Abstract
Phosphorelay systems are important mediators of signal transduction during bacterial adaptation to new environments. Previously we described the vieSAB operon, encoding a putative three-protein component phosphorelay involved in regulating Vibrio cholerae virulence gene expression. At least part of the regulatory activity of VieSAB is exerted through the cyclic diguanylate (c-di-GMP)-degrading activity of the putative response regulator VieA. So far no direct evidence that VieSAB encodes a phosphorelay system exists. In addition, the role VieS plays in modulating VieA activity remains unclear. To address these questions, we expressed and purified VieA and a soluble cytoplasmic portion of VieS and used them in autophosphorylation and phosphotransfer assays. These assays showed that VieS has kinase activity in vitro and is able to selectively phosphorylate VieA. A phenotypic comparison revealed that deletion of vieS results in increased biofilm production comparable to that seen for deletion of vieA, whereas motility was decreased only slightly in the ΔvieS mutant compared to the profound defect observed in a ΔvieA mutant. We also found that the ΔvieS strain has a lower level of vieA transcript and, similar to a ΔvieA mutant, an increased intracellular level of c-di-GMP. Further analysis using site-directed vieA mutants showed that some of the phenotypes observed were due to the phosphorylation status of VieA. The evidence presented in this report is the first to link VieS and VieA biochemically and genetically, lending support to the hypothesis that these proteins function together in a signaling system.
Bacterial two-component signal transduction systems are structurally and functionally diverse, and much remains to be learned about their mechanisms of action. One such system is encoded by the Vibrio cholerae vieSAB operon, which was first identified in a screen for genes induced during colonization of the infant mouse small intestine (8). Further study of the locus revealed that it encodes a putative hybrid sensor kinase, VieS, and two response regulators, VieA and VieB (19). The predicted VieS protein sequence shares homology with the Bordetella pertussis hybrid sensor kinase BvgS (2). Analysis of VieA determined that it shares a high degree of similarity to response regulators like CheY (19, 31). The predicted VieB sequence lacks any recognizable DNA binding domain but contains both a phosphoreceiver domain and a tetratricopeptide repeat motif thought to be involved in protein-protein interactions (5).
Given the homology of VieS and VieA to bacterial signaling system components, it was hypothesized that the role of VieS is to sense host-specific signals and, by phosphorylating VieA, initiate changes in the expression of genes important for colonization. However, so far a coherent model of how the VieSAB phosphorelay functions is lacking. Genetic analysis revealed that vieA mutants had more-pronounced phenotypes than either vieS or vieB mutants. Among the phenotypes noted for vieA mutants are increased biofilm production and decreased motility in vitro (28, 36). These phenotypes were found to be a direct result of loss of VieA phosphodiesterase (PDE) activity toward the secondary messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) (28, 34, 36). Based on the homology of VieS to bacterial sensor kinases and the critical enzymatic role that VieA plays in regulating virulence genes, a model was proposed where, upon receiving a yet-unknown signal, VieS autophosphorylates and transfers this phosphate to VieA. The phosphorylated form of VieA then acts as a positive transcriptional regulator of vieSAB, increasing the levels of VieS, VieA, and VieB until the activating signal is removed. In support of this model, VieA was shown by transcriptional profiling using DNA microarrays to positively regulate the entire vieSAB locus (4). The role of VieB in this regulatory scheme, if any, remains unknown, since mutations in vieB have thus far lacked detectable phenotypes. Once upregulated, VieA would then act to deplete the intracellular level of c-di-GMP and as a result promote the expression of virulence genes and suppress genes required for biofilm formation. Arguing against this model were findings that a ΔvieS mutant did not exhibit the same phenotypes associated with a ΔvieA mutation. This finding suggested that VieA may act independently of VieS.
Herein we reexamine the relationship between VieS and VieA in order to resolve some of these experimental discrepancies and to gain a better understanding of the role of the VieSAB proteins in the biology of V. cholerae. We provide evidence that VieS is the cognate sensor kinase of VieA and that VieS contributes to the c-di-GMP-mediated regulation of biofilm and motility by controlling expression of VieA.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this work are listed in Table 1. Unless indicated otherwise, strains were grown with aeration in Luria-Bertani (LB) broth at 37°C or 30°C. A morpholinepropanesulfonic acid (MOPS) (30)-based chemically defined minimal medium supplemented with 0.75 mM KH2PO4, 0.5% glycerol, trace metals (1 ml/liter of 5% MgSO4, 0.5% MnCl2·4H2O, 0.5% FeCl3, 0.4% nitrilotriacetic acid) (7), and 25 mM l-asparagine, l-arginine, l-glutamate, and l-serine was prepared as previously described (25). Cultures were grown in the presence of appropriate antibiotics: streptomycin (100 μg/ml) and/or ampicillin (Amp) (50 μg/ml).
TABLE 1.
Strains used in this work
| Strain | Description | Reference/source |
|---|---|---|
| AC50 | V. cholerae O395 classical biotype | 35 |
| AC61 | V. cholerae O395 classical biotype, lacZ::res-tet-res | 8 |
| AC1101 | V. cholerae O395 classical biotype, ΔvieS | This work |
| AC-V1212 | V. cholerae O395 classical biotype, vieA ΔHTH | 36 |
| AC-V1386 | V. cholerae O395 classical biotype, ΔvieA | 36 |
| AC-V1487 | V. cholerae O395 classical biotype, VieA D52A | 36 |
| AC-V1596 | V. cholerae O395 classical biotype, VieA E170A | 36 |
| AC1817 | E. coli DH5α(pMMB67EH::vieA His6) | 34 |
| AC4000 | E. coli DH5α(pMalc2e::′VieS) | This work |
| AC4001 | E. coli DH5α(pMMB67EH::vieA D52A-His6) | This work |
| AC4002 | E. coli DH5α(pMALc2E::vieS D1018A) | This work |
| AC4003 | E. coli DH5α(pMALc2E::vieS H1142Q) | This work |
Construction of ΔvieS strain.
