Abstract
We have previously identified the phyC gene of Bacillus amyloliquefaciens FZB45, encoding extracellular phytase, as a member of the PhoP regulon, which is expressed only during phosphate starvation. Its σA-dependent promoter is positively and negatively regulated by the phosphorylated PhoP response regulator in a phosphate-dependent manner (O. Makarewicz, S. Dubrac, T. Msadek, and R. Borriss, J. Bacteriol. 188:6953-6965, 2006). Here, we provide experimental evidence that the transcription of phyC underlies a second control mechanism exerted by the global transient-phase regulator protein, AbrB, which hinders its expression during exponential growth. Gel mobility shift and DNase I footprinting experiments demonstrated that AbrB binds to two different regions in the phyC promoter region that are separated by about 200 bp. One binding site is near the divergently orientated yodU gene, and the second site is located downstream of the phyC promoter and extends into the coding region of the phyC gene. Cooperative binding to the two distant binding regions is necessary for the AbrB-directed repression of phyC transcription. AbrB does not affect the transcription of the neighboring yodU gene.
Several Bacillus species produce phytases, extracellular degradative enzymes, which release free phosphates from myo-inositol hexakisphosphate, the main storage form of phosphate in plants. While the monocistronic phyC gene is silent in the laboratory strain Bacillus subtilis 168, probably due to the absence of functional PhoP binding sites of its transcriptional activator, PhoP, the phyC gene of Bacillus amyloliquefaciens FZB45 (phyCFZB45) is well expressed (8). In vitro and in vivo studies with phyCFZB45 promoter lacZ reporter gene fusions expressed in the heterologous but closely related and genetically amenable Bacillus subtilis 168 host revealed that the phosphorylated response regulator PhoP∼P affects the phyCFZB45 expression in a bifunctional manner. The phyC gene becomes activated by PhoP∼P under phosphate starvation due to the binding of the response regulator to the PhoP box, positioned between −32 and −49. However, rising concentrations of PhoP∼P cause the binding of the response regulator to a second site located near the −10 region, thereby reducing the efficiency of transcription by displacing the RNA polymerase (9).
In addition, our earlier studies indicated that the expression of phyCFZB45 underlies a second level of control. We observed that the induction of phytase under phosphate starvation is delayed until the transition from the exponential to the stationary growth phase (9). A mutant strain bearing a deletion of the response regulator Spo0A, which governs the initiation of sporulation, was unable to express phyCFZB45 even under low-phosphate conditions (O. Makarewicz, unpublished observation). Spo0A is known to downregulate the global transition state regulator AbrB (6, 12, 17), which results in the relief of expression of numerous genes involved in functions such as the production of antibiotics (10, 11), the formation of biofilms (4, 7), the development of competence (6), the initiation of sporulation (11, 23), the production of extracellular proteases and other degradative enzymes (13), cannibalistic behavior (6), and, in Bacillus anthracis, the expression of toxin genes (15). During the transition state, phosphorylated Spo0A∼P relieves AbrB-mediated repression by enhanced binding to the P2 promoter of abrB, thus lowering its transcription (12). On the other hand, during exponential growth AbrB indirectly controls the expression of Spo0A by repressing the transcription of the alternative sigma factor sigH, which is necessary for the full transcription of spo0A (22). In spite of the partial antagonism of both regulators, we decided to carry out lacZ reporter studies in a Spo0A- and AbrB-deficient background. Like other global transcriptional regulators shown to affect phytase gene expression (PhoP and Spo0A), AbrB is a highly conserved protein in Firmicutes. The 94-amino-acid AbrB proteins from B. subtilis and B. amyloliquefaciens are distinguished by only a single amino acid substitution. Since genetic studies are not feasible in FZB45 due to its low transformation frequency, we decided to use the related B. subtilis 168 as a heterologous host for in vivo studies with the phyCFZB45 promoter fragment.
