next, the mean cell lengths of the wild type and mutants or of the mutant populations of actively growing cells were compared in pairwise two-sample t tests assuming equal variances (alpha level of 0.05) (Microsoft Excel [Microsoft, Inc.]). The differences between the means analyzed were considered statistically significant if the P value was <0.05.
RESULTS
CheA1 and CheY1 play a minor role in chemotaxis.
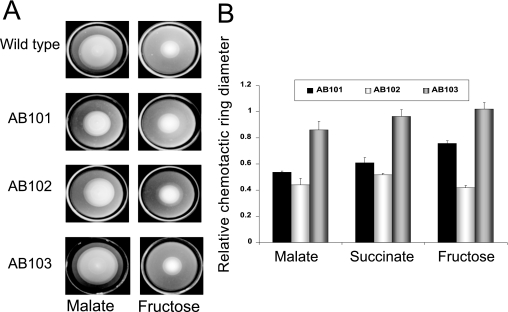
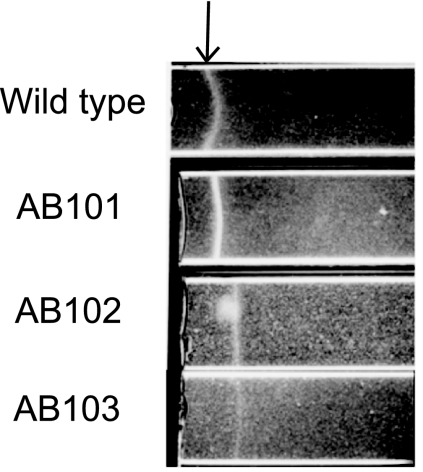
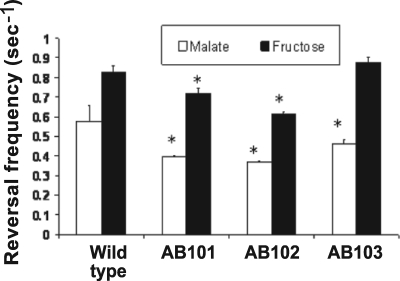
The chemotaxis pathway in A. brasilense has been identified in a cosmid library of A. brasilense Sp7 by functional complementation of generally nonchemotactic mutants generated by chemical mutagenesis (here referred to as Che1) (21). Although the exact gene(s) mutated in these mutants (IZ15 and IZ21) (21) has not been previously characterized, each mutant is locked into a specific motility pattern (highly reversing for IZ15 and smooth swimming for IZ21), consistent with a defect in chemotaxis signal transduction. However, we previously found that a strain carrying a che1 insertion mutation (strain BS110, generated by a Campbell mutation) disrupting the entire operon had a random swimming pattern similar to that of the wild-type strain and a minor (if any) defect in chemotaxis in soft agar plates (39). This atypical result suggested that the primary function of the Che1 pathway may not be control of the motility pattern (chemotaxis) (39). To extend these observations and clarify the function of the che1 operon in chemotaxis and aerotaxis, we have characterized the motility and tactic behaviors of strains carrying mutations in cheA1 (strain AB101) and cheY1 (strain AB102) as well as a strain carrying a newly constructed deletion-insertion mutation that disrupts the entire che1 operon (strain AB103) (Fig. 1). We have found that strains carrying mutations in cheA1, cheY1, and che1 have a defect in chemotaxis in soft agar plates but are not fully defective (Fig. 2A and B). The chemotactic defect of strains AB101 and AB102, as well as that of BS104 [carrying a Δ(cheB1-cheR1)::Km mutation] (39) could be complemented by expressing the corresponding genes from a plasmid (see Fig. S1 in the supplemental material), indicating that the chemotactic defects are not due to polar effects. Interestingly, all mutant strains were capable of forming typical “chemotactic” rings on all substrates tested. This contrasts with the phenotypes of the original nonchemotactic mutants (IZ15 and IZ21), which were complemented by this operon (21) (see Fig. S2A in the supplemental material). All strains were capable of forming an aerotactic band in the spatial gradient assay (Fig. 3). Although equivalent cell concentrations were placed in the capillaries, strain AB101 formed an aerotactic band significantly faster than the wild-type strain (less than 1 min, versus about 2 min for the wild-type strain), resulting in the formation of an aerotactic band with a greater density of motile cells. As shown previously, the time required for the formation of the aerotactic band by A. brasilense cells in the capillary assay is reproducible and consistent (about 2 min) because it depends on the ability of cells to efficiently sense the oxygen gradient and navigate toward its optimum concentration (1, 4, 48). Strain AB102 also formed an aerotactic band, but the band formed slightly slower (about 5 min) and farther away from the meniscus than the band formed by the wild type or AB101 (Fig. 3). Strain AB103 was the most affected in aerotaxis: the band formed farther away from the meniscus and it also formed significantly slower (if at all) (about 15 to 20 min), resulting in a low density of motile cells within the “band.” It is noteworthy that cells from all strains were actively motile within the band as well as outside of it. If the primary function of the che1 operon is chemotaxis, then mutants lacking any component of the excitation pathway (such as CheA and CheY homologs) should have a locked smooth-swimming motility pattern, similar to equivalent mutants of E. coli (40, 42). To test this hypothesis, we determined the steady-state (with a constant concentration of an attractant) swimming pattern of free-swimming cells by using motion tracking analysis (Fig. 4). Consistent with our previous observations of the motility pattern in the wild-type A. brasilense and the BS104 strain (39), we found that the steady-state motility pattern of strains carrying mutations in cheA1, cheY1, and che1 was dependent upon the substrate present: the probability of changes in the swimming direction (reversal frequency) was higher when cells were grown on fructose than when cells were grown on malate. However, strains carrying mutations in cheA1 (strain AB101) and cheY1 (strain AB102) tended to swim with a statistically significant (P < 0.05) “smoother” motility pattern (changed swimming direction less frequently) than the wild-type cells under all conditions. Noticeably, in both the semisoft agar plate assay and in analysis of the motility pattern under steady-state conditions, AB101 and AB102 had more pronounced defects than AB103, which carries a mutation encompassing the che1 cluster. The AB103 strain had a motility pattern similar to that of AB101 and AB102 in the presence of malate, but it did not have a distinct motility pattern in the presence of fructose. Taken together, these data strongly suggest that the function of the Che1 pathway in chemotaxis may be indirect.