The V. cholerae classical biotype O395 ΔvieS was constructed as previously described (37), except AC50 was used as the parental strain (35).
Purification and expression of MBP-′VieS.
The cytoplasmic portion of VieS was amplified by PCR from V. cholerae strain AC61 genomic DNA with the oligonucleotides SCDF (5′-CGCGGATCCTTACGCAGCTCCGAACAAG-3′) and SCDR (5′-ACGCGTCGACTTATTCGCTCTGATACTGATG-3′). The SCDF primer introduced a BamHI restriction site and the SCDR primer a SalI site (underlined). After amplification, the fragment was digested with BamHI and SalI (New England Biolabs, Ipswich, MA) and purified using the Stratagene PCR product purification kit (Stratagene, Agilent Technologies, Santa Clara, CA). The double-digested PCR product was ligated into a pMALc2E expression vector (New England Biolabs, Ipswich, MA), which was prepared by double digestion with BamHI and SalI. The ligation product was electroporated into Escherichia coli DH5α. The electroporated cells were plated on LB agar plates supplemented with Amp and incubated for 16 h at 37°C. Amp-resistant colonies were grown in LB broth with Amp at 37°C with aeration and subjected to plasmid extraction via an alkaline lysis miniprep kit (Qiagen, Valencia, CA). The resulting plasmids were confirmed by sequencing with the oligonucleotides pMALc2EF (5′-AAGGTGAAATCATGCCGAAC-3′) and pMALc2ER (5′-GTTTTCCCAGTCACGACGTT-3′). The correct pMALc2E::′VieS clone, in which the ′VieS fragment is in frame with the maltose-binding protein (MBP), was stored at −80°C in 30% glycerol.
pMALc2E::′VieS was expressed by adding 1 mM isopropyl-β-d-thiogalactopyranoside to a 1-liter LB culture of cells grown to mid-exponential phase (optical density at 600 nm = 0.5) at 37°C with aeration. After a 3-h induction, the cells were pelleted at 10,000 × g for 10 min at 4°C. Lysis of pellets by sonication, purification by amylose resin chromatography, and elution with maltose in low-salt buffer were done following the manufacturer's protocol (New England Biolabs, Ipswich, MA). The protein concentration was determined by measuring absorbance at 280 nm, and protein integrity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. The protein preparation was stored at −80°C in buffer containing 50 mM Tris-HCl [pH 7.6], 50 mM KCl, 2 mM dithiothreitol, and 10% glycerol.
We cloned, expressed, and purified the VieS phosphoreceiver point mutant (D1018A) and the histidine phosphotransferase point mutant (H1142Q) as MBP fusions using the same procedure outlined above, with the exception that we used a splicing-by-overlap-extension strategy (15) to generate the mutated ′VieS constructs that were cloned into the pMALc2E expression vectors. The upstream and downstream products carrying the desired mutations were amplified with the following primer sets: (for D1018A) upstream fragment, SCDF and OLJ604 (5′-CGAAAAGCTCTCGCAACATCCTGAGCAGTATGATTTGTTGATTACCAATTGCCA-3′); downstream fragment, OLJ065 (5′-TATGGCAATTGGTAATCAACAAATCATACTGCTCAGGATGTTGCGAGAGCTTTT-3′) and SCDR; (for the H1142Q) upstream fragment, SCDF and OLJ066 (5′-TAGTGGCTGAGTCACTTGCTCAAGATATCGCTTTACTCAATCAGCCAGACTGTGACGTCAAAGCACTGGCACAGCGAATCAAAGGGGCAG-3′); downstream fragment, OLJ067 (5′-CCGGCTGCCCCTTTGATTCGCTGTGCCAGTGCTTTGACGTCACAGTCTGGCTGATTGAGTAAAGCGATATCTTGAGCAAGTGACTCAGCCACTACC-3′) and SCDR; The resulting products were then combined using the standard splicing-by-overlap-extension approach as previously described (15). Once joined, the fragments were cloned into the pMALc2E system, following the procedure outlined above.
Generation of vieA His6 and vieA D52A-His6 expression vectors.
The vieA and vieA(D52A) alleles were amplified by PCR from V. cholerae strains AC61 and AC1487 and cloned into expression plasmid pMMB67EH as previously described (34). The resulting plasmids were confirmed by DNA sequencing. The VieA-His6 and VieA D52A-His6 proteins were expressed and purified as previously described (34).
Phosphorylation assays.
Phosphorylation reactions were carried out in 50 mM Tris-HCl (pH 7.6), 50 mM KCl, 2 mM dithiothreitol, and 10% glycerol. Unless otherwise noted, 5 mM MnCl2 was used as the divalent cation in phosphorylation experiments. The concentration of radiolabeled [γ-32P]ATP (specific activity, 10 mCi/ml; Perkin Elmer, Boston, MA) was 30 μM. Purified MBP-′VieS, VieA, and VieA D52A were used in phosphorylation reactions at 1-μM final concentrations. The reactions were carried out at 30°C for the appropriate times. To terminate the reaction, sample buffer was added to a final concentration of 0.08 M Tris-HCl (pH 6.8), 0.025 M EDTA, 2% SDS, and 10% glycerol. The reaction products were separated by SDS-PAGE at a constant voltage of 200 V for 2 h. After electrophoresis, gels were mounted onto blotting paper 703 (VWR international, Westchester PA) and exposed to a high-resolution phosphor screen (Molecular Dynamics, Amersham Biosciences, Pittsburg, PA) for 4 h. Screens were analyzed and quantitated with a Storm PhosphorImager equipped with Image Quant software (Molecular Dynamics, Amersham Biosciences, Pittsburg PA).
PDE activity assay.