Here, we present experimental evidence that in addition to the control exerted by the PhoPR system phyCFZB45, transcription is directly repressed by the transition state regulator AbrB during exponential growth. Identical responses were found when using the AbrB protein isolated from B. subtilis and the mutant protein AbrBQ82K, corresponding to the B. amyloliquefaciens AbrB (EU549819).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani (LB) medium and low-phosphate medium, described previously (9). When appropriate, antibiotics were added in the following concentrations: for Escherichia coli, 100 mg/liter of ampicillin (Ap) and 50 mg/liter of kanamycin (Km); and for B. subtilis, 5 mg/liter of chloramphenicol (Cm) and 5 mg/liter of Km.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE44 ΔlacU169 (Φ80dlacZΔM15) hsdR17 recA1 gyrA96 thi-1relA1 | Lab strain |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| ECAbrB | F−ompT hsdSB(rB− mB−) gal dcm (DE3)::pPHOP | This work |
| Bacillus amyloliquefaciens strain | ||
| FZB45 | wt | FZB Berlin |
| Bacillus subtilis | ||
| MF1 | trpC2 pheA1 rpoCHis6 Neor | M. Fujita |
| 168 | trpC2 | Laboratory sock |
| 1S13 | trpC2 spo0A1 | BGSC |
| OM61 | trpC2 amy::lacZ-phyC (495 bp) cat (Cmr) | This work |
| OM61Km | trpC2 amy::lacZ-phyC (495 bp) cat:aphA3 (Kmr) | This work |
| OM71 | trpC2 amy::lacZ-phyC (316 bp) cat | This work |
| OM545 | trpC2 amy::lacZ-phyC (355 bp) cat | This work |
| OM57 | trpC2 amy::lacZ-phyC (176 bp)Cmr | This work |
| OM64 | trpC2 spo0A1 amy::lacZ-phyC (495 bp) cat | This work |
| OM64Km | trpC2 spo0A1 amy::lacZ-phyC (495 bp) cat:aphA3 | This work |
| OM613 | trpC2 abrB::Cmr amy::lacZ-phyC (495 bp) cat:aphA3 | This work |
| OM612 | trpC2 spo0A1 abrB::lacZ-phyC (495 bp) cat | This work |
| OM74 | trpC2 spo0A1 amy::lacZ-phyC (316 bp) cat | This work |
| OM45 | trpC2 spo0A1 amy::lacZ-phyC (355 bp) cat | This work |
| OM457 | trpC2 spo0A1 amy::lacZ-phyC (176 bp) cat | This work |
| EB21 | trpC2 amy::lacZ-phyCabrB2 (495 bp) cat | This work |
| EB24 | trpC2 spo0A1 amy::lacZ-phyCabrB2 (495 bp) cat | This work |
| EB31 | trpC2 amy::lacZ-phyCabrB31 (495 bp) cat | This work |
| EB34 | trpC2 spo0A1 amy::lacZ-phyCabrB (495 bp) cat | This work |
| EB41 | trpC2 amy::lacZ-phyCabr312 (495 bp) cat | This work |
| EB44 | trpC2 spo0A1 amy::lacZ-phyCabrB32 (495 bp) cat | This work |
| Plasmids | ||
| pDG268 | Integrative vector amy::lacZ cat bla (Apr) | C. Antoniewski |
| pOM6 | pDG268/phyC from −287 (Om01) to +208 (Om09) → 495-bp insert | This work |
| pOM7 | pDG268/phyC from −287 (Om01) to +29 (Om14) → 316-bp insert | This work |
| pCUT5 | pDG268/phyC from −147 (Cut5) to +208 (Om09) → 355-bp insert | This work |
| pCUT57 | pDG268/phyC from −147 (Cut5) to +29 (Om14) → 176-bp insert | This work |
| pEB2 | pOM6, substitution of AbrB site 1 (eb2) | This work |
| pEB3 | pOM6, substitution of AbrB site 1 (eb3) | This work |
| pEB4 | pOM6, substitution of AbrB site 1 (eb4) | This work |
| pEB5 | pOM6, substitution of AbrB site 2 (eb5) | This work |
| pEB6 | pOM6, substitution of AbrB site 2 (eb6) | This work |
| pEB23 | pOM6, double substitution of AbrB site 1 (eb2 and eb3) | This work |
| pEB36 | pOM6, double substitution of AbrB sites 1 (eb3) and 2 (eb6) | This work |
| pET15b | Expression vector, T7lac lacI bla | Novagen |
| pABRB | pET15b/abrB of B. subtilis → 354-bp insert | This work NdeI/XhoI site |
| pABRBQ82K | pET15b/abrB of B. subtilis → 354-bp insert, substitution C:A (−245*) | This work NdeI/XhoI site |
| pPHOP | pET15b/phoP of B. subtilis → 728-bp insert | 11 |
| pPHOR231 | pET28b(+)/C-terminal domain of phoR of B. subtilis → 1,051-bp insert | 11 |
| pECE73 | Cmr::Kmr | M. Steinmetz/R. Rich |
*, position of the substitution of abrB is indicated relative to translation start. Neor; neomycin resistance.
DNA manipulations and general methods.
Isolation of plasmid and chromosomal DNA, restriction endonuclease digestion, agarose gel electrophoresis, PCR, and transformation of E. coli and B. subtilis were performed as described previously (16).
Construction of plasmids and bacterial strains.
Specific DNA fragments were amplified from the phyC promoter region of B. amyloliquefaciens FZB45 by use of the primer pairs listed in Table S1 in the supplemental material. Strains containing promoter-lacZ fusions derived from B. amyloliquefaciens FZB45 were constructed as previously described (9). Mutagenesis of the AbrB binding sites eb2, eb3, and eb4 was performed using the splicing by overlapping extension method. First, PCR with plasmid pOM6 and the following primer pairs were used to generate overlapping fragments: AbrB2for/Om09 and Om01/AbrB2rev, AbrB3for/Om09 and Om01/AbrB3rev, and Abr4for/Om09 and Om01/AbrB4rev. The fragments were fused in a second PCR in which 10 cycles were run without primers by use of 200 ng of the overlapping fragments and a further 15 cycles were run in the presence of the additional primers Om01 and Om09. The double substitution for eb24 was performed as for eb2 but by use of pEB4 as the template. The resulting PCR products were cloned into the EcoRI and BamHI sites of plasmid pDG268 and the mutations were verified by sequencing using sequencing primer Cy5-Om16. Two-base pair substitutions of the AbrB binding sites eb5, eb6, and eb36 were introduced by using the QuikChange XL site-directed mutagenesis kit (Stratagene). Plasmid pOM61 was used as the template for mutagenizing eb5 and eb6 and pEB3 for eb36. The primers used were AbrB5for/AbrB5for and AbrB6for/AbrB6rev. After linearization by XhoI, the resulting plasmids were transformed into competent cells prepared from B. subtilis 168 and 1S13 (spo0A derivative). Plasmids and resulting strains are compiled in Table 1. A schematic representation of the fusions obtained is shown in Fig. 1.