FIG. 2.
che1 mutant strains are impaired but not null in chemotaxis. (A) Chemotaxis of the wild-type A. brasilense strain Sp7 and strains carrying mutations in the che1 operon in the soft agar plate assay with malate or fructose as the sole carbon source. Representative plates are shown for each strain and condition. (B) The average chemotactic ring diameters are expressed relative to that of the wild-type strain (defined as 1). Error bars represent standard deviations from the means, calculated from at least three repetitions. The chemotactic ring diameters of the mutants were found to be significantly different from those of the wild type with all chemoeffectors tested, except for AB103 on succinate and fructose.
FIG. 3.
che1 mutant strains have different aerotaxis defects. Aerotaxis of the wild-type A. brasilense and the che1 mutant strains in the capillary tube assay is shown. The arrow indicates the position of the center of the aerotactic band. Cell suspensions of similar density were used in the assay. The picture was taken after 10 min of incubation at room temperature.
FIG. 4.
che1 mutant strains are affected in their steady-state swimming pattern. The steady-state swimming pattern is represented by the reversal frequency (probability of changes in the swimming direction) of free-swimming cells grown in minimal MMAB medium containing either 10 mM malate or 10 mM fructose as the sole carbon source. Reversal frequencies are expressed as the number of events per second. Error bars represent standard deviations from the means, calculated from at least 90 cells. Statistically significant differences (P < 0.05) from values for the wild type are indicated by asterisks.
The lack of a null chemotactic phenotype in any of the Che1 pathway mutants prompted us to reexamine the original mutants IZ15 and IZ21, from the complementation of which the Che1 operon was originally isolated (21). These mutant derivative strains have locked high-reversal (IZ15) and smooth-swimming (IZ21) motility phenotypes on all substrates tested (21). Consistent with previous observations, these mutants also failed to spread much beyond the inoculation point, and they did not form any chemotactic ring on semisoft agar plates, even under conditions where the wild-type parental strain and the AB101, AB102, AB103, and BS104 strains formed sharp chemotactic rings (see Fig. S2A in the supplemental material). While a cosmid carrying the entire operon except for the very end of the cheR1 gene could complement both IZ15 and IZ21 (21), we have found that the IZ15 mutant could be complemented by expressing cheB1-cheR1 from a plasmid, while IZ21 was not complemented by expressing either cheA1, cheY1, or cheB1-cheR1 (see Fig. S2B in the supplemental material). These data suggest that the IZ15 mutant strain may carry a mutation in either cheB1 or another gene that may be suppressed through the expression of cheB1-cheR1. Sequencing cheB1 and cheR1 from the IZ15 and IZ21 genomes did not reveal any mutation, supporting the latter hypothesis that the complementation of the IZ15 defect by a plasmid expressing cheB1-cheR1 is a multicopy suppression. On the other hand, the genetic defect responsible for the IZ21 (21) phenotype (complemented by expressing the full Che1 cluster from a cosmid) remains to be determined. These results are consistent with the hypothesis that Che1 may have an indirect role in chemotaxis, perhaps via CheB1.
Che1 mutants have different cell lengths under specific growth conditions.
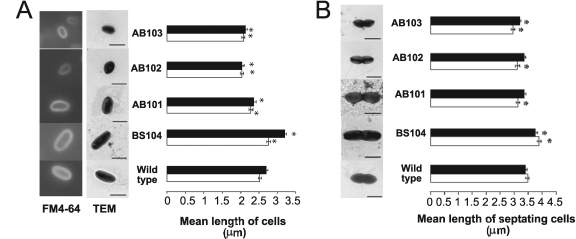
As mentioned above, strains carrying mutations in genes of the Che1 pathway had modest defects in chemotaxis, prompting us to hypothesize that this operon controls other cellular functions. Thus, we systematically analyzed the wild-type and the che1 mutant strains by transmission electron microscopy. Interestingly, we found that most cells in an actively growing culture of the BS104 strain appeared to be longer than cells in similar cultures of the wild-type strain, suggesting that mutations in che1 affected cell morphology under these growth conditions (low OD and standard shaking conditions). We compared the cell lengths of wild-type and mutant cells grown with different substrates by using transmission electron microscopy and by staining cells taken directly from cultures with the vital fluorescent membrane dye FM4-64 (Fig. 5A). Similar results were obtained in all experiments for the BS110 strain (carrying a mutation disrupting Che1 function by Campbell insertion [39]) and the AB103 strain [carrying a Δ(cheA1-cheR1)::Cm mutation], indicating that the constructs used to make each mutant strain were not responsible for the phenotypes observed. We found that in populations of actively growing cells, strains lacking functional cheA1, cheY1, and che1 tended to be shorter than the wild-type cells, whereas cells lacking functional cheB1-cheR1 tended to be longer (Fig. 5A). Furthermore, cells lacking functional cheY1 and che1 were significantly shorter than cells of the other mutant strains, including AB101 (P < 0.05 in pairwise t tests). The biased cell length distribution of the mutant strains could be complemented to wild-type length values in trans by a cosmid carrying the wild-type che1 cluster (21) (data not shown) or specific che1 genes expressed from plasmids (Table 1). There was a similar bias toward shorter or longer cells for a subpopulation of cells undergoing septation (Fig. 5B). Cells that tended to be longer divided at a longer cell length relative to the wild type, and shorter cells divided at a shorter cell length. Given that the doubling times of strains carrying mutations in che1 were similar to that of the wild type, we concluded that the Che1 pathway must modulate cell length independently of cell division. This observation is consistent with the fact that the timings of chromosome replication and cytokinesis are independent from each other, as well as being independent of cell length at division in bacteria (19). This finding also suggests that the Che1 pathway modulates cell length independently of the events leading to cell size increase and growth during cell cycle progression. The changes in cell length as detected here were relatively small but were statistically significant and experimentally reproducible. The differences were especially significant when comparing the BS104 strain, mutated in cheB1-cheR1, with the wild type or any of the other mutants. Our experimental approach for measuring cell size parameters was carefully designed to guard against unintentional bias of locating and measuring unusually long and short cells (see Materials and Methods). Interestingly, differences in cell length between the wild type and the mutant strains as described above were best observed in actively growing cultures maintained in the early exponential phase of growth (OD600 of <0.5) and were less pronounced at other growth stages. These subtle differences in the cell lengths of individual and septating cells were also best observed at lower cell density in cultures in the early exponential phase of growth, suggesting that this effect of the che1 mutations prevailed in the early stages of the cell cycle.