In order to determine what effect phosphorylation has on VieA's PDE activity, phosphorylated VieS was incubated for 30 min with VieA, following the same protocol as outlined above for the phosphorylation assay with the exception that nonradioactive ATP was used. To determine that VieS phosphorylated VieA, a parallel experiment was carried out under identical conditions except that radiolabeled ATP was used. Following the generation of VieA-P, the rate of hydrolysis of radiolabeled c-di-GMP was measured by two-dimensional thin-layer chromatography analysis as previously described (34).
RNA isolation.
RNA was isolated from bacterial pellets by TRIzol (Molecular Research Center, Cincinnati, OH) and phenol-chloroform extraction as previously described (9). To remove DNA, the RNA was treated with DNase I for 1 h, following the DNA-Free kit manufacturer's protocol (Applied Biosystems/Ambion, Austin, TX).
Quantitative reverse transcriptase PCR (RT-PCR).
The iScript cDNA system kit (Bio-Rad, Hercules, CA) was used to generate randomly primed cDNA pools from 200 ng of RNA from each sample, following the manufacturer's protocol. The cDNA samples were subjected to quantitative PCR with vieA-specific oligonucleotides no. 96 (5′-GATATTCGAATGCCGCAGAT-3′) and no. 97 (5′-TTTTGAAACCCCTCGACAAC-3′) using the Brilliant Sybr Green PCR mix and MXP3005 thermocycler (Stratagene/Agilent Technologies, Santa Clara, CA). Amplification included 40 cycles of the following steps: 0.5 min at 95°C, 1 min at 55°C, and 0.5 min at 72°C. Results were calculated by the comparative cycle threshold method (user bulletin no. 2; Applied Biosystems [http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf]), where the amount of target mRNA is normalized to an internal control (rpoB) amplified as previously described (29). At least two independent experiments were done.
Quantitation of intracellular c-di-GMP.
The vieS and vieA mutant strains, as well as the wild-type control, were grown to mid-exponential phase in MOPS minimal medium. Cells were pelleted and resuspended in fresh MOPS medium modified to contain 0.25 mM KH2PO4 and 100 μCi/ml [32P]orthophosphate (Perkin Elmer Boston, MA). The resuspended pellets were incubated for 1 h at 30°C with aeration. After incubation, 1 M formic acid was added to extract nucleotides as previously described (6). Sample aliquots (0.5 μl each) were spotted on polyethyleneimine-cellulose thin-layer chromatography plates (Selecto Scientific, Suwanee, GA) developed, air dried, and autoradiographed as previously described (36). The spot with Rf values corresponding to those of c-di-GMP (36) was quantitated with Image Quant software (Molecular Dynamics, Amersham Biosciences, Pittsburg PA). The amount of labeled c-di-GMP was calculated as a percentage of total labeled GTP, GDP, GMP, and ppGpp. The change in the c-di-GMP level was calculated by setting the wild-type value to 1. Results are presented as the means and standard deviations from three to six biological replicates.
Motility assay.
Chemotaxis plates were composed of MOPS minimal medium plus 0.3% agar. Single colonies from overnight streaks on LB were used to inoculate the chemotaxis plates. The plates were incubated at 30°C for 36 h, at which point the swim diameters were measured. The findings were validated in four independent experiments.
Biofilm assay.
Strains grown to stationary phase in MOPS minimal medium were diluted 1:50 in fresh MOPS minimal medium plus appropriate antibiotics in borosilicate tubes. After incubation for 96 h at room temperature without aeration, the medium was aspirated and adherent biofilms were stained with 1 ml 0.5-mg/ml crystal violet for 30 min at room temperature. Samples were then washed extensively in deionized water, and the bound crystal violet was solubilized with 1 ml of 100% ethanol. The solubilized crystal violet was quantified by measuring the absorbance at 570 nm. Results are representative of three independent experiments.
Statistical analysis.
The two-tailed Student t test was used to analyze the data when indicated. The statistical analysis software used was Graphpad Prism, version 5.0 (Graphpad Software, San Diego, CA).
RESULTS
VieS is a functional histidine kinase.
Through bioinformatic analysis, we determined that the vieSAB locus encodes a putative three-component signal transduction system in which VieS is the inner membrane-localized periplasmic sensor and VieA and VieB have conserved features corresponding to those of response regulators (19). The VieS sequence shows the presence of conserved domains that suggested it belongs to the BvgS family of bacterial hybrid sensor kinases (2). The N-terminal portion of VieS consists of two putative periplasmic amino acid binding domains flanked by two transmembrane domains. Following the second transmembrane domain, VieS contains three tandemly arranged conserved domains. The first domain, HisKa, is the dimerization and phosphoacceptor domain of bacterial protein kinases and contains a conserved histidine residue (H677) predicted to be the site of phosphorylation. Spanning residues 785 to 887 is a histidine kinase domain belonging to a family of bacterial ATPases known to catalyze histidine phosphoamide bond formation (12). Residues 968 to 1084 constitute a phosphoreceiver domain, characteristic of bacterial response regulators (33, 39), with the conserved residue D1018 serving as the predicted target of phosphorylation by the HisKa domain. Finally, VieS contains a conserved histidine phosphotransferase domain (Hpt) at the C terminus, predicted to mediate the transfer of phosphate to its cognate response regulator and also to serve as the recognition site between the sensor and its cognate response regulator (21, 27).
Based on the homologies described above, we hypothesized that VieS acts as a periplasmic sensor of amino acids or small peptides which, upon activation, autophosphorylates and mediates phosphotransfer to the cognate response regulator VieA. In order to facilitate in vitro analysis of the VieS sensor kinase, we constructed a translational fusion comprised of the C-terminal cytoplasmic portion of VieS fused to MBP. Studies conducted with similar systems have shown that the cytoplasmic portion of bacterial sensor kinases, when expressed as translational fusions with other proteins, often retain kinase activity in vitro (17).