FIG. 1.
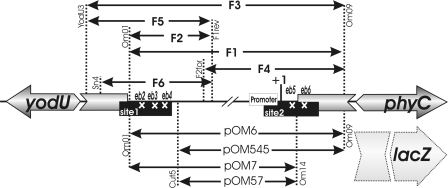
Schematic representation of the yodU-phyC intergenic region of B. amyloliquefaciens FZB45. The position of the phyCFZB45 promoter and the initiation point of phyC transcription (+1) are indicated. The two AbrB binding regions (sites 1 and 2) are indicated as filled boxes. The positions of the mutated binding areas (eb2, eb3, eb4, eb5, and eb6) within regions 1 and 2 are marked by white crosses. Double-headed arrows indicate DNA fragments amplified from parts of the entire phyC promoter region. DNA fragments used for lacZ fusions are listed at bottom, while DNA fragments used for DNase I footprinting, gel shift, and in vitro transcription are listed at top. DNA primers used for amplifying the respective DNA fragments are also shown at the vertical dotted lines.
Exchange of the antibiotic-resistant markers was accomplished with the plasmid pECE73 (BGSC). It was transformed into strains OM61 and OM64 to generate strains OM61KM (wild type [wt]) and OM64KM (spo0A derivative), in which the Cm resistance cassette was replaced by the Km resistance cassette. To obtain the clones OM612 (spo0A abrB) and OM613 (abrB), OM61KM and OM64KM were transformed with chromosomal DNA isolated from the abrB-negative B. subtilis mutant strain JH12586 (pheA1 trpC2 abrB::cat) generously supplied by Tarek Msadek, Institut Pasteur. The abrB::Cm genotype was verified by PCR using primers AbrB1 and AbrB4. To construct the expression plasmid pABRB, the abrB gene was amplified using primers AbrB3 and AbrB4. The PCR product was cloned into the NdeI and BamHI sites of pET15b. To obtain the AbrBQ82K protein, corresponding to AbrB of B. amyloliquefaciens, the Q amino acid residue of B. subtilis AbrB (AbrBsub) was replaced by K by use of the QuikChange XL site-directed mutagenesis kit with primers AbrBQ82Kfor and AbrBQ82Krev and template pABRB DNA. The sequences of the primers used in this study are compiled in Table S1in the supplemental material.
Overexpression and purification of proteins.
PhoP, PhoR231-His6, and RNA polymerase were overexpressed and purified as described previously (1, 5, 9, 14). The His6-AbrB proteins were overexpressed in E. coli C41 (BL21) according to the protocol of Novagen. The cells were lysed by sonication in buffer A, consisting of 50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride. His6-AbrBsub and His6-AbrBQ82K were purified by Ni-agarose chromatography and dialyzed against 10 mM Tris-HCl (pH 7.5), 300 mM NaCl, and 50% glycerol. Protein concentrations were determined at 280 nm.
Enzyme assays.
The assays for alkaline phosphatase (APase) and β-galactosidase were performed as described previously (9), except for a slight modification of the β-galactosidase assay: a 100-μl cell suspension was mixed with 800 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 1 mM dithiothreitol, 0.5 U/ml benzonase, 100 μg/ml Cm, 0.3 mg/ml lysozyme, 0.005% Triton X-100) and incubated for 10 min at 30°C.
DNase I footprinting assay.
DNase I footprinting experiments were performed essentially as described previously (9). Two separate DNA fragments, F6 and F4, covering the extended phyC promoter region (Fig. 1), were amplified using the primer pairs Sn4/F1rev and F2for/Om09, respectively, and purified with the QIAquick PCR purification kit (Qiagen). The PCR products were labeled either at the coding or at the noncoding strand as described previously (9). The DNA-binding reactions were performed for 20 min at room temperature in binding buffer with different AbrB concentrations (0, 1.2, 2.4, 4.8, 9.6, 14.4, and 20 μM). The labeled F6 fragment (100,000 cpm) and 5 nM of the nonlabeled F4 fragment and vice versa were used.
Gel shift assay.
5′ γ-32P-labeled DNA fragments corresponding to the phyC promoter region were synthesized using the primer pairs Om01/[γ32P]Om09 (F1), [γ32P]Om01/F1rev (F2), and F2for/[γ32P]Om09 (F4) and then purified with the QIAquick PCR purification kit (Qiagen). The binding reaction was carried out in binding buffer [20 mM Tris-HCl buffer (pH 8), 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, and 0.1 mg/ml poly(dI-dC) as a competitive nonspecific DNA] with labeled DNA (∼5 nM) and various AbrB concentrations (0.25 μM to 2 μM) for 20 min at room temperature. The entire phyC promoter fragment (pOM6 [Fig. 1]) and the appA gene of E. coli were used for competition experiments. Concentrations of AbrB (1 μM) and labeled phyC (5 nM) were kept constant, and the amounts of the competitor DNA varied between 1 nM and 5 nM. The reaction mixtures were then separated on 6% nondenaturing polyacrylamide gels in 1× Tris-borate-EDTA buffer at 100 V.