FIG. 5.
che1 mutant strains have different cell lengths. (A) Mean cell lengths from actively growing populations of the wild-type A. brasilense and the strains carrying mutations in genes of the che1 operon. Cells were grown in minimal medium supplemented with 10 mM malate (white columns) or 5 mM malate and 5 mM fructose (black columns) as the carbon sources. Cell length values were measured on transmission electron micrographs or on images obtained by fluorescence microscopy after staining the cells with the membrane fluorescent dye FM4-64 along the length of individual cells, as specified in Materials and Methods. (B) Mean cell lengths of septating cells from actively growing cultures of the wild-type A. brasilense and of strains carrying mutations in genes of the che1 operon. Cells were grown in minimal medium supplemented with 10 mM malate (white bars) or 5 mM malate and 5 mM fructose (black bars) as the carbon sources. The cell length values for individual cells were obtained from transmission electron micrographs or fluorescent images as described for panel A. Cells were identified as undergoing septation during division by the presence of a clear constriction located at midcell. In both panels A and B, the asterisks indicate statistically significant differences from the mean cell length value of the wild-type strain as determined by a t test (P < 0.05). Representative images of cells stained with the fluorescent membrane dye FM4-64 or transmission electron micrographs are shown on the left side of each graph as typical examples of nonproliferating (A) or septating (B) cells grown on malate and fructose (5 mM each). In both panels, the error bars represent the standard deviations from the means. The scale bars in the transmission electron microscopy pictures represent 2 μm.
TABLE 1.
Complementation of che1 mutant strains for cell length
| Strain | Mean cell length (μm) ± SDa |
|---|---|
| Sp7(pRK415) | 2.61 ± 0.0055 |
| AB101(pRK415) | 2.35 ± 0.0038 |
| AB101(pRK-cheA1) | 2.71 ± 0.0061 |
| BS104(pRK415) | 2.84 ± 0.0085 |
| BS104(pRK-cheB1R1) | 2.50 ± 0.0106 |
| AB102(pRK415) | 2.46 ± 0.0045 |
| AB102(pRK-cheY1) | 2.56 ± 0.0047 |
Mean cell length was determined for cells grown in MMAB with malate (5 mM) and fructose (5 mM).
Changes in cell length do not affect growth rate.
Changes in the average cell length of the mutants in the presence of different substrates are correlated with the growth rate, consistent with the fact that cells growing faster tend to be larger (19) (see Table S1 in the supplemental material). However, differences in cell length between the strains could not be attributed to differences in growth rate because the doubling times of the mutant strains were not different from that of the wild type (see Table S1 in the supplemental material). Further comparison of the growth rates and the cell length in actively growing populations of cells revealed that at higher growth rates, the average cell length increased for wild-type cells and some (but not all) mutant cells (Fig. 5; see Table S1 in the supplemental material). When comparing the length of cells grown on malate with that of cells grown on malate and fructose, the differences in the average cell length between the two growth conditions were found to be statistically significant (t test, alpha level of 0.05) for the wild-type strain (P = 0.0043) and BS104 (P = 9.02e−8) but not for the AB101, AB102, AB103, or BS110 strain. This result indicates that although the average length of wild-type cells can change under different nutrient conditions, the cell lengths of strains carrying mutations in cheA1, cheY1, and che1 remained constant regardless of the growth conditions. This implies that strains carrying mutations in cheA1, cheY1, and che1 cannot sense and/or transduce the signal(s) that leads to an increased cell length at higher growth rates, as opposed to the wild type or a strain carrying a mutation disrupting both cheB1 and cheR1. Altogether, these data indicate that the Che1 pathway may contribute to the ability to regulate changes in cell length under conditions of optimal growth (low OD and low aeration).
The changes in cell length are predicted to affect nutrient uptake in shorter but not longer cells.
In order to assess whether the subtle changes in cell length could affect the physiology of the cells, we used a previously developed mathematical model (7, 43) to compare the rates of diffusive nutrient uptake of the mutant strains (see Fig. S3 in the supplemental material). The model predicted that the changes in cell length would not significantly affect nutrient uptake in the BS104 strain but would result in a significant decrease in the nutrient uptake abilities of the shorter cells (strains AB101, AB10, and AB103). These predictions also imply that the Che1 pathway could function to reduce cell length in order to decrease nutrient uptake.
Che1 mutants have a different ability to clump under certain growth conditions.