We proceeded to test the ability of purified MBP-′VieS to autophosphorylate in vitro using radiolabeled ATP and varying the concentration of Mg2+ (Fig. 1A). Our results showed that Mg2+ failed to stimulate autophosphorylation. Usually Mg2+ stimulates histidine kinase activity in vitro, although there is evidence of other divalent cations being used (12, 13, 23). Accordingly, we tested various concentrations of Mn2+ and Ca2+. Our experiment showed that autophosphorylation of MBP-′VieS was instead stimulated by the presence of Mn2+ or Ca2+, with the latter resulting in higher levels of autokinase activity (Fig. 1A).
FIG. 1.
VieS autophosphorylation is stimulated by Ca2+ and Mn2+ and exhibits second-order kinetics. (A) Autophosphorylation of MBP-′VieS occurs in the presence of Mn2+ or Ca2+ but not Mg2+. The cytoplasmic portion of VieS was fused to Escherichia coli MBP and purified. MBP-′VieS was incubated for 30 min at 30°C in phosphorylation buffer in the presence of 30 μM [γ-32P]ATP and the divalent cations indicated. The samples were subjected to SDS-PAGE and subsequent autoradiography for 12 h. (B) Autophosphorylation of MBP-VieS is not inhibited by Mg2+. Autophosphorylation of MBP-′VieS was carried out as described above in the presence of 5 mM Mn2+ and increasing concentrations of Mg2+ to determine the effect that the presence of both cations would have on the kinase activity of MBP-′VieS. (C) Phosphorylation of MBP-′VieS at twofold serial dilutions. Autophosphorylation was carried out as described above in the presence of 10 mM Ca2+. (D) The results from panel B were quantitated and plotted (solid circles) compared with theoretical values obtained if MBP-′VieS followed perfect second-order kinetics (open squares) or first-order kinetics (dashed line).
Given that Mg2+ failed to stimulate MBP-′VieS activity, we investigated if Mg2+ had the ability to inhibit the autokinase activity of MBP-′VieS. We performed an autophosphorylation assay in vitro using a fixed concentration of Mn2+ (5 mM) and added increasing concentration of Mg2+ (Fig. 1B). The results showed no significant difference between the sample containing no Mg2+ and samples containing Mg2+. Taken together, these results suggest that Mg2+ is not the preferred divalent cation for in vitro MBP-′VieS activity and rule out the possibility that Mg2+ acts as an inhibitor of MBP-′VieS kinase activity.
VieS autophosphorylation follows second-order kinetics.
It is known that at least some bacterial sensor kinases function as dimers, whereby one monomer catalyzes the phosphoamide bond formation at the conserved histidine residue on the opposing monomer and vice versa (3, 10). Thus, we tested whether MBP-′VieS dimerized prior to autophosphorylation. To this end, we devised a kinetic experiment where we subjected serially diluted MBP-′VieS to a standard autophosphorylation assay and quantitated the amount of radiolabeled MBP-′VieS formed in each reaction (Fig. 1C). We predicted that if MBP-′VieS was active as a dimer, it would follow second-order reaction kinetics and thus a two-fold decrease in the concentration would result in a decrease in activity proportional to the square of the MBP-′VieS concentration. Indeed, the experimental results precisely overlapped the theoretical second-order kinetic values at each dilution (Fig. 1D) and did not match the expected results for first-order kinetics (Fig. 1D). Thus, we conclude that MBP-′VieS phosphorylation follows second-order kinetics and requires dimerization prior to autokinase activity.
VieA is selectively phosphorylated by VieS.
The function of many bacterial sensor kinases is to translate an extracellular stimulus into an appropriate intracellular adaptive response through the phosphorylation-induced transcriptional activity of a downstream response regulator. The vieS gene is located adjacent to a putative response regulator gene, vieA. Examination of the predicted sequence of VieA shows that the protein contains three conserved domains. The N-terminal domain is homologous to bacterial phosphoreceiver domains, with a conserved predicted phosphorylation site at residue D52 (33, 39). The second domain is an EAL domain that has c-di-GMP PDE activity (34). Finally, VieA has a putative C-terminal helix-turn-helix (HTH) domain similar to the LuxR and NarL DNA binding HTH domain (1).
Given the adjacent location of vieS and vieA on the large chromosome of V. cholerae and the presence of a putative phosphoreceiver domain within VieA, we hypothesized that VieA is the cognate response regulator for VieS and that VieS would phosphorylate VieA at the conserved D52 residue. To test this, we prelabeled purified MBP-′VieS using [γ-32P]ATP and then added purified VieA. We obtained samples at various time points and subjected them to SDS-PAGE and autoradiography analysis. As early as 15 s, we observed the appearance of a radiolabeled band at the appropriate molecular weight for VieA (Fig. 2A). This signal continued to intensify over the time span examined. That VieS is able to rapidly phosphorylate VieA is consistent with kinetic preference of VieS for VieA (32).
FIG. 2.
VieS phosphorylates VieA at the D52 residue. (A) One micromolar MBP-′VieS was prelabeled by incubation for 30 min at 30°C in phosphorylation buffer in the presence of 30 μM [γ-32P]ATP. VieA was added to a 1 μM concentration, and samples were taken at the various times indicated. The samples were subjected to SDS-PAGE and autoradiography for 4 h. (B) MBP-′VieS was incubated with 1 μM purified V. cholerae CheY-3 protein. Samples were taken at the various times indicated and subjected to SDS-PAGE and autoradiography. (C) One micromolar CheA2 was incubated with 1 μM CheY-3 in phosphorylation buffer and radiolabeled ATP as described above. After a 30-min incubation, the sample was subjected to SDS-PAGE and autoradiography. A band corresponding to CheY-3-P was detected at the expected molecular mass of approximately 14 kDa. (D) MBP-′VieS was incubated with VieA or the VieA D52A phosphoreceiver point mutant in phosphorylation buffer. No detectable VieA-P was observed in the reaction containing MBP-′VieS and VieA D52A. (E) One micromolar of the purified MBP-′VieS receiver D1018A point mutant and the Hpt H1142Q point mutant were assayed for their ability to phosphorylate VieA by incubation with 1 μM purified VieA in phosphorylation buffer at 30°C for 30 min. A band corresponding to VieA-P was observed only in the wild-type VieS protein, indicating that both residues are required for proper phosphorelay and phosphorylation of VieA.