In vitro transcription assay.
Three linear templates were amplified using the following primer pairs: YodU3/OM09, yielding product F3; F2for/Om09, yielding product F4; and Yod3/F1rev, yielding product F5 (Fig. 1). Pfu DNA polymerase and chromosomal DNA of B. amyloliquefaciens FZB45 were used for PCR. The in vitro transcription was performed using 10 nM DNA, 0.25 μM previously phosphorylated PhoP∼P (9), 0.1 μM RNA polymerase, and various concentrations of AbrB or AbrBQ82K (0.25 μM to 2 μM) in transcription buffer (20 mM Tris-HCl, pH 8, 10 mM NaCl, 10 mM MgCl2, 50 mM KCl, 1 mM CaCl2, 0.02 mM EDTA, 1 mM dithiothreitol, and 2% glycerol). The reaction mixtures were incubated at 37°C for 20 min, and then the reactions were stopped by the addition of 5 μl stop solution (95% deionized formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol); finally, products were separated on a 6% polyacrylamide gel containing 7 M urea.
Sequence determination.
All substitutions were confirmed by sequence analysis. The Thermo Sequenase Cy5 dye terminator kit (Amersham Biosciences) was used to perform the sequencing reactions. The samples were run on ALFexpress II (Amersham Biosciences) using ReproGel high resolution (Amersham Biosciences) and analyzed by DS Gene (Accelrys) and NCBI BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
RESULTS
Effects of spo0A and abrB mutations on the phyC promoter activity.
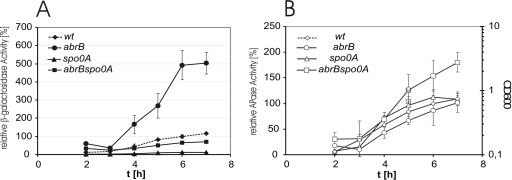
To analyze how the antagonistically acting transcriptional regulators AbrB and Spo0A affect phytase expression in vivo, we fused the extended FZB45 phyC promoter DNA sequence ranging from −289 to +221 with the lacZ reporter gene as described previously (9). The resulting construct, pOM6, consisted of the whole yodU-phyC intergenic region and the 5′ part of the phyC coding region (Fig. 1). Linearized plasmid pOM6 was ectopically integrated into the amyE site of the heterologous host B. subtilis 168 and its respective mutant strains (see Materials and Methods). We analyzed the phyC promoter-driven reporter activities in low-phosphate medium in the genetic backgrounds of B. subtilis wt and the ΔabrB, Δspo0A, and ΔabrB spo0A mutants. The β-galactosidase activity was abolished in the spo0A mutant but was five times enhanced in the abrB mutant. The introduction of the abrB mutation in the spo0A background restored 80% of the β-galactosidase activity compared to what was seen for the wt (Fig. 2A). In contrast, PhoP-dependent expression of the APase was not dramatically altered in the wt and in the respective mutant strains (Fig. 2B). From these results, we conclude that AbrB negatively affects the expression of the phyC gene and that Spo0A antagonizes this effect. Since AbrB acts as a positive regulator of the phoPR operon (19), the phyC expression is only partially restored in the double mutant.
FIG. 2.
Effects of abrB, spo0A, and abrB spo0A mutations on the expression of phyC(pOM6)::lacZ fusions. (A) Relative β-galactosidase activities were measured from cells grown in low-phosphate medium. The activity of the wt cells grown for 6 h was defined as 100%. (B) The APase activity was measured under the same conditions.
AbrB binds to two different sites within the phyC promoter region.
To investigate whether AbrB interacts directly with the phyC promoter sequence, we expressed and isolated the AbrB protein from B. subtilis and the AbrBQ82K protein, corresponding to the B. amyloliquefaciens AbrB protein (see Materials and Methods). Sequence comparison revealed that the abrB gene products derived from B. subtilis 168 and B. amyloliquefaciens FZB45 are nearly identical in sequence except for one Gln→Lys substitution at position 82.
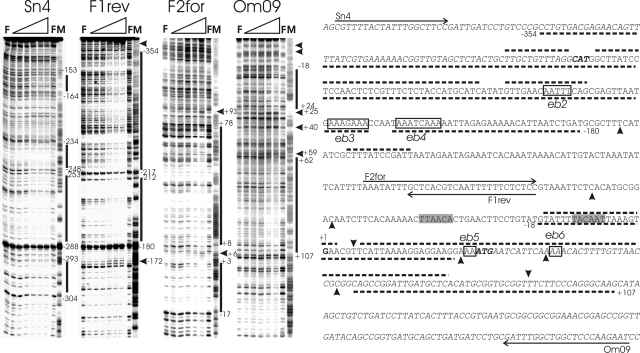
Two binding sites of AbrBsub at the phyCFZB45 promoter separated by a spacer region of about 185 bp were identified by DNase I footprinting, despite the fact that the protection patterns of the coding and the noncoding strands were slightly different. To obtain sufficient resolution in the DNase I footprinting assay, we used two different phyCFZB45 fragments: F6, amplified by primers Sn4 and F1rev, covering the upstream phyCFZB45 promoter sequence ranging from −392 to −76; and F4, amplified by primers F2for and Om09, covering the core promoter region and the 5′part of the phyC FZB45 coding region ranging from −104 to +221 (Fig. 1). One of the two AbrB binding sites (site 2), ranging from −18 to approximately +107, overlaps with the −10 region, the transcription start, and extends into the phyC coding region. The second site, site 1, is located upstream of the phyC core promoter, covering the sequence from −180 to approximately −354 relative to the phyC transcription start. Both regions bound by AbrB appeared to be heterogeneous, with protected areas flanked or interrupted by hypersensitive sites (Fig. 3).