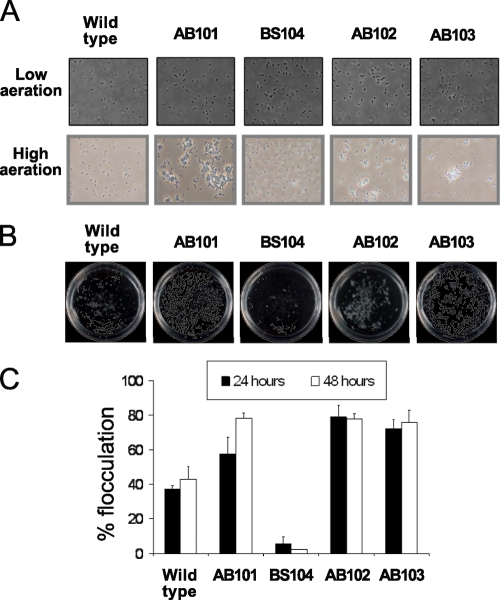
In addition to the effect of mutations in che1 genes on cell length, we have also observed that the AB101, AB102, AB103, and BS110 strains, but not the wild-type or BS104 strain, were capable of cell-to-cell aggregation and clumping under conditions of growth at high aeration (Fig. 6A). Clumps were not observed under conditions of growth at low aeration for any of the strains tested (Fig. 6A), indicating that this behavior was induced under conditions of high aeration and that it was not constitutive. Under these conditions of growth at high aeration, the AB101, AB102, AB103, and BS110 strains still exhibited different cell lengths, and they also formed clumps due to cell-to-cell contacts initiated at the nonflagellated poles. The cell clumps were very dynamic in that some cells were capable of leaving the clumps. We have analyzed clumped cells by light microscopy, transmission electron microscopy, scanning electron microscopy, and fluorescence microscopy using various fluorescent lectins (data not shown), but we were unable to detect any specific extracellular bacterial structure(s), such as pili, polysaccharide fibrils, or holdfasts, that could be invoked to mediate such interactions between cells. Clumping and cell-to-cell interactions have been described previously and have been studied in detail in A. brasilense because it is one of the first observable steps in flocculation, a cellular differentiation event that occurs when cells are grown under conditions of nutrient stress at high aeration (13). In order to determine if the clumping observed was related to flocculation, we compared the wild-type strain and the chemotaxis mutant strains for the ability to flocculate and found that strains carrying mutations in cheA1, cheY1, and che1 flocculated not only sooner but also quantitatively more than the wild-type strain (Fig. 6B and C). We also observed that these mutant strains formed clumps earlier and in greater numbers than the wild type under these conditions (about 6 h after inoculation into the flocculation medium, versus about 9 h for the wild type). Conversely, BS104 flocculated very poorly even after prolonged incubation, suggesting that this strain is significantly delayed rather than impaired in flocculation (Fig. 6B and C). Taken together, these data indicate that the Che1 pathway modulates the propensity of cells for clumping under conditions of high aeration and that this behavior directly affects the timing of flocculation.
FIG. 6.
The che1 mutants are affected in the ability to clump and to flocculate under specific growth conditions. (A) Actively growing cells of A. brasilense do not clump under conditions of low aeration (top), whereas cells of AB101, AB102, and AB103, but not those of the wild type or BS104, clump under these conditions (bottom). Pictures were from actively growing cultures in MMAB with 5 mM malate and 5 mM fructose as the carbon sources, observed by phase-contrast microscopy. Magnification, ×400. (B) Visible aggregates of flocculated cells. Petri dishes (60 by 15 mm) containing flocculated liquid cultures are shown. (C) Quantitative analysis of flocculation. The data represent the fraction of flocculated cells relative to the total number of cells in the culture, after gentle homogenization, expressed as a percentage as detailed in Materials and Methods. The error bars represent the standard deviations from the means.
Che1 mutants are affected in the production of EPS.
The ability of A. brasilense cells to aggregate, to form large clumps, and to flocculate in liquid cultures under conditions of unbalanced growth was previously correlated with changes in the concentration and composition of EPS (13, 35). The production of Congo red-binding EPS has been correlated with the ability of cells to flocculate under certain conditions (24). Therefore, we compared strains carrying mutations in che1 genes to the wild-type strain for the ability of colonies to bind the Congo red dye (Fig. 7A). We noticed dramatic differences in the appearance of colonies (both color and morphology) formed by the che1 mutant strains in comparison with the wild type (Fig. 7A). Four-day-old colonies of the wild type and of the BS104 strain bound Congo red moderately and mostly at the center of the colony, whereas AB101 and AB102 bound more Congo red homogeneously throughout the colony (Fig. 7A). In contrast, 4-day-old colonies of AB103 (and BS110 [not shown]) did not bind Congo red under these conditions. When comparing 1-week-old colonies grown on medium containing Congo red, we observed that all colonies, including those of AB103, bound the Congo red dye included in the plates. These observations suggest that the mutants are affected in the amount and/or timing of production of at least some Congo red-binding EPS. Consistent with the hypothesis that the che1 mutant strains are impaired in the production of EPS, we found that the surfaces of 1-week-old colonies were different (Fig. 7A). The surfaces of colonies of AB101, and to a lesser extent that of AB102, were wrinkled and appeared “dried” in aspect. In contrast, colonies of AB103 were smooth and appeared “wet” under the same conditions, while colonies of the wild type and of the BS104 strain were similar and appeared intermediate between those of strains carrying mutations disrupting che1 or cheA1 (Fig. 7A). Comparing the total amount of EPS produced by each strain revealed that AB102, AB103, and BS104 produced significantly less total EPS under these conditions, while AB101 produced EPS in amounts equivalent to that extracted from the wild-type strain (Fig. 7B). However, the amount of EPS produced by the different strains did not correlate with a particular Congo red-binding phenotype on plates or with the ability of any of the mutants to clump or to flocculate. We have also directly tested the hypothesis that EPS could be responsible for the clumping behavior by attempting to induce clumping by adding EPS isolated from clumped and flocculated cells in suspensions of free-swimming cells. However, none of the isolated EPS fractions could induce the clumping behavior. This result does not rule out the possibility that the EPS may modulate clumping, since it is possible that the structure of the EPS could be involved in this behavior; however, the EPS structure may have been compromised by the extraction procedure. Collectively, these data suggest that it is most likely the nature, composition, and/or structure of the EPS, rather than the total amount of EPS produced alone, that contributes to the different cell surface properties and associated clumping abilities of the che1 mutants.