Prolonged incubation of a sensor kinase with a noncognate response regulator can result in nonspecific phosphorylation of the response regulator (18, 32). Therefore, we investigated whether VieS is able to phosphorylate the noncognate response regulator CheY-3 from V. cholerae. To do this we incubated MBP-′VieS and CheY-3 (gift from Susan M. Butler-Wu) and took samples at various time points to determine the levels of both phosphorylated species. After incubating both VieS and CheY-3 for an extended period of 30 min, we were able to observe a faint band corresponding to phosphorylated CheY-3 and an intense higher-molecular-weight band corresponding to MBP-′VieS (Fig. 2B). In contrast, incubation of CheY-3 with its cognate sensor kinase, CheA-2 (gift from Susan M. Butler-Wu), resulted in robust phosphorylation (Fig. 2C). Thus, CheY-3 could be phosphorylated by its cognate sensor kinase but only poorly by VieS. Taken together, these results suggest that MBP-′VieS is the cognate sensor kinase for VieA.
Having demonstrated that MBP-′VieS is able to phosphorylate VieA, we tested whether the D52 residue of VieA is the site of phosphorylation. For this, we purified a VieA D52A point mutant and incubated it with prelabeled MBP-′VieS in a standard phosphorylation reaction (Fig. 2D). As expected, MBP-′VieS was able to phosphorylate VieA but not the VieA D52A point mutant, indicating that this is the conserved phosphorylation site of VieA.
In order to determine which residues were involved in the VieS-to-VieA phosphorelay, we tested purified point mutants of MBP-′VieS. The D1018A mutant has a mutation in the conserved aspartate within the receiver domain of VieS, while the H1142Q mutant has a mutation in the conserved histidine within the Hpt domain of VieS. We performed a standard phosphorylation assay as described above in the presence of 1 μM VieA. After a 30-min incubation, we subjected the samples to SDS-PAGE and subsequent autoradiography. Results showed that at equimolar amounts of protein, the VieS D1018A mutant had a highly elevated steady-state phosphorylation level compared to that of wild-type VieS whereas the H1142Q mutant was not able to be phosphorylated (Fig. 2E). As expected, the H1142Q mutant was unable to transfer phosphate to VieA. However, the same was also true for the D1018A mutant, indicating that this residue within VieS is required for phosphotransfer to VieA.
VieS is involved in regulating vieA transcript level.
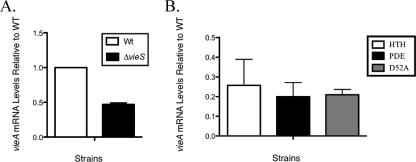
In order to adapt to the surrounding environment, bacterial signaling systems are precisely regulated. It has been demonstrated in the homologous BvgAS system that upon sensing the appropriate signal, BvgS phosphorylates BvgA and changes the affinity of BvgA for specific promoter promoter elements (10). This change ultimately results in the regulation of genes required for host colonization. As previously stated, analysis of the predicted protein sequence of VieA revealed a conserved HTH domain associated with the LuxR family of response regulators. A transcriptional profiling study indicated that VieA acted as a positive regulator of both vieS and vieB, since a vieA null mutant had approximately four- and fivefold reductions in vieS and vieB transcript, respectively, compared to the wild-type levels (4). These observations were consistent with the role of VieA as a positive regulator of vieSAB. Given our data that showed phosphorylation of VieA by VieS, we investigated the link between VieS and vieA transcriptional regulation. For this, the VieS null mutant was grown in MOPS minimal medium containing amino acids previously shown to trigger virulence gene induction (7). Late-exponential-phase V. cholerae was isolated, and the levels of vieA transcript were measured by quantitative RT-PCR. Results revealed that compared to the wild type, the vieS mutant had a twofold decrease in the vieA transcript level (Fig. 3A). These results suggest that VieS plays a role in the regulation of vieA expression.
FIG. 3.
VieS affects the level of vieA expression. (A) V. cholerae wild-type (WT) and ΔvieS strains were grown in MOPS minimal medium supplemented with 25 mM Asn, Arg, Glu, and Ser (NRES) and 0.75 mM KH2PO4 to late exponential phase. Total RNA was isolated, and a cDNA pool was prepared by random-priming RT-PCR. Quantitative RT-PCR was used to quantitate the level of vieA transcript using gene-specific primers. Results were normalized to an internal control, rpoB, and presented as quantities relative to that for the wild type. The mean and standard deviation from technical replicates are shown. An independent experiment gave similar results. (B) VieA mutant strains were also assayed for vieA transcript level upon stimulation with NRES as described for panel A. The VieA ΔHTH (HTH) mutant was assayed along with the PDE E170A (PDE) mutant and the phosphoreceiver D52A mutant (D52A). The mean and standard deviation for three biological replicates are shown.
To further pinpoint the roles VieS and VieA may have in regulating vieA expression, we measured transcript levels of vieA in a series of VieA mutants (Fig. 3B). The VieA ΔHTH mutant contained an in-frame deletion of the putative DNA binding HTH domain. We also tested the VieA E170A mutant, which contained a point mutation within the EAL domain, known to be responsible for VieA′s PDE activity (34). We investigated the role phosphorylation of VieA had on its regulatory activity by using the VieA D52A phosphoreceiver point mutant. We found that in all cases the level of vieA transcript was significantly lower than that of the wild type. That the VieA D52A point mutant also showed decreased transcript levels comparable to those of the other VieA mutants suggests that phosphorylation is important for proper VieA autoregulation. Furthermore, the fact that the level of vieA transcript is lower in the D52A mutant than in the vieS null mutant suggests that in the absence of VieS, VieA may be phosphorylated to an extent by another sensor kinase. Finally, the comparable levels of vieA transcript between the HTH and D52A mutants suggest that phosphorylation affects the DNA binding ability of VieA. Interestingly, the VieA E170A PDE mutant also showed a decreased level of vieA transcript. This suggests that vieA is regulated in some manner by the level of c-di-GMP in the cell.