FIG. 3.
DNase I footprinting analysis of AbrBsub at the phyCFZB45 promoter region. The footprints of the coding and noncoding strands obtained with fragments F6 (Sn4 and F1rev) and F4 (F2for and Om09) in the presence of increasing concentrations of AbrB can be seen in the four panels on the left. The AbrB concentrations were 0 μM (for lanes F) and 0.5 μM, 1 μM, 2 μM (missing for fragment F4), 4 μM, 6 μM, and 8 μM (from left to right for the lanes marked with gradients). M indicates the corresponding A+G (Maxam-Gilbert) sequencing reaction. The protected and hypersensitive sites are marked with bars and arrowheads, respectively. The corresponding sequence of the phyC intergenic region is shown on the right. Protected areas of AbrB are delineated by dotted lines, and hypersensitive sites are shown by filled arrowheads (top, coding strand; bottom, noncoding strand). The phyC and yodU coding regions are indicated in italic letters, and the −10 and −35 promoter sequences are shaded. Binding sites within AbrB binding regions 1 and 2 (eb2, eb3, eb4, eb5, and eb6) are framed and labeled.
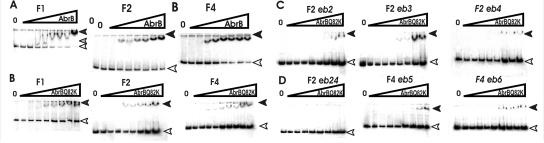
Gel retardation studies performed with AbrBsub and AbrBQ82K corroborated the existence of the two AbrB binding sites revealed by DNase I footprinting analysis. Again, we used different DNA fragments either bearing both putative recognition areas (F1) or containing only one of the two binding sites (F2 and F4). The sizes and positions of the fragments are shown in Fig. 1. Both AbrB proteins were able to shift the F1 fragment in a concentration-dependent manner. At low AbrB concentrations of between 0.375 μM and 0.875 μM, an unstable DNA-protein complex was visible. A more stable and larger complex was detected at higher concentrations of AbrB, i.e., those exceeding 1 μM (Fig. 4A and B). The addition of the phyC promoter DNA as a cold competitor to the DNA-protein complex resulted in the dissociation of the complex and the release of unbound labeled DNA. This effect did not occur when nonspecific competitor DNA derived from the E. coli appA gene was used (see Fig. S1 in the supplemental material). AbrB was also able to shift fragments F2 and F4, which contained only one of the two AbrB binding sites. However, the affinities of the AbrB proteins were slightly reduced in those fragments, suggesting that AbrB might possess a higher affinity to the phyC promoter when occupying both sites present in the F1 fragment simultaneously.
FIG. 4.
Gel retardation mobility shift assays in the presence of increasing concentrations of AbrB and AbrBQ82K. AbrB concentrations used were 0 (for lanes 0) and 0.25, 0.5, 0.75, 1, 1.25, 1.5, and 2 μM (from left to right for the lanes marked with gradients). (A) Gel shift analysis performed with DNA fragments of different lengths bound to AbrB. (B) Assay corresponding to that for panel A but performed with AbrBQ82K. Fragment F1 harbors the entire phyC promoter region with both AbrB binding regions, fragment F2 harbors region 1, and fragment F4 harbors region 2. (C and D) Effect of mutations eb2, eb3, eb4, and eb24 introduced into AbrB binding region 1 on the mobility of the F2 fragment and effect of mutations eb5 and eb6 within AbrB binding region 2 on the mobility of the F4 fragment. White arrowheads, free DNA; filled arrowheads, protein-DNA complex.
Three AT-rich elements, eb2, eb3, and eb4, located within the AbrB binding region 1 (Fig. 1), were replaced by the corresponding GC-rich sequences (Fig. 3 and 2). Gel retardation experiments performed with the mutated F2 and F4 fragments revealed decreased affinity to AbrBQ82K. The F2 fragment bearing the mutations eb2, eb3, and eb4 began to interact with AbrBQ82K at concentrations above 1 μM, while the nonmutated fragments were shifted as early as 0.5 μM (Fig. 4C). AbrBQ82K did not shift the F2 fragment eb24, in which two putative AbrB binding sites within binding region 1, eb2 and eb4, have been eliminated (Fig. 4D). Similarly, the F4 fragments bearing the mutated binding sites eb5 and eb6, located within binding region 2, shifted less in the presence of AbrBQ82K than the corresponding wt sequences, underlining the importance of both regions for AbrB binding.
TABLE 2.