FIG. 7.
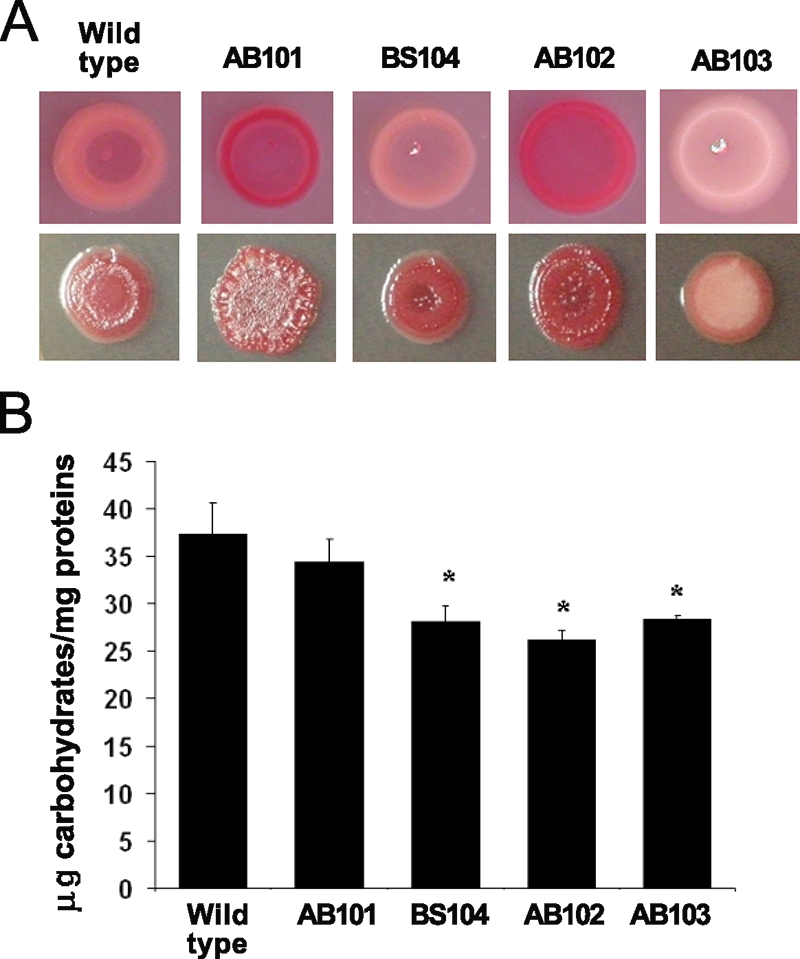
The che1 mutants are affected in the production of EPS. (A) Colony morphologies of the wild type and the che1 mutants observed on solid agar plates containing Congo red on minimal medium (top) or TY medium (bottom), after 4 days of incubation. Although the exact patterns of Congo red binding varied with the growth conditions, the patterns produced by the mutants were always different from those produced by the wild type. The colony morphology was also different under different conditions and between the wild type and some of the mutant strains (AB101 and AB102 in particular). (B) Quantitation of the EPS produced by the wild-type A. brasilense strain and the che1 mutants. The extraction and quantification of EPS were performed as described in Materials and Methods. The error bars represent the standard deviations from the means for each sample. Asterisks indicate statistically significant differences compared to the wild-type strain (P < 0.05).
DISCUSSION
The che1 cluster was initially identified by functional complementation of two generally nonchemotactic mutants (IZ15 and IZ21) that each had a distinct swimming pattern (21). Here, we confirmed that the Che1 pathway contributes to chemotaxis, and we also show that this control is likely to be indirect. We further demonstrate that the Che1 pathway of A. brasilense functions to modulate the propensity of cells for clumping, likely in response to changes in oxygen concentrations, and that this pathway can also bias the cell length, suggesting a link between these two cellular processes.
Evidence for a complex role of Che1 in chemotaxis.
Our data indicate an indirect role for the Che1 pathway in chemotaxis, perhaps mediated by the adaptation proteins CheB1 and/or CheR1. The following lines of evidence support this hypothesis: (i) a mutant lacking the entire che1 cluster is capable of chemotaxis and it is affected only subtly in its swimming pattern; (ii) strains with mutations in cheB1 and/or cheR1 have the strongest chemotaxis and aerotaxis phenotypes (39); (iii) strains carrying mutations in cheB1 and/or cheR1, but not strains with mutations in cheA1 or cheY1, have a distinct motility pattern (39) (Fig. 3); (iv) the motility patterns of the different che1 mutant strains are more pronounced on malate than on fructose, consistent with our previous findings that CheB1-dependent demethylation of chemoreceptors is required in response to malate but not in response to fructose (39); and (v) the IZ15 mutant (21) can be complemented by expressing CheB1 and CheR1 from a plasmid. Interestingly, the CheBII adaptation protein from the che2 operon of Rhizobium leguminosarum bv. viciae was also suggested to indirectly modulate chemotaxis mediated by the che1 operon, which is essential for all chemotactic behaviors in this species (30). The role of the Che1 pathway of A. brasilense in aerotaxis is puzzling since each che1 mutant is differently affected, consistent with an indirect role of the Che1 pathway in mediating this behavior. Previously, we have shown that a strain carrying a mutation in both cheB1 and cheR1 (BS104) was impaired in aerotaxis, while strains carrying a mutation in only cheB1 or cheR1 were null for aerotaxis (39). Our finding that the AB103 strain had an impaired aerotactic phenotype mostly similar to that of BS104 (39) lends further support to this hypothesis.