VieS regulates motility and biofilm formation by altering intracellular c-di-GMP level.
The secondary messenger c-di-GMP is an important signaling molecule in bacterial gene regulation (16). In V. cholerae, fluctuation of c-di-GMP levels has been linked to regulation of genes involved in virulence, biofilm formation, and motility (20, 28, 36). VieA has been shown to regulate the c-di-GMP level through EAL domain-dependent PDE activity (34, 36). Based on our finding of reduced vieA transcript level in the vieS mutant (Fig. 3), we hypothesized that in this strain the level of c-di-GMP would be elevated. To investigate this, we quantitated total intracellular c-di-GMP in the wild type and vieS and various vieA mutant strains under minimal medium growth supplemented with amino acids known to stimulate virulence gene expression (7). The results showed that the vieS null strain had a highly elevated intracellular level of c-di-GMP, comparable to that observed in a vieA null strain (Table 2). These results suggest a strong link between VieS and c-di-GMP regulation. To investigate the role that VieA autoregulation had in modulating the c-di-GMP level, we also quantitated the intracellular c-di-GMP level in a vieA ΔHTH mutant. Results showed that this deletion caused a threefold increase in the c-di-GMP level, suggesting that transcriptional regulation plays an important role in regulating the intracellular VieA level and thus indirectly altering the intracellular c-di-GMP level. Unexpectedly, when we investigated the intracellular c-di-GMP level in the VieA D52A strain, we found that the concentration of c-di-GMP was unaffected (ratio of 1.1 relative to that of the wild type). This suggested that the inability to phosphorylate VieA had no effect on the global level of c-di-GMP. Finally, we measured the in vitro PDE activity of VieA compared to that of VieA-P and observed no difference in activity (data not shown).
TABLE 2.
Quantitation of total intracellular c-di-GMP for V. cholerae strains
| Genotypea | % c-di-GMP (±SD)b,d | Fold change in c-di-GMP (P value)c,d |
|---|---|---|
| WT | 0.069 (0.01) | 1 |
| ΔvieA | 0.56 (0.10) | 8.1 (<0.0004) |
| vieA(E170A) | 0.96 (0.29) | 14 (<0.005) |
| vieA ΔHTH | 0.24 (0.01) | 3.3 (<0.0001) |
| ΔvieS | 0.75 (0.10) | 11 (<0.0001) |
Strains were grown to mid-exponential growth phase in MOPS minimal medium containing radiolabeled orthophosphate. WT, wild type.
Two-dimensional thin-layer chromatography quantitation of radiolabeled guanine nucleotides was carried out to determine levels of c-di-GMP.
c-di-GMP levels are expressed as n-fold increases relative to the wild-type level of c-di-GMP normalized to total cell guanine nucleotide levels.
Student's t test was used to determine statistical significance.
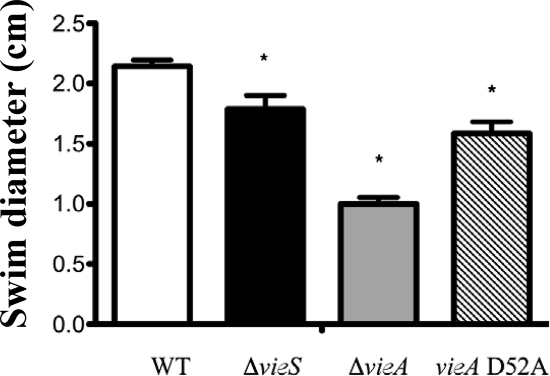
Because we observed a similar-magnitude increase of c-di-GMP in the vieS and vieA null mutants during growth under phosphate-limiting conditions, we hypothesized that measuring motility under the same conditions would uncover similar motility defects for both mutants. However, the motility defect for the vieS mutant was not as pronounced as that seen for the vieA mutant (Fig. 4) but was comparable to that seen for the VieA D52A mutant. This suggests that even though the c-di-GMP levels are comparably elevated in the two null mutants, other functions attributable to VieA that are lost in the ΔvieA strain affect the motility phenotype. Additionally, it also suggests that phosphorylation of VieA does contribute to motility.
FIG. 4.
The vieS null strain exhibits a slight motility/chemotaxis defect. The V. cholerae wild type (WT) and ΔvieS, ΔvieA, and vieA D52A mutants were stabbed into MOPS minimal medium supplemented with 25 mM Asn, Arg, Glu, and Ser. After 36 h at 30°C, the swim diameters were measured from three independent stabs; means and standard deviations are shown. Asterisks indicate significant reductions in swim diameter compared to that for the WT (P < 0.05 using Student's t test).
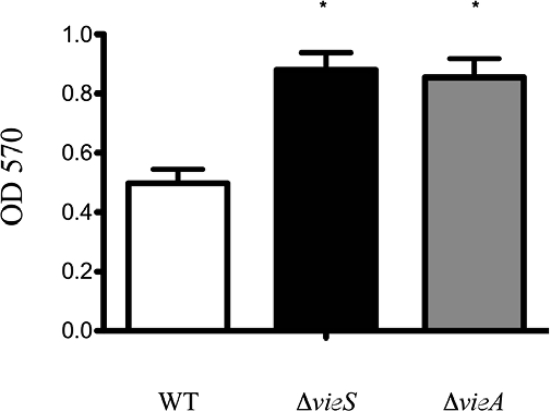
Given the strong link between the c-di-GMP level and biofilm formation (11, 20, 36), we hypothesized that the increased c-di-GMP level observed in the vieS null strain would result in a concomitant increase in biofilm production. To test this hypothesis, we measured biofilm formation under static conditions in MOPS minimal medium. Results showed an increase in biofilm production in the vieS mutant comparable to that of the vieA mutant (Fig. 5). Thus, VieS negatively regulates biofilm production in a manner similar to that of VieA, likely through control of the c-di-GMP level.