β-Galactosidase activities of phyC promoter-lacZ fusions expressed in the wt (B. subtilis 168) and the spo0A mutant (B. subtilis 1S13)
| Plasmid | Promoter range/substitutiona | % Relative promoter activity (± SD) for:
|
|
|---|---|---|---|
| wt | spo0A mutant | ||
| pOM6 | −287 to +208 | 100 (± 10) | 5 (± 0.5) |
| pOM545 | −147 to +208 | 370 (± 20) | 350 (± 20) |
| pOM7 | −287 to +29 | 410 (±20) | 420 (± 15) |
| pOM57 | −147 to +29 | 375 (± 25) | 470 (± 20) |
| pEB2 | −241 to −236 (AATTT : CCCCC) | 207 (± 15) | 247 (± 17) |
| pEB3 | −223 to −216 (AAAGAAA : CCCGCCC) | 326 (± 23) | 356 (± 25) |
| pEB4 | −211 to −203 (AAATCAAA : CCCTCCCC) | 372 (± 25) | 302 (± 20) |
| pEB5 | +25 to +27 (AA : CC) | 98 (± 7) | 103 (± 7) |
| pEB6 | +40 to +42 (AA : CC) | 311 (± 22) | 278 (± 19) |
| pEB24 | −241 to −236 (AATTT : CCCCC) | 278 (± 20) | 417 (± 19) |
| −211 to −203 (AAATCAAA : CCCTCCCC) | |||
| pEB36 | +40 to +42 (AA : CC) | 263 (± 13) | 378 (± 15) |
| −223 to −216 (AAAGAAA : CCCGCCC) | |||
The nucleotide substitutions of the mutations eb2, eb3, eb4, eb5, and eb6 are indicated with underlining.
Mapping of functional AbrB binding sites.
To map functional regions important for the AbrB interactions, we introduced deletions at the upstream and downstream termini of the phyCFZB45 promoter region and fused them with the lacZ reporter gene. The resulting integrative plasmids, pOM545, pOM7, and pOM57, missing either of AbrB binding sites 1 and 2 (Fig. 1), were ectopically integrated as single copies into the chromosomes of B. subtilis 168 and its spo0A derivative, 1S13. The lacZ reporter activities were measured in low-phosphate medium under phyC-inducing conditions. Both the upstream and downstream truncations yielded similar effects. Each deletion resulted in a fourfold increase in the reporter gene activity compared to what was seen for the full-length promoter fragment, pOM6 (Table 2). Double truncations at both sites did not further augment the activity of the reporter gene. β-Galactosidase activity remained in the same range as that determined for the single truncations. Interestingly, the repressive effect on phyC expression observed for pOM6 in the spo0A background was completely abolished (Table 2). This indicates that simultaneous binding of AbrB to both sites is necessary for full repression.
These findings were corroborated by the results obtained with lacZ fusion strains bearing nucleotide substitutions within one of the AbrB binding regions, i.e., regions 1 (eb2, eb3, eb4, and eb24) and 2 (eb5 and eb6), or within both of them (eb36). The substitutions of the AT-rich sequences located either between −241 and −203 (AbrB binding region 1) or between +8 and +78 (AbrB binding region 2) by the corresponding GC-rich sequences relieved the inhibition of phyCFZB45 expression by AbrB. The activity of the lacZ reporter gene was at least two times enhanced in the mutant strains. The effect of the mutations introduced within the AbrB binding region was more pronounced in the Δspo0A background. Despite the continuous production of AbrB in the absence of Spo0A, phyCFZB45 promoter activity was found to be enhanced by three to five times (Table 2), suggesting reduced repressor affinity for AbrB binding sites 1 and 2. Remarkably, our attempt to relieve the AbrB-dependent inhibitory effect by introducing a corresponding substitution, eb5, in AbrB binding region 2 resulted in comparably reduced reporter gene activities in both wt and spo0A mutants. We assume that altering the nucleotide sequence at +25 and +26 affects translation initiation at the Shine-Dalgarno sequence, which is in close vicinity to the substituted sequence. On the other hand, the introduction of a mutated sequence within binding site eb6 (+40 and +41) resulted in a complete abolition of the repressive effect. No dependence on the genetic background (wt or spo0A) was registered (Table 2).
Both AbrB binding sites are required for efficient transcriptional repression.
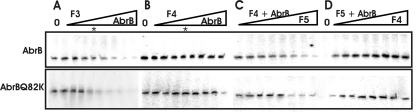
Genetic and in vitro analyses revealed that the repressive effect of AbrB on phyC gene expression is due to the binding of AbrB to the phyCFZB45 promoter region. In vitro transcription performed in the presence of 0.2 μM phosphorylated PhoP together with F3, the full-length DNA fragment harboring both AbrB binding sites (Fig. 1), confirmed this idea. Rising AbrB concentrations gradually repressed the synthesis of the 221-bp phyC gene fragment. A faint second transcript, most likely indicating weak transcription of the divergently orientated yodU gene, was also visible. The transcript was more strongly expressed when using fragment F5 in the transcription assay. In any case, AbrB did not affect its expression.