Several mechanisms can be invoked to explain how the CheB1 and/or CheR1 protein, but not other Che1 pathway components, could mediate chemotaxis. One possibility is that CheB1 and CheR1 together with other adaptation proteins, including some directly involved in controlling chemotaxis, contribute to the overall methylation status of chemotaxis transducers and thus fine-tune the sensitivity of the chemotactic response. In this scenario, in the absence of components of the Che1 pathway, there is a change in the sensitivity of the chemotactic response, and an impaired rather than a null chemotactic phenotype is expected. Our observation that strains carrying mutations in cheB1 and cheR1 have the most severe defect in chemotaxis (39) and that several che1 mutant strains, but not AB103 or BS110 (both lacking a functional Che1 pathway), are generally affected but are not null for chemotaxis would fit such a model. An alternative possibility is that the methylation status of chemoreceptors and/or the activity of CheB1 and CheR1 affects the activity of another protein(s) that directly regulates chemotaxis. Candidates for such functions are CheC and CheD homologs, which have been studied in most detail in Bacillus subtilis (33). CheC was shown to have a dual role as a phosphatase for P∼CheY as well as in adaptation by interacting with CheB and affecting its activity on chemoreceptors in a methylation-independent process (33). CheC has also been proposed to bind directly to the switch of the flagellar motor and to affect the motility pattern (37). It is interesting to note that methylation-independent adaptation was previously shown to be important in chemotaxis in A. brasilense (39) and that the genome of A. brasilense encodes two CheC homologs (http://genome.ornl.gov/microbial/abra), whose function remains to be determined. CheD modifies chemoreceptors by deamidation and can modulate their sensitivity and/or signal transduction abilities (34). There are two homologs of CheD encoded within the A. brasilense genome (http://genome.ornl.gov/microbial/abra). A third possibility, which does not exclude the previous two hypotheses, is that the Che1 pathway contributes directly to chemotaxis but competes with another chemotaxis pathway(s) for the flagellar motor targets and thus provides a redundant function. In the latter alternative, another pathway(s) or protein(s) is expected to compensate for the lack of activity of Che1 on the flagellar motors. There are three other chemotaxis-like operons in addition to Che1, as well as an additional, genetically unlinked CheY homolog, within the A. brasilense genome (http://genome.ornl.gov/microbial/abra). Any of the possibilities described above, alone or in combination, could possibly account for our data. Insights into the functions of other chemotaxis clusters and adaptation proteins should refine these hypotheses.
Che1 modulates changes in cell morphology.
The effect of che1 mutations on the clumping behavior depended on the growth conditions, whereas the effects on cell length were observed under all conditions at low OD. The changes in cell length were predicted to significantly reduce nutrient uptake in shorter cells (strains carrying mutations in cheA1, cheY1, and che1) but not in longer cells (the strain carrying mutations in both cheB1 and cheR1), suggesting that the Che1 pathway may function to modulate cell length as an adaptive response to optimize nutrient uptake. The effects on clumping were observed under conditions of growth with high (but not low) aeration, suggesting a role for oxygen (or a related parameter such as intracellular energy) in this process. Azospirillum spp. are microaerophilic in that they generate maximum intracellular energy at low oxygen concentrations, and they actively seek environments with low oxygen tensions by aerotaxis (48). Interestingly, when motile cells of A. brasilense accumulate within an aerotactic band, cells in front of the aerotactic band experience higher oxygen concentrations and these cells tend to form clumps that are dynamic, with some cells joining the clumps as they leave the optimal conditions of the aerotaxis band (4, 48). These clumps are similar to those formed by strains carrying mutations in cheA1, cheY1, and che1 under conditions of high aeration. Given that cell clumping is observed only under conditions of growth with high aeration (growth with vigorous shaking, front of an aerotaxis band, and growth under flocculation conditions), this behavior may represent an adaptive response to high oxygen concentrations, which are known to decrease the energy levels in A. brasilense (48). One possibility is that in clumped cells, the cell surfaces involved in cell-to-cell clumping are no longer (or less) available for exchanges with the surrounding medium, regardless of cell size or shape, thus providing a possible advantage to limiting oxygen diffusion under these conditions. It also suggests that the changes in the cell length observed under conditions where A. brasilense generates maximum energy (48) (low aeration and low OD) may be an alternative response to clumping and that both behaviors would represent adaptive responses in order to optimize metabolism. However, we cannot rule out the possibility that the changes in cell length are secondary, i.e., that changes in cell length are morphological manifestations of other primary effects not detected here. Irrespective of that, the observation that the Che1 pathway affects (even if only indirectly) the cell length in A. brasilense, as well as clumping, is intriguing, as it suggests that these two functions are linked.