FIG. 5.
The vieS null strain exhibits an increase in biofilm production. ΔvieA and ΔvieS V. cholerae mutants were tested for their ability to form biofilm in a MOPS minimal medium supplemented with 0.75 mM KH2PO4 and 25 mM Asn, Arg, Glu, and Ser amino acids. After incubation at 30°C for 96 h in glass borosilicate culture tubes, the samples were subjected to the standard crystal violet staining for biofilm. The means and standard deviations of three independent experiments are shown. Asterisks indicate significant increases in biofilm formation compared to WT results (P < 0.05 using Student's t test).
DISCUSSION
There is increasing evidence that links c-di-GMP regulation to environmental sensors. One example is the Pseudomonas aeruginosa wsp locus, which is thought to function in a similar fashion to chemoreceptor signaling systems with the exception that instead of affecting flagellar rotation, it is believed to affect levels of c-di-GMP via a conserved GGDEF domain within the WspR response regulator (14). In this study, we investigated the V. cholerae VieS/VieA sensory and signal transduction system and showed how it impacts the c-di-GMP level and associated phenotypes. We showed that VieS is a functional sensor kinase that selectively targets the autoregulatory response regulator and c-di-GMP PDE, VieA, for phosphorylation. Moreover, we established a link between the VieS/VieA phosphorelay and regulation of motility and biofilm formation. Our data support a model whereby VieS activity leads to decreased c-di-GMP as a direct result of signaling through VieA. Support for this model comes from work on a homologous system in P. aeruginosa encoded by rocS1RA1/sadSRA, which is predicted to be part of two-component signaling system that regulates biofilm production through affecting the c-di-GMP level (11).
We first investigated the role of VieS as a sensor kinase by purifying a cytoplasmic portion of the enzyme and testing in vitro autokinase activity. Instead of the common Mg2+ requirement, we found that both Mn2+ and especially Ca2+ were able to stimulate the autokinase activity of VieS. This requirement was interesting, since the overwhelming majority of bacterial histidine kinases studied to date autophosphorylate in vitro in the presence of Mg2+. Although it is tempting to speculate that Mn2+ or Ca2+ may be the divalent cation preferred by VieS in vivo, it should be pointed out that the in vivo concentrations of Mn2+ and Ca2+ in E. coli are estimated to be ∼0.1 mM and ∼0.2 μM, respectively, which are below the concentrations needed to stimulate robust VieS activity in vitro (24, 26). If the concentrations of these cations in V. cholerae are similar to those in E. coli, then it is unlikely that they function to stimulate VieS in vivo. It is possible that Mn2+ and Ca2+ are inducing a conformational change in MBP-′VieS in vitro that mimics the activated state of VieS in vivo. Such a scenario has been proposed for the Mn2+-stimulated Myxococcus xanthus histidine kinase FrzE (23). Since MBP-′VieS lacks the two transmembrane domains found naturally in VieS, it is possible that Mn2+ or Ca2+ may induce a conformational change that is normally induced by the physiological activator of VieS when the full protein is properly localized within the inner membrane.
Studies of the B. pertussis BvgAS and Bacillus subtilis Kin/Spo phosphorelay systems have shown that sensor kinases, such as BvgS and Kin histidine kinases, form dimers prior to autophosphorylation (10, 40). Given the similarity in domain structure between VieS and these sensor kinases, we predicted that VieS would also require dimerization for activity. Our data showing second-order kinetics of an MBP-′VieS chimera is in agreement with this hypothesis.
Prior to this work, the relationship between VieS and VieA was unclear. Our results showed that the MBP-′VieS construct was capable of phosphorylating VieA at the conserved D52 residue within the conserved phosphoreceiver domain. Studies done with Caulobacter crescentus demonstrated that sensor proteins show kinetic preference toward their respective cognate response regulator (32). Our data suggest that VieA is the cognate response regulator for VieS, since we observed that VieS had a kinetic preference toward VieA compared to the noncognate CheY-3 response regulator. These results are consistent with the adjacent location of vieS and vieA in an apparent operon, and they support our model of VieS/VieA interaction as a sensor kinase/response regulator pair.
Until this study, a role for phosphorylation in the regulation of c-di-GMP levels in V. cholerae had not been demonstrated. An increase in the VieA concentration as a result of VieS-to-VieA phosphotransfer and subsequent VieA autoregulation would be expected to affect the intracellular level of c-di-GMP. This is because VieA is known to function as a PDE which degrades c-di-GMP, thus promoting motility and virulence gene expression while repressing biofilm genes (34, 36). Consistent with this model, we found that the intracellular level of c-di-GMP was elevated in both vieS and vieA null strains.
These changes in the c-di-GMP level resulted in corresponding changes in motility and biofilm formation. However, the magnitude of these changes was not always consistent. Notably, although the vieS deletion resulted in only a twofold drop in vieA transcription and a corresponding minor reduction in motility, the concentration of c-di-GMP was nevertheless greatly increased and biofilm formation was increased to levels similar to that observed for a vieA deletion. The fact that we observed a motility decrease in the VieA D52A mutant similar to that for the vieS deletion strain suggests that phosphorylation of VieA by VieS is important for regulating motility and thus establishes a biological role for VieA-P. The modest decrease in vieA expression in the vieS null background may be a result of residual VieA phosphorylation and subsequent autoregulation from other phosphodonors, such as acyl phosphate. Such a scenario has already been shown for response regulators in some gram-negative bacteria (22). Another possibility is that there may be another kinase that can phosphorylate VieA and activate it. This has already been described for the E. coli osmosensor EnvZ (38) and could explain the phenotype discrepancies between ΔvieS and ΔvieA strains. Furthermore, our finding that the VieA D52A mutant strain had a lower level of vieA transcript than the vieS null strain supports the idea that in the absence of VieS, other sensor kinases or inorganic phosphates may act on VieA to phosphorylate it.