No inhibition of phyC transcription by AbrB was detected when fragment F4 was used as the template. F4 contains only promoter-proximal AbrB binding site 2 (Fig. 1), again suggesting that direct contact of AbrB with one binding area is not sufficient to inhibit phyC expression. Interestingly, the repressing effect exerted by AbrB was gradually restored in the presence of increasing amounts of DNA fragment F5, containing promoter-distal AbrB binding site 1, which is in close vicinity to the divergently orientated yodU gene (Fig. 1). On the other hand, the addition of increasing amounts of fragment F4 to the F5 template did not restore the inhibitory effect of AbrB, which was registered when the entire fragment was incubated with increasing AbrB concentrations. No differences of the effects exerted by AbrBsub and AbrBQ82K on in vitro transcription were detected (Fig. 5).
FIG. 5.
In vitro transcription analysis of different phyC fragments. The transcription reactions were carried out in 20-μl volumes in transcription buffer by use of 200 fmol of the template, 0.25 μM PhoP∼P, and 0.1 μM RNA polymerase (B. subtilis). The F3 fragment, harboring the entire phyC promoter region with AbrB binding sites (A), and the F4 fragment, harboring the promoter proximal AbrB binding site 2 (B), were transcribed in the presence of various AbrB or AbrBQ82K concentrations as described for Fig. 4. The concentration of 1 μM AbrB or AbrBQ82K is indicated by an asterisk. (C) In vitro transcription of F4 in the presence of 1 μM AbrB and increasing concentrations of the complementary F5 DNA fragment (Fig. 1). (D) In vitro transcription of fragment F5, which does not contain the phyC coding region, in the presence of 1 μM AbrB or AbrBQ82K and increasing concentrations of the complementary fragment F4. The DNA fragments were added at amounts from 0 fmol to 400 fmol in 50-fmol steps.
DISCUSSION
Using an array of in vivo and in vitro experiments, we have shown here that the growth phase-dependent expression of the FZB45 phyC gene is directly controlled by the global transition state regulator AbrB, a transcription factor known to control the expression of more than 60 different genes during late exponential and early stationary phase (18). We detected the repression of the phyCFZB45 promoter activity in the spo0A mutant, while phyC FZB45 gene transcription was found to be strikingly enhanced in the abrB-negative background. In the abrB spo0A double mutant, the phyCFZB45 promoter-driven expression of the lacZ reporter was nearly completely restored, suggesting that phyC underlies repression by AbrB. DNase I footprinting demonstrated that AbrB binds to two remote regions flanking the core phyC promoter which are separated by about 200 bp. One binding region, site 1, lies near the mainly silent yodU gene and is located approximately between −300 and −180 relative to the phyC transcription initiation site. This area consists of three AT-rich core binding sites of 15 bp, 18 bp, and 27 bp in size. The second AbrB binding region, site 2, overlaps the +1 transcription initiation site and extends into the phyC coding region. It consists of two core binding sites of about 21 and 23 bp (Fig. 3).
Although AbrB interacts with numerous DNA targets, no general consensus sequence for AbrB binding sites could be defined. Local variations of the DNA helix configurations (e.g., minor groove width and degree of propeller twisting, etc.) contribute to the differential binding proclivities of AbrB (3). Here, we demonstrated that the replacement of local AT-rich sequence stretches within the AbrB binding region by GC-rich oligonucleotides decreases affinity toward AbrB. Simultaneously, the repressing effect of AbrB on phyCFZB45 gene transcription was abolished. This effect was especially pronounced in spo0A mutants, in which AbrB is permanently expressed (Table 2). The functional form of AbrB has been described as a homotetramer rather than as a homodimer (2, 20). DNA-binding and dimerization functions are located in the N-terminal domain (AbrBN), and the C-terminal domain is responsible for the tendency towards multimerization (2, 3, 20, 21). In this study, we have not addressed the oligomeric state of AbrB, but gel filtration suggested that AbrB could form oligomers containing more than four subunits (see Fig. S2 in the supplemental material), at least in vitro.
In its tetrameric state, AbrB possesses two oppositely orientated DNA-binding sites at its N terminus. This fact allows us to speculate that the two remote AbrB recognition sites may be bound and held together simultaneously by the AbrB tetramer. However, in spite of the large distance between the two sites, it seems that AbrB molecules of a higher oligomeric state, or, more likely, multiple AbrB tetramers, are involved in the binding. In addition, given the extensive nature of the AbrB protected sites in DNase I footprinting (Fig. 3), it is unlikely that one AbrB tetramer causes such an effect. The full functionality of AbrB is accomplished only if both AbrB target sites are occupied. This idea is supported by the following experimental data. (i) Leftward and rightward deletions of the promoter fragment fused with the lacZ reporter gene relieved AbrB-dependent repression. Similarly, mutation of each of the two AbrB binding regions relieved the regulatory effect exerted by AbrB. Only if both binding sites were available was phyCFZB45 promoter expression repressed. (ii) Gel retardation experiments revealed that the affinity of AbrB for the phyCFZB45 promoter fragment was diminished in DNA fragments consisting of only one binding region. A similar observation was made when one of the two AbrB binding regions had been mutated. (iii) The in vitro transcription of phyC in the presence of PhoP∼P and AbrB confirmed that no repression occurs if shortened phyC DNA fragments containing only one of the AbrB binding sites are used as templates. Remarkably, by providing the promoter-distal AbrB binding region 1 by use of a separate DNA fragment, the inhibitory effect of AbrB on phyC transcription could be restored (Fig. 5C).