Based on the E. coli paradigm for chemotaxis, the CheA1 and CheY1 homologs are expected to comprise the excitation pathway, with CheY1 as the signaling output (31, 41, 42). If CheY1 is the signaling output of the pathway described in this study, then strains carrying mutations in cheA1, cheY1, and che1 are expected to have similar phenotypes and should no longer be capable of transducing signals and causing an adequate cellular response. Indeed, we found that these strains (AB101, AB102, AB103, and BS110) are biased toward shorter cell lengths and toward a higher propensity to clump under conditions of high aeration, suggesting that CheY1 provides the signaling output for this pathway. In E. coli chemotaxis, CheB and CheR comprise an adaptation system that allows the cells to return to a prestimulus motility pattern (6, 11, 38). By analogy, we propose that cells of the BS104 strain, which lacks functional CheB1 and CheR1, receive the positive signal for cell elongation or the negative signal for clumping but do not adapt, yielding a population of cells significantly biased toward longer cells or free-swimming, nonclumping cells, respectively. The different clumping behaviors of strains with mutations in the che1 cluster would also explain their different flocculation phenotypes, since clumping is a prerequisite to flocculation (13). Taken together, our data strongly support the notion that the main output activity of the che1 operon in A. brasilense results in a positive signal for cell elongation and a negative signal for clumping (under conditions of high aeration). The sugar composition of EPS produced by A. brasilense was shown to change depending on the stage of growth and the growth conditions (3). Furthermore, it was recently shown that A. brasilense constitutively produces an outer membrane lectin that interacts with an unidentified oligosaccharide structure found in arabinose-rich EPS, which is specifically produced in aggregating A. brasilense cells, suggesting that this lectin may be required for flocculation (3, 14, 32). Here we show that strains with mutations in che1 are affected in their ability to produce Congo red-binding EPS, suggesting that the Che1 pathway affects the production of some EPS. Thus, although the exact mechanism responsible for the clumping behavior is still unknown, we hypothesize that it involves changes in cell surface properties such as changes in the structure and/or composition of EPS. Interestingly, the effect of the Che1 pathway of A. brasilense in modulating the cell surface properties (cell length and possibly clumping via EPS production) is reminiscent of the effect of the che2 chemotaxis-like signal transduction pathway, which regulates flagellum biogenesis in Rhodospirillum centenum (8), and the Dif pathway, which modulates fibril polysaccharide biogenesis in Myxococcus xanthus (5, 46). Similar to the case for Che1 of A. brasilense, the exact mechanism(s) by which these chemotaxis-like signal transduction pathways affect nonchemotactic cellular properties remains to be determined. While we have yet to identify the molecular targets of the Che1 pathway, our results suggest that this pathway may affect molecular machinery responsible for concomitantly regulating cell elongation (positive regulation) and clumping behavior, likely via production of specific EPS (negative regulation).
Potential implications of the role of Che1 in modulating changes in cell surface properties.
Why would A. brasilense use a Che-like signal transduction pathway to coordinate such dynamic changes in cell surface properties? Several features of information processing by Che-like pathways make them particularly well suited for the regulation of dynamic cellular behaviors. First, multiple cues sensed by independent putative chemoreceptors may be integrated into a single cellular response by interaction with a particular chemotaxis-like pathway. Thus, chemotaxis-like responses are initiated when a set of conditions, rather than a single trigger, are met. Second, the activity of CheB and CheR homologs on chemoreceptors modulates chemosensory sensitivity so that a response to concentration changes is initiated under a broad range of background conditions. Although we have not identified a particular chemoreceptor for the Che1 pathway, we have shown that CheB1 and CheR1 are functional and essential for at least some behavioral responses (39), indicating that these proteins function to modulate sensory input. Furthermore, we have described phenotypes of strains carrying mutations in che1 genes that are dependent on growth conditions, suggesting that environmental input is required to modulate the observed responses. Comparative genomics indicate that many bacterial genomes encode multiple chemotaxis-like pathways, including some that were shown to regulate cellular behaviors that are a priori not directly related to flagellar motility (45). In addition, as shown here and elsewhere (21, 30), orthologous chemotaxis-like operons may modulate different cellular functions in closely related bacterial species. This observation thus suggests that these signal transduction systems evolve rapidly, as they are likely to provide significant advantages to the organisms that must constantly seek optimum conditions for growth in ever-changing environments. The function of Che1 in modulating dynamic changes in cell morphology expands the range of cellular functions modulated by such versatile signal transduction pathways.
Supplementary Material
Acknowledgments
We thank Annie Lin for the initial characterization of the Congo red phenotypes of some of the mutants and for help with the some of the chemotaxis assays. We thank M. Lidström for the gift of the pCM184 plasmid and Matt Russell, Beth Mullin, Igor Zhulin, and anonymous reviewers for insightful comments on the manuscript.
This work was supported by an NSF career award (MCB-0622277) and generous start-up funds from the University of Tennessee, Knoxville, to G.A.