Our work demonstrates that VieS is involved in the regulation of motility and biofilm production via control of VieA autoregulation and subsequent alteration of the intracellular level of c-di-GMP. Additional work will be required to elucidate the function of VieB and how it may interact with VieS and/or VieA. Understanding the VieSAB signal transduction system will contribute not only to our knowledge of V. cholerae biology but also more generally to that of how the important secondary messenger c-di-GMP is regulated in response to environmental signals.
Acknowledgments
We thank Ernesto Muñoz-Elias for assistance with the manuscript, as well as helpful suggestions and discussions.
This work was funded by the Howard Hughes Medical Institute and the National Institute of Allergy and Infectious Diseases (AI45746).
Footnotes
Published ahead of print on 1 August 2008.
REFERENCES
- 1.Antunes, L. C., R. B. Ferreira, C. P. Lostroh, and E. P. Greenberg. 14 December 2007. A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. doi: 10.1128/JB. 01443-07. [DOI] [PMC free article] [PubMed]
- 2.Beier, D., H. Deppisch, and R. Gross. 1996. Conserved sequence motifs in the unorthodox BvgS two-component sensor protein of Bordetella pertussis. Mol. Gen. Genet. 252169-176. [DOI] [PubMed] [Google Scholar]
- 3.Beier, D., and R. Gross. 2006. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9143-152. [DOI] [PubMed] [Google Scholar]
- 4.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 743633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blatch, G. L., and M. Lässle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21932-939. [DOI] [PubMed] [Google Scholar]
- 6.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 2579759-9769. [PubMed] [Google Scholar]
- 7.Callahan, L. T., R. C. Ryder, and S. H. Richardson. 1971. Biochemistry of Vibrio cholerae virulence. II. Skin permeability factor/cholera enterotoxin production in a chemically defined medium. Infect. Immun. 4611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilli, A., and J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, P. A., and A. M. Jones. 2003. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 11367-373. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, P. A., and S. Stibitz. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 1017-23. [DOI] [PubMed] [Google Scholar]
- 12.Gamble, R. L., M. L. Coonfield, and G. E. Schaller. 1998. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc. Natl. Acad. Sci. USA 957825-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haydel, S. E., N. E. Dunlap, and W. H. Benjamin. 1999. In vitro evidence of two-component system phosphorylation between the Mycobacterium tuberculosis TrcR/TrcS proteins. Microb. Pathog. 26195-206. [DOI] [PubMed] [Google Scholar]
- 14.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 10214422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8528-535. [PubMed] [Google Scholar]
- 16.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40385-4007. [DOI] [PubMed] [Google Scholar]
- 17.Kaspar, S., R. Perozzo, S. Reinelt, M. Meyer, K. Pfister, L. Scapozza, and M. Bott. 1999. The periplasmic domain of the histidine autokinase CitA functions as a highly specific citrate receptor. Mol. Microbiol. 33858-872. [DOI] [PubMed] [Google Scholar]
- 18.Laub, M. T., E. G. Biondi, and J. M. Skerker. 2007. Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 423531-548. [DOI] [PubMed] [Google Scholar]
- 19.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 1802298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60331-348. [DOI] [PubMed] [Google Scholar]
- 21.Matsushika, A., and T. Mizuno. 1998. The structure and function of the histidine-containing phosphotransfer (HPt) signaling domain of the Escherichia coli ArcB sensor. J. Biochem. 124440-445. [DOI] [PubMed] [Google Scholar]
- 22.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 26931567-31572. [PubMed] [Google Scholar]
- 23.McCleary, W. R., and D. R. Zusman. 1990. Purification and characterization of the Myxococcus xanthus FrzE protein shows that it has autophosphorylation activity. J. Bacteriol. 1726661-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Médicis, E. D., J. Paquette, J. J. Gauthier, and D. Shapcott. 1986. Magnesium and manganese content of halophilic bacteria. Appl. Environ. Microbiol. 52567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1702575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naseem, R., S. R. Davies, H. Jones, K. T. Wann, I. B. Holland, and A. K. Campbell. 2007. Cytosolic Ca2+ regulates protein expression in E. coli through release from inclusion bodies. Biochem. Biophys. Res. Commun. 36033-39. [DOI] [PubMed] [Google Scholar]
- 27.Perraud, A. L., B. Kimmel, V. Weiss, and R. Gross. 1998. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal Hpt domains of the sensor proteins. Mol. Microbiol. 27875-887. [DOI] [PubMed] [Google Scholar]
- 28.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 28212860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinones, M., H. H. Kimsey, and M. K. Waldor. 2005. LexA cleavage is required for CTX prophage induction. Mol. Cell 17291-300. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sanders, D. A., B. L. Gillece-Castro, A. M. Stock, A. L. Burlingame, and D. E. Koshland. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 26421770-21778. [PubMed] [Google Scholar]
- 32.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock, J. B., A. M. Stock, and J. M. Mottonen. 1990. Signal transduction in bacteria. Nature 344395-400. [DOI] [PubMed] [Google Scholar]
- 34.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 28033324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 842833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tischler, A. D., S. H. Lee, and A. Camilli. 2002. The Vibrio cholerae vieSAB locus encodes a pathway contributing to cholera toxin production. J. Bacteriol. 1844104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhamme, D. T., J. C. Arents, P. W. Postma, W. Crielaard, and K. J. Hellingwerf. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 14869-78. [DOI] [PubMed] [Google Scholar]
- 39.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 3211741-11753. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L., C. Fabret, K. Kanamaru, K. Stephenson, V. Dartois, M. Perego, and J. A. Hoch. 2001. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J. Bacteriol. 1832795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]