What are the physiological consequences of our finding that AbrB negatively affects phyC gene expression in vivo and in vitro? Gene expression in response to phosphate depletion during vegetative growth is a well-known phenomenon. Genes which meet this criterion and which are also dependent on the PhoP-PhoR two-component system have been defined as members of the Pho regulon. We have shown previously that the phyC gene also belongs to this group (9). However, repression exerted by the transition state global regulator AbrB hinders an adequate response to phosphate depletion during vegetative growth. If we assume that the main function of phytase lies in its ability to overcome phosphate limitation by making an additional source of this important nutrient available, we must revise the simple model that the limitation of phosphate leads directly to the expression of the target gene via activation of the PhoPR two-component signal transduction system. The AbrB-dependent reduction of phyC gene expression even in the presence of the phosphorylated response regulator suggests that lowering the growth rate is a basic necessity for the action of PhoPR on phyC gene expression. As a positive regulator of the phoPR operon, AbrB leads to the synthesis of sufficient amounts of the phoPR gene products before transition to the stationary growth phase. This ensures that the activated response regulator PhoP∼P is ready to act immediately when growth reduction due to phosphate limitation occurs and AbrB expression is relieved by Spo0A-P.
Supplementary Material
Acknowledgments
Financial support given within the framework of the genomic network sponsored by BMBF, the German ministry for education and research, and by the Deutsche Forschungsgemeinschaft, DFG (grant BO 1113/9-1 to R.B.), is gratefully acknowledged.
We thank Masaya Fujita and Tarek Msadek for providing the Bacillus strains. We also thank Christiane Müller for technical support and sequencing. Alexandra Koumoutsi and Kelvin Eckert are thanked for critical reading of the manuscript.
Footnotes
Published ahead of print on 1 August 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Asai, K., M. Fujita, F. Kawamura, H. Takahashi, Y. Kobayashi, and Y. Sadaie. 1998. Restricted transcription from sigma H or phosphorylated spo0A dependent promoters in the temperature-sensitive secA341 mutant of Bacillus subtilis. Biosci. Biotechnol. Biochem. 621707-1713. [DOI] [PubMed] [Google Scholar]
- 2.Benson, L. M., J. L. Vaughn, M. A. Strauch, B. G. Bobay, R. Thompson, S. Naylor, and J. Cavanagh. 2002. Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Anal. Biochem. 306222-227. [DOI] [PubMed] [Google Scholar]
- 3.Bobay, B. G., L. Benson, S. Naylor, B. Feeney, A. C. Clark, M. B. Goshe, M. A. Strauch, R. Thompson, and J. Cavanagh. 2004. Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry 4316106-16118. [DOI] [PubMed] [Google Scholar]
- 4.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 9811621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita, M., and Y. Sadaie. 1998. Rapid isolation of RNA polymerase from sporulating cells of Bacillus subtilis. Gene 221185-190. [DOI] [PubMed] [Google Scholar]
- 6.Hahn, J., M. Roggiani, and D. Dubnau. 1995. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 1773601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Idriss, E. E., O. Makarewicz, A. Farouk, K. Rosner, R. Greiner, H. Bochow, T. Richter, and R. Borriss. 2002. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 1482097-2109. [DOI] [PubMed] [Google Scholar]
- 9.Makarewicz, O., S. Dubrac, T. Msadek, and R. Borriss. 2006. Dual role of the PhoP∼P response regulator: Bacillus amyloliquefaciens FZB45 phytase gene transcription is directed by positive and negative interactions with the phyC promoter. J. Bacteriol. 1886953-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marahiel, M. A., M. M. Nakano, and P. Zuber. 1993. Regulation of peptide antibiotic production in Bacillus. Mol. Microbiol. 7631-636. [DOI] [PubMed] [Google Scholar]
- 11.Perego, M., and J. A. Hoch. 1991. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J. Bacteriol. 1732514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2689-699. [DOI] [PubMed] [Google Scholar]
- 13.Phillips, Z. E., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pragai, Z., N. E. Allenby, N. O'Connor, S. Dubrac, G. Rapoport, T. Msadek, and C. R. Harwood. 2004. Transcriptional regulation of the phoPR operon in Bacillus subtilis. J. Bacteriol. 1861182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 871801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauch, M. A., M. Perego, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 31203-1209. [DOI] [PubMed] [Google Scholar]
- 19.Sun, G., S. M. Birkey, and F. M. Hulett. 1996. Three two-component signal-transduction systems interact for Pho regulation in Bacillus subtilis. Mol. Microbiol. 19941-948. [DOI] [PubMed] [Google Scholar]
- 20.Vaughn, J. L., V. Feher, S. Naylor, M. A. Strauch, and J. Cavanagh. 2000. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB. Nat. Struct. Biol. 71139-1146. [DOI] [PubMed] [Google Scholar]
- 21.Vaughn, J. L., V. A. Feher, C. Bracken, and J. Cavanagh. 2001. The DNA-binding domain in the Bacillus subtilis transition-state regulator AbrB employs significant motion for promiscuous DNA recognition. J. Mol. Biol. 305429-439. [DOI] [PubMed] [Google Scholar]
- 22.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis σH factor. J. Bacteriol. 173521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuber, P., and R. Losick. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 1692223-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.