Footnotes
Published ahead of print on 18 July 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexandre, G., S. E. Greer, and I. B. Zhulin. 2000. Energy taxis is the dominant behavior in Azospirillum brasilense. J. Bacteriol. 1826042-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signalling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 997060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahat-Samet, E., S. Castro-Sowinski, and Y. Okon. 2004. Arabinose content of extracellular polysaccharide plays a role in cell aggregation of Azospirillum brasilense. FEMS Microbiol. Lett. 237195-203. [DOI] [PubMed] [Google Scholar]
- 4.Barak, I., I. Nur, Y. Okon, and Y. Henis. 1982. Aerotactic response of Azospirillum brasilense. J. Bacteriol. 152643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellenger, K., X. Ma. W. Shi, and Z. Yang. 2002. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J. Bacteriol. 1845654-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, H. C., and D. A. Brown. 1972. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature 239500-504. [DOI] [PubMed] [Google Scholar]
- 7.Berg, H. C., and E. M. Purcell. 1977. Physics of chemoreception. Biophys. J. 20193-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berleman, J. E., and C. E. Bauer. 2005. A Che-like signal transduction cascade involved in controlling flagella biosynthesis in Rhodospirillum centenum. Mol. Microbiol. 551390-1402. [DOI] [PubMed] [Google Scholar]
- 9.Berleman, J. E., and C. E. Bauer. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol. Microbiol. 561457-1466. [DOI] [PubMed] [Google Scholar]
- 10.Blackhart, B. D., and D. R. Zusman. 1985. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. USA 828767-8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borkovitch, K. A., L. A. Alex, and M. I. Simon. 1992. Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci.USA 896756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 13.Burdman, S., E. Jurkevitch, M. E. Soria-Diaz, A. M. Serrano, and Y. Okon. 1998. Aggregation of Azospirillum brasilense: effects of chemical and physical factors and involvement of extracellular components. Microbiology 1441989-1999. [DOI] [PubMed] [Google Scholar]
- 14.Burdman, S., E. Jurkevitch, M. E. Soria-Diaz, A. M. Serrano, and Y. Okon. 2000. Extracellular polysaccharide composition of Azospirillum brasilense and its relation to cell aggregation. FEMS Microbiol. Lett. 189259-264. [DOI] [PubMed] [Google Scholar]
- 15.Dennis, J., and G. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25772-776. [DOI] [PubMed] [Google Scholar]
- 16.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 14426-430. [DOI] [PubMed] [Google Scholar]
- 17.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and Fred Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28350-356. [Google Scholar]
- 18.Enos-Berlage, J. L., and L. L. McCarter. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 1825513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 6752-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauwaerts, D., G. Alexandre, S. K. Das, J. Vanderleyden, and I. B. Zhulin. 2002. A major chemotaxis gene cluster in Azospirillum brasilense and relationships between chemotaxis operons in alpha-proteobacteria. FEMS Microbiol. Lett. 20861-67. [DOI] [PubMed] [Google Scholar]
- 22.Hickman, J. W., T. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, Z. J., H. Gest, and C. E. Bauer. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 1795720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katupitiya, S., J. Millet, M. Vesk, L. Viccars, A. Zeman, Z. Lidong, C. Elmerich, and I. R. Kennedy. 1995. A mutant of Azospirillum brasilense Sp7 impaired in flocculation with a modified colonization pattern and superior nitrogen fixation in association with wheat. Appl. Environ. Microbiol. 611987-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katzen, F., A. Becker, M. V. Ielmini, C. G. Oddo, and L. Ielpi. 1999. New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 65278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70191-197. [DOI] [PubMed] [Google Scholar]
- 27.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 1002008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. BioTechniques 166175-176. [DOI] [PubMed] [Google Scholar]
- 29.Marx, C. J., and M. E. Lidström. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 331062-1067. [DOI] [PubMed] [Google Scholar]
- 30.Miller, L. D., C. K. Yost, M. F. Hynes, and G. Alexandre. 2007. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63348-362. [DOI] [PubMed] [Google Scholar]
- 31.Montrone, M., M. Eisenbach, D. Oesterhelt, and W. Marwan. 1998. Regulation of switching frequency and bias of the flagellar motor by CheY and fumarate. J. Bacteriol. 1803375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mora, P., F. Rosconi, L. Franco, Franguas, and Susana-Sowinski. 2008. Azospirillum brasilense Sp7 produces an outer-membrane lectin that specifically binds to surface-exposed extracellular polysaccharide produced by the bacterium. Arch. Microbiol. 189519-524. [DOI] [PubMed] [Google Scholar]
- 33.Rosario, M. M., J. R. Kirby, D. A. Bochar, and G. W. Ordal. 1995. Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry 213823-3831. [DOI] [PubMed] [Google Scholar]
- 34.Rosario, M. M., and G. W. Ordal. 1996. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol. Microbiol. 21511-518. [DOI] [PubMed] [Google Scholar]
- 35.Sadasivan, L., and C. A. Neyra. 1985. Flocculation in Azospirillum brasilense and Azospirillum lipoferum: exopolysaccharides and cyst formation. J. Bacteriol. 163716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Saulmon, M. M., E. Karatan, and G. W. Ordal. 2004. Effect of loss of CheC and other adaptational proteins on chemotactic behaviour in Bacillus subtilis. Microbiology 150581-589. [DOI] [PubMed] [Google Scholar]
- 38.Segall, J. E., S. M. Block, and H. C. Berg. 1986. Temporal comparisons in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 838987-8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens, B. B., S. N. Loar, and G. Alexandre. 2006. Role of CheB and CheR in the complex chemotactic and aerotactic pathway of Azospirillum brasilense. J. Bacteriol. 1884759-4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson, R. V., L. A. Alex, and M. I. Simon. 1994. Histidine and aspartate phosphorylation: two-component systems and the limits of homology. Trends Biochem. 19485-490. [DOI] [PubMed] [Google Scholar]
- 41.Szurmant, H., and G. H. Ordal. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 51024-1037. [DOI] [PubMed] [Google Scholar]
- 43.Wagner, J. K., S. Setayeshgar, L. A. Sharon, J. P. Reilly, and Y. V. Brun. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc. Natl. Acad. Sci. USA 10311772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26369-376. [DOI] [PubMed] [Google Scholar]
- 45.Wuichet, K., R. P. Alexander, and I. B. Zhulin. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 4223-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Z., D. Geng, H. Xu, m. H. B. Kaplan, and W. Shi. 1998. A new set of chemotaxis homologues is essential for Myxococcus xanthus social motility. Mol. Microbiol. 301123-1130. [DOI] [PubMed] [Google Scholar]
- 47.Zhulin, I. B. 2001. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv. Microbiol. Physiol. 45157-198. [DOI] [PubMed] [Google Scholar]
- 48.Zhulin, I. B., V. A. Bespalov, M. S. Johnson, and B. L. Taylor. 1996. Oxygen taxis and proton motive force in Azospirillum brasilense. J. Bacteriol. 1785199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.